α-Arrestins and Their Functions: From Yeast to Human Health
Abstract
1. Introduction
2. Conserved Aspects of α-Arrestin Biology
3. Nature of α-Arrestins—A Lesson from Yeast
3.1. Function of Yeast α-Arrestins
3.2. Regulation of α-Arrestins
3.3. α-Arrestin–Substrate Interaction
4. Non-Arrestin Rsp5 Adaptors
5. Function of Human α-Arrestin Family
5.1. Unique Roles of Human α-Arrestins
5.2. α-Arrestins as Regulators of GPCRs
5.3. α-Arrestins in Cancer Research
5.4. The Role of α-Arrestins in Cellular and Tissue Metabolism
5.5. α-Arrestins as Immune Response Regulators
5.6. α-Arrestins in Brain and Neurodegenerative Diseases
6. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Wilden, U.; Hall, S.W.; Kühn, H. Phosphodiesterase Activation by Photoexcited Rhodopsin Is Quenched When Rhodopsin Is Phosphorylated and Binds the Intrinsic 48-KDa Protein of Rod Outer Segments. Proc. Natl. Acad. Sci. USA 1986, 83, 1174–1178. [Google Scholar] [CrossRef]
- Alvarez, C.E. On the Origins of Arrestin and Rhodopsin. BMC Evol. Biol. 2008, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Rajagopal, S. The β-Arrestins: Multifunctional Regulators of G Protein-Coupled Receptors. J. Biol. Chem. 2016, 291, 8969–8977. [Google Scholar] [CrossRef]
- Luttrell, L.M.; Gesty-Palmer, D. Beyond Desensitization: Physiological Relevance of Arrestin-Dependent Signaling. Pharm. Rev. 2010, 62, 305–330. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; McClatchy, D.B.; Shukla, A.K.; Zhao, Y.; Chen, M.; Shenoy, S.K.; Yates, J.R.; Lefkowitz, R.J. Functional Specialization of Beta-Arrestin Interactions Revealed by Proteomic Analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 12011–12016. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Benovic, J.L. Differential Roles of Arrestin-2 Interaction with Clathrin and Adaptor Protein 2 in G Protein-Coupled Receptor Trafficking. J. Biol. Chem. 2002, 277, 30760–30768. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-O.; Kommaddi, R.P.; Shenoy, S.K. Distinct Roles for β-Arrestin2 and Arrestin-Domain-Containing Proteins in Β2 Adrenergic Receptor Trafficking. EMBO Rep. 2013, 14, 164–171. [Google Scholar] [CrossRef]
- Simonin, A.; Fuster, D. Nedd4-1 and Beta-Arrestin-1 Are Key Regulators of Na+/H+ Exchanger 1 Ubiquitylation, Endocytosis, and Function. J. Biol. Chem. 2010, 285, 38293–38303. [Google Scholar] [CrossRef]
- Shukla, A.K.; Kim, J.; Ahn, S.; Xiao, K.; Shenoy, S.K.; Liedtke, W.; Lefkowitz, R.J. Arresting a Transient Receptor Potential (TRP) Channel: Beta-Arrestin 1 Mediates Ubiquitination and Functional down-Regulation of TRPV4. J. Biol. Chem. 2010, 285, 30115–30125. [Google Scholar] [CrossRef]
- Lee, S.; Park, S.; Lee, H.; Han, S.; Song, J.-M.; Han, D.; Suh, Y.H. Nedd4 E3 Ligase and Beta-Arrestins Regulate Ubiquitination, Trafficking, and Stability of the MGlu7 Receptor. Elife 2019, 8, e44502. [Google Scholar] [CrossRef]
- Lin, C.H.; MacGurn, J.A.; Chu, T.; Stefan, C.J.; Emr, S.D. Arrestin-Related Ubiquitin-Ligase Adaptors Regulate Endocytosis and Protein Turnover at the Cell Surface. Cell 2008, 135, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Boase, N.A.; Kelly, J.M. A Role for CreD, a Carbon Catabolite Repression Gene from Aspergillus Nidulans, in Ubiquitination. Mol. Microbiol. 2004, 53, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Nikko, E.; Pelham, H.R.B. Arrestin-Mediated Endocytosis of Yeast Plasma Membrane Transporters. Traffic 2009, 10, 1856–1867. [Google Scholar] [CrossRef]
- Novoselova, T.V.; Zahira, K.; Rose, R.-S.; Sullivan, J.A. Bul Proteins, a Nonredundant, Antagonistic Family of Ubiquitin Ligase Regulatory Proteins. Eukaryot. Cell 2012, 11, 463–470. [Google Scholar] [CrossRef]
- Wondafrash, D.Z.; Nire’a, A.T.; Tafere, G.G.; Desta, D.M.; Berhe, D.A.; Zewdie, K.A. Thioredoxin-Interacting Protein as a Novel Potential Therapeutic Target in Diabetes Mellitus and Its Underlying Complications. Diabetes Metab. Syndr. Obes. 2020, 13, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Batista, T.M.; Dagdeviren, S.; Carroll, S.H.; Cai, W.; Melnik, V.Y.; Noh, H.L.; Saengnipanthkul, S.; Kim, J.K.; Kahn, C.R.; Lee, R.T. Arrestin Domain-Containing 3 (Arrdc3) Modulates Insulin Action and Glucose Metabolism in Liver. Proc. Natl. Acad. Sci. USA 2020, 117, 6733–6740. [Google Scholar] [CrossRef] [PubMed]
- Domingues, A.; Jolibois, J.; Marquet de Rougé, P.; Nivet-Antoine, V. The Emerging Role of TXNIP in Ischemic and Cardiovascular Diseases; A Novel Marker and Therapeutic Target. Int. J. Mol. Sci. 2021, 22, 1693. [Google Scholar] [CrossRef]
- Tsubaki, H.; Tooyama, I.; Walker, D.G. Thioredoxin-Interacting Protein (TXNIP) with Focus on Brain and Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 9357. [Google Scholar] [CrossRef]
- Oka, S.; Masutani, H.; Liu, W.; Horita, H.; Wang, D.; Kizaka-Kondoh, S.; Yodoi, J. Thioredoxin-Binding Protein-2-like Inducible Membrane Protein Is a Novel Vitamin D3 and Peroxisome Proliferator-Activated Receptor (PPAR)Gamma Ligand Target Protein That Regulates PPARgamma Signaling. Endocrinology 2006, 147, 733–743. [Google Scholar] [CrossRef]
- Chen, Y.; Ning, J.; Cao, W.; Wang, S.; Du, T.; Jiang, J.; Feng, X.; Zhang, B. Research Progress of TXNIP as a Tumor Suppressor Gene Participating in the Metabolic Reprogramming and Oxidative Stress of Cancer Cells in Various Cancers. Front. Oncol. 2020, 10, 568574. [Google Scholar] [CrossRef]
- Mohankumar, K.M.; Currle, D.S.; White, E.; Boulos, N.; Dapper, J.; Eden, C.; Nimmervoll, B.; Thiruvenkatam, R.; Connelly, M.; Kranenburg, T.A.; et al. An in Vivo Screen Identifies Ependymoma Oncogenes and Tumor-Suppressor Genes. Nat. Genet. 2015, 47, 878–887. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Hirsch, J.A.; Velez, M.-G.; Gurevich, Y.V.; Gurevich, V.V. Transition of Arrestin into the Active Receptor-Binding State Requires an Extended Interdomain Hinge. J. Biol. Chem. 2002, 277, 43961–43967. [Google Scholar] [CrossRef] [PubMed]
- Aubry, L.; Guetta, D.; Klein, G. The Arrestin Fold: Variations on a Theme. Curr. Genom. 2009, 10, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Aubry, L.; Klein, G. True Arrestins and Arrestin-Fold Proteins: A Structure-Based Appraisal. Prog. Mol. Biol. Transl. Sci. 2013, 118, 21–56. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, 412–419. [Google Scholar] [CrossRef]
- Baile, M.G.; Guiney, E.L.; Sanford, E.J.; MacGurn, J.A.; Smolka, M.B.; Emr, S.D. Activity of a Ubiquitin Ligase Adaptor Is Regulated by Disordered Insertions in Its Arrestin Domain. Mol. Biol. Cell 2019, 30, 3057–3072. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Kumar, S. Nedd4 and Nedd4-2: Closely Related Ubiquitin-Protein Ligases with Distinct Physiological Functions. Cell Death Differ. 2010, 17, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Belgareh-Touzé, N.; Léon, S.; Erpapazoglou, Z.; Stawiecka-Mirota, M.; Urban-Grimal, D.; Haguenauer-Tsapis, R. Versatile Role of the Yeast Ubiquitin Ligase Rsp5p in Intracellular Trafficking. Biochem. Soc. Trans. 2008, 36 Pt 5, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Staub, O.; Dho, S.; Henry, P.; Correa, J.; Ishikawa, T.; McGlade, J.; Rotin, D. WW Domains of Nedd4 Bind to the Proline-Rich PY Motifs in the Epithelial Na+ Channel Deleted in Liddle’s Syndrome. EMBO J. 1996, 15, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.J.; Zhou, X.Z.; Shen, M.; Lu, K.P. Function of WW Domains as Phosphoserine- or Phosphothreonine-Binding Modules. Science 1999, 283, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, K.; Foot, N.J.; Anand, S.; Dalton, H.E.; Chaudhary, N.; Collins, B.M.; Mathivanan, S.; Kumar, S. Regulation of the Divalent Metal Ion Transporter via Membrane Budding. Cell Discov. 2016, 2, 16011. [Google Scholar] [CrossRef] [PubMed]
- Soung, Y.H.; Ford, S.; Yan, C.; Chung, J. The Role of Arrestin Domain-Containing 3 in Regulating Endocytic Recycling and Extracellular Vesicle Sorting of Integrin Β4 in Breast Cancer. Cancers 2018, 10, 507. [Google Scholar] [CrossRef]
- Puca, L.; Chastagner, P.; Meas-Yedid, V.; Israël, A.; Brou, C. A-Arrestin 1 (ARRDC1) and β-Arrestins Cooperate to Mediate Notch Degradation in Mammals. J. Cell Sci. 2013, 126 Pt 19, 4457–4468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, C.; Gao, K.; Wang, D.; Mao, J.; An, J.; Xu, C.; Wu, D.; Yu, H.; Liu, J.O.; et al. The Ubiquitin Ligase Itch Regulates Apoptosis by Targeting Thioredoxin-Interacting Protein for Ubiquitin-Dependent Degradation. J. Biol. Chem. 2010, 285, 8869–8879. [Google Scholar] [CrossRef]
- GTEx Consortium. Human Genomics. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.-J.; Hu, J.; Esnault, S.; Dozmorov, I.; Malter, J.S. RNA Seq Profiling Reveals a Novel Expression Pattern of TGF-β Target Genes in Human Blood Eosinophils. Immunol. Lett. 2015, 167, 1–10. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Guo, X.; Zhu, Z.; Kong, X.; Yu, F.; Wang, Q. Identification of Differentially Expressed Genes and Signaling Pathways in Chronic Obstructive Pulmonary Disease via Bioinformatic Analysis. FEBS Open Bio 2019, 9, 1880–1899. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tian, D.; Chen, X.; Tang, Z.; Li, K.; Huang, Z.; Fu, Y.; Feng, Y.; Yang, Z. ARRDC3 as a Diagnostic and Prognostic Biomarker for Epithelial Ovarian Cancer Based on Data Mining. Int. J. Gen. Med. 2021, 14, 967–981. [Google Scholar] [CrossRef]
- Shea, F.F.; Rowell, J.L.; Li, Y.; Chang, T.-H.; Alvarez, C.E. Mammalian α Arrestins Link Activated Seven Transmembrane Receptors to Nedd4 Family E3 Ubiquitin Ligases and Interact with β Arrestins. PLoS ONE 2012, 7, e50557. [Google Scholar] [CrossRef]
- Patwari, P.; Chutkow, W.A.; Cummings, K.; Verstraeten, V.L.R.M.; Lammerding, J.; Schreiter, E.R.; Lee, R.T. Thioredoxin-Independent Regulation of Metabolism by the Alpha-Arrestin Proteins. J. Biol. Chem. 2009, 284, 24996–25003. [Google Scholar] [CrossRef]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (ARMMs) at Plasma Membrane by Recruitment of TSG101 Protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef]
- Saxena, G.; Chen, J.; Shalev, A. Intracellular Shuttling and Mitochondrial Function of Thioredoxin-Interacting Protein. J. Biol. Chem. 2010, 285, 3997–4005. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Choe, J.-Y.; Park, K.-Y. TXNIP-Mediated Nuclear Factor-ΚB Signaling Pathway and Intracellular Shifting of TXNIP in Uric Acid-Induced NLRP3 Inflammasome. Biochem. Biophys. Res. Commun. 2019, 511, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global Analysis of Protein Localization in Budding Yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- MacGurn, J.A.; Hsu, P.-C.; Smolka, M.B.; Emr, S.D. TORC1 Regulates Endocytosis via Npr1-Mediated Phosphoinhibition of a Ubiquitin Ligase Adaptor. Cell 2011, 147, 1104–1117. [Google Scholar] [CrossRef]
- Guiney, E.L.; Klecker, T.; Emr, S.D. Identification of the Endocytic Sorting Signal Recognized by the Art1-Rsp5 Ubiquitin Ligase Complex. Mol. Biol. Cell 2016, 27, 4043–4054. [Google Scholar] [CrossRef]
- Ivashov, V.; Zimmer, J.; Schwabl, S.; Kahlhofer, J.; Weys, S.; Gstir, R.; Jakschitz, T.; Kremser, L.; Bonn, G.K.; Lindner, H.; et al. Complementary α-Arrestin-Ubiquitin Ligase Complexes Control Nutrient Transporter Endocytosis in Response to Amino Acids. Elife 2020, 9, e58246. [Google Scholar] [CrossRef]
- Prosser, D.C.; Pannunzio, A.E.; Brodsky, J.L.; Thorner, J.; Wendland, B.; O’Donnell, A.F. α-Arrestins Participate in Cargo Selection for Both Clathrin-Independent and Clathrin-Mediated Endocytosis. J. Cell Sci. 2015, 128, 4220–4234. [Google Scholar] [CrossRef]
- Alvaro, C.G.; O’Donnell, A.F.; Prosser, D.C.; Augustine, A.A.; Goldman, A.; Brodsky, J.L.; Cyert, M.S.; Wendland, B.; Thorner, J. Specific α-Arrestins Negatively Regulate Saccharomyces Cerevisiae Pheromone Response by down-Modulating the G-Protein-Coupled Receptor Ste2. Mol. Cell. Biol. 2014, 34, 2660–2681. [Google Scholar] [CrossRef]
- Nikko, E.; Sullivan, J.A.; Pelham, H.R.B. Arrestin-like Proteins Mediate Ubiquitination and Endocytosis of the Yeast Metal Transporter Smf1. EMBO Rep. 2008, 9, 1216–1221. [Google Scholar] [CrossRef]
- Savocco, J.; Nootens, S.; Afokpa, W.; Bausart, M.; Chen, X.; Villers, J.; Renard, H.-F.; Prévost, M.; Wattiez, R.; Morsomme, P. Yeast α-Arrestin Art2 Is the Key Regulator of Ubiquitylation-Dependent Endocytosis of Plasma Membrane Vitamin B1 Transporters. PLoS Biol. 2019, 17, e3000512. [Google Scholar] [CrossRef] [PubMed]
- Kozu, F.; Shirahama-Noda, K.; Araki, Y.; Kira, S.; Niwa, H.; Noda, T. Isoflurane Induces Art2-Rsp5-dependent Endocytosis of Bap2 in Yeast. FEBS Open Bio 2021, 11, 3090–3100. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.F.; Apffel, A.; Gardner, R.G.; Cyert, M.S. α-Arrestins Aly1 and Aly2 Regulate Intracellular Trafficking in Response to Nutrient Signaling. Mol. Biol. Cell 2010, 21, 3552–3566. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.P.; Hawbaker, S.; Chiang, A.; Jordahl, E.M.; Anaokar, S.; Nikiforov, A.; Bowman, R.W.; Ziegler, P.; McAtee, C.K.; Patton-Vogt, J.; et al. Alpha-Arrestins Aly1/Art6 and Aly2/Art3 Regulate Trafficking of the Glycerophosphoinositol Transporter Git1 and Impact Phospholipid Homeostasis. Biol. Cell 2022, 114, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, R.; Kamiya, M.; Takahara, T.; Maeda, T. Endocytosis of the Aspartic Acid/Glutamic Acid Transporter Dip5 Is Triggered by Substrate-Dependent Recruitment of the Rsp5 Ubiquitin Ligase via the Arrestin-like Protein Aly2. Mol. Cell. Biol. 2010, 30, 5598–5607. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.F.; Huang, L.; Thorner, J.; Cyert, M.S. A Calcineurin-Dependent Switch Controls the Trafficking Function of α-Arrestin Aly1/Art6. J. Biol. Chem. 2013, 288, 24063–24080. [Google Scholar] [CrossRef]
- Wawrzycka, D.; Sadlak, J.; Maciaszczyk-Dziubinska, E.; Wysocki, R. Rsp5-Dependent Endocytosis and Degradation of the Arsenite Transporter Acr3 Requires Its N-Terminal Acidic Tail as an Endocytic Sorting Signal and Arrestin-Related Ubiquitin-Ligase Adaptors. Biochim. Biophys. Acta Biomembr. 2019, 1861, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Hsieh, W.-C.; Hanna, C.B.; Hsu, C.-C.; Pearson, M., II; Tao, W.A.; Aguilar, R.C. The Na+ Pump Ena1 Is a Yeast Epsin-Specific Cargo Requiring Its Ubiquitylation and Phosphorylation Sites for Internalization. J. Cell Sci. 2020, 133, jcs245415. [Google Scholar] [CrossRef]
- Nishimura, A.; Tanahashi, R.; Takagi, H. The Yeast α-Arrestin Art3 Is a Key Regulator for Arginine-Induced Endocytosis of the High-Affinity Proline Transporter Put4. Biochem. Biophys. Res. Commun. 2020, 531, 416–421. [Google Scholar] [CrossRef]
- Wu, A.L.; Hallstrom, T.C.; Moye-Rowley, W.S. ROD1, a Novel Gene Conferring Multiple Resistance Phenotypes in Saccharomyces Cerevisiae. J. Biol. Chem. 1996, 271, 2914–2920. [Google Scholar] [CrossRef]
- Llopis-Torregrosa, V.; Ferri-Blázquez, A.; Adam-Artigues, A.; Deffontaines, E.; van Heusden, G.P.H.; Yenush, L. Regulation of the Yeast Hxt6 Hexose Transporter by the Rod1 α-Arrestin, the Snf1 Protein Kinase, and the Bmh2 14-3-3 Protein. J. Biol. Chem. 2016, 291, 14973–14985. [Google Scholar] [CrossRef] [PubMed]
- Becuwe, M.; Vieira, N.; Lara, D.; Gomes-Rezende, J.; Soares-Cunha, C.; Casal, M.; Haguenauer-Tsapis, R.; Vincent, O.; Paiva, S.; Léon, S. A Molecular Switch on an Arrestin-like Protein Relays Glucose Signaling to Transporter Endocytosis. J. Cell Biol. 2012, 196, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Hovsepian, J.; Defenouillère, Q.; Albanèse, V.; Váchová, L.; Garcia, C.; Palková, Z.; Léon, S. Multilevel Regulation of an α-Arrestin by Glucose Depletion Controls Hexose Transporter Endocytosis. J. Cell Biol. 2017, 216, 1811–1831. [Google Scholar] [CrossRef] [PubMed]
- Paiva, S.; Vieira, N.; Nondier, I.; Haguenauer-Tsapis, R.; Casal, M.; Urban-Grimal, D. Glucose-Induced Ubiquitylation and Endocytosis of the Yeast Jen1 Transporter. J. Biol. Chem. 2009, 284, 19228–19236. [Google Scholar] [CrossRef]
- Fujita, S.; Sato, D.; Kasai, H.; Ohashi, M.; Tsukue, S.; Takekoshi, Y.; Gomi, K.; Shintani, T. The C-Terminal Region of the Yeast Monocarboxylate Transporter Jen1 Acts as a Glucose Signal–Responding Degron Recognized by the α-Arrestin Rod1. J. Biol. Chem. 2018, 293, 10926–10936. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.-C.; MacGurn, J.A.; Emr, S.D. Deubiquitinating Enzymes Ubp2 and Ubp15 Regulate Endocytosis by Limiting Ubiquitination and Degradation of ARTs. Mol. Biol. Cell 2017, 28, 1271–1283. [Google Scholar] [CrossRef]
- O’Donnell, A.F.; McCartney, R.R.; Chandrashekarappa, D.G.; Zhang, B.B.; Thorner, J.; Schmidt, M.C. 2-Deoxyglucose Impairs Saccharomyces Cerevisiae Growth by Stimulating Snf1-Regulated and α-Arrestin-Mediated Trafficking of Hexose Transporters 1 and 3. Mol. Cell. Biol. 2015, 35, 939–955. [Google Scholar] [CrossRef]
- Becuwe, M.; Léon, S. Integrated Control of Transporter Endocytosis and Recycling by the Arrestin-Related Protein Rod1 and the Ubiquitin Ligase Rsp5. eLife 2014, 3, e03307. [Google Scholar] [CrossRef]
- Tamayo Rojas, S.A.; Schmidl, S.; Boles, E.; Oreb, M. Glucose-Induced Internalization of the S. Cerevisiae Galactose Permease Gal2 Is Dependent on Phosphorylation and Ubiquitination of Its Aminoterminal Cytoplasmic Tail. FEMS Yeast Res. 2021, 21, foab019. [Google Scholar] [CrossRef]
- Yofe, I.; Weill, U.; Meurer, M.; Chuartzman, S.; Zalckvar, E.; Goldman, O.; Ben-Dor, S.; Schütze, C.; Wiedemann, N.; Knop, M.; et al. One Library to Make Them All: Streamlining Yeast Library Creation by a SWAp-Tag (SWAT) Strategy. Nat. Methods 2016, 13, 371–378. [Google Scholar] [CrossRef]
- Khanday, F.A.; Saha, M.; Bhat, P.J. Molecular Characterization of MRG19 of Saccharomyces Cerevisiae. Implication in the Regulation of Galactose and Nonfermentable Carbon Source Utilization. Eur. J. Biochem. 2002, 269, 5840–5850. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, C.; van der Merwe, G. Regulation of Hxt3 and Hxt7 Turnover Converges on the Vid30 Complex and Requires Inactivation of the Ras/CAMP/PKA Pathway in Saccharomyces Cerevisiae. PLoS ONE 2012, 7, e50458. [Google Scholar] [CrossRef] [PubMed]
- Herrador, A.; Herranz, S.; Lara, D.; Vincent, O. Recruitment of the ESCRT Machinery to a Putative Seven-Transmembrane-Domain Receptor Is Mediated by an Arrestin-Related Protein. Mol. Cell. Biol. 2010, 30, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Herrador, A.; Livas, D.; Soletto, L.; Becuwe, M.; Léon, S.; Vincent, O. Casein Kinase 1 Controls the Activation Threshold of an α-Arrestin by Multisite Phosphorylation of the Interdomain Hinge. Mol. Biol. Cell 2015, 26, 2128–2138. [Google Scholar] [CrossRef] [PubMed]
- Smardon, A.M.; Kane, P.M. Loss of Vacuolar H+-ATPase Activity in Organelles Signals Ubiquitination and Endocytosis of the Yeast Plasma Membrane Proton Pump Pma1p. J. Biol. Chem. 2014, 289, 32316–32326. [Google Scholar] [CrossRef] [PubMed]
- Marqués, M.C.; Zamarbide-Forés, S.; Pedelini, L.; Llopis-Torregrosa, V.; Yenush, L. A Functional Rim101 Complex Is Required for Proper Accumulation of the Ena1 Na+-ATPase Protein in Response to Salt Stress in Saccharomyces Cerevisiae. FEMS Yeast Res. 2015, 15, fov017. [Google Scholar] [CrossRef]
- O’Donnell, A.F. The Running of the Buls: Control of Permease Trafficking by α-Arrestins Bul1 and Bul2. Mol. Cell. Biol. 2012, 32, 4506–4509. [Google Scholar] [CrossRef]
- Hovsepian, J.; Albanèse, V.; Becuwe, M.; Ivashov, V.; Teis, D.; Léon, S. The Yeast Arrestin-Related Protein Bul1 Is a Novel Actor of Glucose-Induced Endocytosis. Mol. Biol. Cell 2018, 29, 1012–1020. [Google Scholar] [CrossRef]
- Talaia, G.; Gournas, C.; Saliba, E.; Barata-Antunes, C.; Casal, M.; André, B.; Diallinas, G.; Paiva, S. The α-Arrestin Bul1p Mediates Lactate Transporter Endocytosis in Response to Alkalinization and Distinct Physiological Signals. J. Mol. Biol. 2017, 429, 3678–3695. [Google Scholar] [CrossRef]
- Crapeau, M.; Merhi, A.; André, B. Stress Conditions Promote Yeast Gap1 Permease Ubiquitylation and Down-Regulation via the Arrestin-like Bul and Aly Proteins. J. Biol. Chem. 2014, 289, 22103–22116. [Google Scholar] [CrossRef]
- Kawai, K.; Moriya, A.; Uemura, S.; Abe, F. Functional Implications and Ubiquitin-Dependent Degradation of the Peptide Transporter Ptr2 in Saccharomyces Cerevisiae. Eukaryot. Cell 2014, 13, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Villers, J.; Savocco, J.; Szopinska, A.; Degand, H.; Nootens, S.; Morsomme, P. Study of the Plasma Membrane Proteome Dynamics Reveals Novel Targets of the Nitrogen Regulation in Yeast. Mol. Cell. Proteom. 2017, 16, 1652–1668. [Google Scholar] [CrossRef] [PubMed]
- Abe, F.; Iida, H. Pressure-Induced Differential Regulation of the Two Tryptophan Permeases Tat1 and Tat2 by Ubiquitin Ligase Rsp5 and Its Binding Proteins, Bul1 and Bul2. Mol. Cell. Biol. 2003, 23, 7566–7584. [Google Scholar] [CrossRef]
- Liu, J.; Sitaram, A.; Burd, C. Regulation of Copper-Dependent Endocytosis and Vacuolar Degradation of the Yeast Copper Transporter, Ctr1p, by the Rsp5 Ubiquitin Ligase. Traffic 2007, 8, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, R.; Matsushita, T.; Nishimura, A.; Takagi, H. Downregulation of the Broad-Specificity Amino Acid Permease Agp1 Mediated by the Ubiquitin Ligase Rsp5 and the Arrestin-like Protein Bul1 in Yeast. Biosci. Biotechnol. Biochem. 2021, 85, 1266–1274. [Google Scholar] [CrossRef]
- Gournas, C.; Saliba, E.; Krammer, E.-M.; Barthelemy, C.; Prévost, M.; André, B. Transition of Yeast Can1 Transporter to the Inward-Facing State Unveils an α-Arrestin Target Sequence Promoting Its Ubiquitylation and Endocytosis. Mol. Biol. Cell 2017, 28, 2819–2832. [Google Scholar] [CrossRef]
- Koteliansky, V.E.; Glukhova, M.A.; Bejanian, M.V.; Surguchov, A.P.; Smirnov, V.N. Isolation and Characterization of Actin-like Protein from Yeast Saccharomyces Cerevisiae. FEBS Lett. 1979, 102, 55–58. [Google Scholar] [CrossRef]
- Ghaddar, K.; Merhi, A.; Saliba, E.; Krammer, E.-M.; Prévost, M.; André, B. Substrate-Induced Ubiquitylation and Endocytosis of Yeast Amino Acid Permeases. Mol. Cell. Biol. 2014, 34, 4447–4463. [Google Scholar] [CrossRef]
- Zhao, Y.; MacGurn, J.A.; Liu, M.; Emr, S. The ART-Rsp5 Ubiquitin Ligase Network Comprises a Plasma Membrane Quality Control System That Protects Yeast Cells from Proteotoxic Stress. eLife 2013, 2, e00459. [Google Scholar] [CrossRef]
- Jones, C.B.; Ott, E.M.; Keener, J.M.; Curtiss, M.; Sandrin, V.; Babst, M. Regulation of Membrane Protein Degradation by Starvation-Response Pathways. Traffic 2012, 13, 468–482. [Google Scholar] [CrossRef]
- Andoh, T.; Hirata, Y.; Kikuchi, A. PY Motifs of Rod1 Are Required for Binding to Rsp5 and for Drug Resistance. FEBS Lett. 2002, 525, 131–134. [Google Scholar] [CrossRef]
- Lu, C.-C.; Ho, S.-T.; Wong, C.-S.; Wang, J.-J.; Tsai, C.-S.; Hu, O.Y.-P.; Chang, S.-Y.; Lin, C.-Y. Pharmacokinetics of Isoflurane: Uptake in the Body. Pharmacology 2003, 69, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Sonner, J.M.; Cantor, R.S. Molecular Mechanisms of Drug Action: An Emerging View. Annu. Rev. Biophys. 2013, 42, 143–167. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, J.; Kikuchi, Y. Rod1, an Arrestin-Related Protein, Is Phosphorylated by Snf1-Kinase in Saccharomyces Cerevisiae. Biochem. Biophys. Res. Commun. 2007, 364, 258–263. [Google Scholar] [CrossRef]
- Muir, A.; Ramachandran, S.; Roelants, F.M.; Timmons, G.; Thorner, J. TORC2-Dependent Protein Kinase Ypk1 Phosphorylates Ceramide Synthase to Stimulate Synthesis of Complex Sphingolipids. eLife 2014, 3, e03779. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.F.; Schmidt, M.C. AMPK-Mediated Regulation of Alpha-Arrestins and Protein Trafficking. Int. J. Mol. Sci. 2019, 20, 515. [Google Scholar] [CrossRef]
- Merhi, A.; André, B. Internal Amino Acids Promote Gap1 Permease Ubiquitylation via TORC1/Npr1/14-3-3-Dependent Control of the Bul Arrestin-like Adaptors. Mol. Cell. Biol. 2012, 32, 4510–4522. [Google Scholar] [CrossRef]
- Megarioti, A.H.; Primo, C.; Kapetanakis, G.C.; Athanasopoulos, A.; Sophianopoulou, V.; André, B.; Gournas, C. The Bul1/2 Alpha-Arrestins Promote Ubiquitylation and Endocytosis of the Can1 Permease upon Cycloheximide-Induced TORC1-Hyperactivation. Int. J. Mol. Sci. 2021, 22, 10208. [Google Scholar] [CrossRef]
- Kakiuchi, K.; Yamauchi, Y.; Taoka, M.; Iwago, M.; Fujita, T.; Ito, T.; Song, S.-Y.; Sakai, A.; Isobe, T.; Ichimura, T. Proteomic Analysis of in Vivo 14-3-3 Interactions in the Yeast Saccharomyces Cerevisiae. Biochemistry 2007, 46, 7781–7792. [Google Scholar] [CrossRef]
- Kahlhofer, J.; Leon, S.; Teis, D.; Schmidt, O. The α-Arrestin Family of Ubiquitin Ligase Adaptors Links Metabolism with Selective Endocytosis. Biol. Cell 2021, 113, 183–219. [Google Scholar] [CrossRef]
- Lee, S.; Ho, H.-C.; Tumolo, J.M.; Hsu, P.-C.; MacGurn, J.A. Methionine Triggers Ppz-Mediated Dephosphorylation of Art1 to Promote Cargo-Specific Endocytosis. J. Cell Biol. 2019, 218, 977–992. [Google Scholar] [CrossRef]
- Wu, N.; Zheng, B.; Shaywitz, A.; Dagon, Y.; Tower, C.; Bellinger, G.; Shen, C.-H.; Wen, J.; Asara, J.; McGraw, T.E.; et al. AMPK-Dependent Degradation of TXNIP upon Energy Stress Leads to Enhanced Glucose Uptake via GLUT1. Mol. Cell 2013, 49, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Waldhart, A.N.; Dykstra, H.; Peck, A.S.; Boguslawski, E.A.; Madaj, Z.B.; Wen, J.; Veldkamp, K.; Hollowell, M.; Zheng, B.; Cantley, L.C.; et al. Phosphorylation of TXNIP by AKT Mediates Acute Influx of Glucose in Response to Insulin. Cell Rep. 2017, 19, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, C.; Shields, S.B.; Williams, C.A.; Winistorfer, S.; Piper, R.C. A Cycle of Ubiquitination Regulates Adaptor Function of the Nedd4-Family Ubiquitin Ligase Rsp5. Curr. Biol. 2020, 30, 465–479.e5. [Google Scholar] [CrossRef] [PubMed]
- Milano, S.K.; Pace, H.C.; Kim, Y.-M.; Brenner, C.; Benovic, J.L. Scaffolding Functions of Arrestin-2 Revealed by Crystal Structure and Mutagenesis. Biochemistry 2002, 41, 3321–3328. [Google Scholar] [CrossRef]
- Busto, J.V.; Elting, A.; Haase, D.; Spira, F.; Kuhlman, J.; Schäfer-Herte, M.; Wedlich-Söldner, R. Lateral Plasma Membrane Compartmentalization Links Protein Function and Turnover. EMBO J. 2018, 37, e99473. [Google Scholar] [CrossRef]
- Bilsland, E.; Hult, M.; Bell, S.D.; Sunnerhagen, P.; Downs, J.A. The Bre5/Ubp3 Ubiquitin Protease Complex from Budding Yeast Contributes to the Cellular Response to DNA Damage. DNA Repair 2007, 6, 1471–1484. [Google Scholar] [CrossRef]
- Frattini, C.; Villa-Hernández, S.; Pellicanò, G.; Jossen, R.; Katou, Y.; Shirahige, K.; Bermejo, R. Cohesin Ubiquitylation and Mobilization Facilitate Stalled Replication Fork Dynamics. Mol. Cell 2017, 68, 758–772.e4. [Google Scholar] [CrossRef]
- Stimpson, H.E.M.; Lewis, M.J.; Pelham, H.R.B. Transferrin Receptor-like Proteins Control the Degradation of a Yeast Metal Transporter. EMBO J. 2006, 25, 662–672. [Google Scholar] [CrossRef]
- Zhu, L.; Sardana, R.; Jin, D.K.; Emr, S.D. Calcineurin-Dependent Regulation of Endocytosis by a Plasma Membrane Ubiquitin Ligase Adaptor, Rcr1. J. Cell Biol. 2020, 219, e201909158. [Google Scholar] [CrossRef]
- Léon, S.; Erpapazoglou, Z.; Haguenauer-Tsapis, R. Ear1p and Ssh4p Are New Adaptors of the Ubiquitin Ligase Rsp5p for Cargo Ubiquitylation and Sorting at Multivesicular Bodies. Mol. Biol. Cell 2008, 19, 2379–2388. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Kee, Y.; Huibregtse, J.M.; Piper, R.C. Hse1, a Component of the Yeast Hrs-STAM Ubiquitin-Sorting Complex, Associates with Ubiquitin Peptidases and a Ligase to Control Sorting Efficiency into Multivesicular Bodies. Mol. Biol. Cell 2007, 18, 324–335. [Google Scholar] [CrossRef]
- MacDonald, C.; Stringer, D.K.; Piper, R.C. Sna3 Is an Rsp5 Adaptor Protein That Relies on Ubiquitination for Its MVB Sorting. Traffic 2012, 13, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.H.Y.; Urban-Grimal, D.; Bugnicourt, A.; Greenblatt, J.F.; Haguenauer-Tsapis, R.; Emili, A. Interaction of the Deubiquitinating Enzyme Ubp2 and the E3 Ligase Rsp5 Is Required for Transporter/Receptor Sorting in the Multivesicular Body Pathway. PLoS ONE 2009, 4, e4259. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.A.; Lewis, M.J.; Nikko, E.; Pelham, H.R.B. Multiple Interactions Drive Adaptor-Mediated Recruitment of the Ubiquitin Ligase Rsp5 to Membrane Proteins in Vivo and in Vitro. Mol. Biol. Cell 2007, 18, 2429–2440. [Google Scholar] [CrossRef] [PubMed]
- Kota, J.; Melin-Larsson, M.; Ljungdahl, P.O.; Forsberg, H. Ssh4, Rcr2 and Rcr1 Affect Plasma Membrane Transporter Activity in Saccharomyces Cerevisiae. Genetics 2007, 175, 1681–1694. [Google Scholar] [CrossRef]
- Li, M.; Rong, Y.; Chuang, Y.-S.; Peng, D.; Emr, S.D. Ubiquitin-Dependent Lysosomal Membrane Protein Sorting and Degradation. Mol. Cell 2015, 57, 467–478. [Google Scholar] [CrossRef]
- Lam, M.H.Y.; Emili, A. Ubp2 Regulates Rsp5 Ubiquitination Activity In Vivo and In Vitro. PLoS ONE 2013, 8, e75372. [Google Scholar] [CrossRef]
- Chen, J.; Saxena, G.; Mungrue, I.N.; Lusis, A.J.; Shalev, A. Thioredoxin-Interacting Protein: A Critical Link between Glucose Toxicity and Beta-Cell Apoptosis. Diabetes 2008, 57, 938–944. [Google Scholar] [CrossRef]
- Anand, S.; Foot, N.; Ang, C.-S.; Gembus, K.M.; Keerthikumar, S.; Adda, C.G.; Mathivanan, S.; Kumar, S. Arrestin-Domain Containing Protein 1 (Arrdc1) Regulates the Protein Cargo and Release of Extracellular Vesicles. Proteomics 2018, 18, e1800266. [Google Scholar] [CrossRef]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. HIV-1 and Ebola Virus Encode Small Peptide Motifs That Recruit Tsg101 to Sites of Particle Assembly to Facilitate Egress. Nat. Med. 2001, 7, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Foot, N.J.; Gonzalez, M.B.; Gembus, K.; Fonseka, P.; Sandow, J.J.; Nguyen, T.T.; Tran, D.; Webb, A.I.; Mathivanan, S.; Robker, R.L.; et al. Arrdc4-Dependent Extracellular Vesicle Biogenesis Is Required for Sperm Maturation. J. Extracell. Vesicles 2021, 10, e12113. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S.K.; McDonald, P.H.; Kohout, T.A.; Lefkowitz, R.J. Regulation of Receptor Fate by Ubiquitination of Activated Beta 2-Adrenergic Receptor and Beta-Arrestin. Science 2001, 294, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S.K.; Xiao, K.; Venkataramanan, V.; Snyder, P.M.; Freedman, N.J.; Weissman, A.M. Nedd4 Mediates Agonist-Dependent Ubiquitination, Lysosomal Targeting, and Degradation of the Beta2-Adrenergic Receptor. J. Biol. Chem. 2008, 283, 22166–22176. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, J.F.; Pan, H.; Lu, Q. Arrestin Domain-Containing Protein 3 Recruits the NEDD4 E3 Ligase to Mediate Ubiquitination of the Beta2-Adrenergic Receptor. EMBO Rep. 2010, 11, 605–611. [Google Scholar] [CrossRef]
- Patwari, P.; Emilsson, V.; Schadt, E.E.; Chutkow, W.A.; Lee, S.; Marsili, A.; Zhang, Y.; Dobrin, R.; Cohen, D.E.; Larsen, P.R.; et al. The Arrestin Domain-Containing 3 Protein Regulates Body Mass and Energy Expenditure. Cell Metab. 2011, 14, 671–683. [Google Scholar] [CrossRef]
- Dores, M.R.; Lin, H.J.; Grimsey, N.; Mendez, F.; Trejo, J. The α-Arrestin ARRDC3 Mediates ALIX Ubiquitination and G Protein-Coupled Receptor Lysosomal Sorting. Mol. Biol. Cell 2015, 26, 4660–4673. [Google Scholar] [CrossRef]
- Komatsu, H.; Fukuchi, M.; Habata, Y. Potential Utility of Biased GPCR Signaling for Treatment of Psychiatric Disorders. Int. J. Mol. Sci. 2019, 20, 3207. [Google Scholar] [CrossRef]
- Martini, M.L.; Ray, C.; Yu, X.; Liu, J.; Pogorelov, V.M.; Wetsel, W.C.; Huang, X.-P.; McCorvy, J.D.; Caron, M.G.; Jin, J. Designing Functionally Selective Noncatechol Dopamine D1 Receptor Agonists with Potent In Vivo Antiparkinsonian Activity. ACS Chem. Neurosci. 2019, 10, 4160–4182. [Google Scholar] [CrossRef]
- Goldberg, S.F.; Miele, M.E.; Hatta, N.; Takata, M.; Paquette-Straub, C.; Freedman, L.P.; Welch, D.R. Melanoma Metastasis Suppression by Chromosome 6: Evidence for a Pathway Regulated by CRSP3 and TXNIP. Cancer Res. 2003, 63, 432–440. [Google Scholar]
- Nakamura, H.; Masutani, H.; Yodoi, J. Extracellular Thioredoxin and Thioredoxin-Binding Protein 2 in Control of Cancer. Semin. Cancer Biol. 2006, 16, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, Q.; Chng, W.-J. TXNIP (VDUP-1, TBP-2): A Major Redox Regulator Commonly Suppressed in Cancer by Epigenetic Mechanisms. Int. J. Biochem. Cell Biol. 2011, 43, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Katsogiannou, M.; Andrieu, C.; Baylot, V.; Baudot, A.; Dusetti, N.J.; Gayet, O.; Finetti, P.; Garrido, C.; Birnbaum, D.; Bertucci, F.; et al. The Functional Landscape of Hsp27 Reveals New Cellular Processes Such as DNA Repair and Alternative Splicing and Proposes Novel Anticancer Targets. Mol. Cell. Proteom. 2014, 13, 3585–3601. [Google Scholar] [CrossRef] [PubMed]
- Draheim, K.M.; Chen, H.-B.; Tao, Q.; Moore, N.; Roche, M.; Lyle, S. ARRDC3 Suppresses Breast Cancer Progression by Negatively Regulating Integrin Beta4. Oncogene 2010, 29, 5032–5047. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Soung, Y.H.; Kashyap, T.; Nguyen, T.; Yadav, G.; Chang, H.; Landesman, Y.; Chung, J. Selective Inhibitors of Nuclear Export (SINE) Compounds Block Proliferation and Migration of Triple Negative Breast Cancer Cells by Restoring Expression of ARRDC3. Oncotarget 2017, 8, 52935–52947. [Google Scholar] [CrossRef]
- Hernández, N.A.; Correa, E.; Avila, E.P.; Vela, T.A.; Pérez, V.M. PAR1 Is Selectively over Expressed in High Grade Breast Cancer Patients: A Cohort Study. J. Transl. Med. 2009, 7, 47. [Google Scholar] [CrossRef]
- Boire, A.; Covic, L.; Agarwal, A.; Jacques, S.; Sherifi, S.; Kuliopulos, A. PAR1 Is a Matrix Metalloprotease-1 Receptor That Promotes Invasion and Tumorigenesis of Breast Cancer Cells. Cell 2005, 120, 303–313. [Google Scholar] [CrossRef]
- Arakaki, A.K.S.; Pan, W.-A.; Lin, H.; Trejo, J. The α-Arrestin ARRDC3 Suppresses Breast Carcinoma Invasion by Regulating G Protein–Coupled Receptor Lysosomal Sorting and Signaling. J. Biol. Chem. 2018, 293, 3350–3362. [Google Scholar] [CrossRef]
- Arakaki, A.K.S.; Pan, W.-A.; Trejo, J. Regulation of GPCR Activation of the Hippo Pathway in Metastatic Breast Cancer. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-A.; Lu, C.-Y.; Cheng, T.-Y.; Pan, S.-H.; Chen, H.-F.; Chang, N.-S. WW Domain-Containing Proteins YAP and TAZ in the Hippo Pathway as Key Regulators in Stemness Maintenance, Tissue Homeostasis, and Tumorigenesis. Front. Oncol. 2019, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, A.K.S.; Pan, W.-A.; Wedegaertner, H.; Roca-Mercado, I.; Chinn, L.; Gujral, T.S.; Trejo, J. α-Arrestin ARRDC3 Tumor Suppressor Function Is Linked to GPCR-Induced TAZ Activation and Breast Cancer Metastasis. J. Cell Sci. 2021, 134, jcs254888. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Sun, X.; Sun, B.; Li, T.; Wu, G.; Li, Y.; Chen, L.; Liu, Q.; Cui, M.; Zhou, Z. ARRDC3 Suppresses Colorectal Cancer Progression through Destabilizing the Oncoprotein YAP. FEBS Lett. 2018, 592, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Shi, Q.; Li, W.; Mu, X.; Peng, J.; Li, M.; Chen, M.; Huang, H.; Wang, C.; Gao, K.; et al. ARRDC1 and ARRDC3 Act as Tumor Suppressors in Renal Cell Carcinoma by Facilitating YAP1 Degradation. Am. J. Cancer Res. 2018, 8, 132–143. [Google Scholar]
- Pearson, J.D.; Huang, K.; Pacal, M.; McCurdy, S.R.; Lu, S.; Aubry, A.; Yu, T.; Wadosky, K.M.; Zhang, L.; Wang, T.; et al. Binary Pan-Cancer Classes with Distinct Vulnerabilities Defined by pro- or Anti-Cancer YAP/TEAD Activity. Cancer Cell 2021, 39, 1115–1134.e12. [Google Scholar] [CrossRef] [PubMed]
- Macheda, M.L.; Rogers, S.; Best, J.D. Molecular and Cellular Regulation of Glucose Transporter (GLUT) Proteins in Cancer. J. Cell Physiol. 2005, 202, 654–662. [Google Scholar] [CrossRef]
- Young, C.D.; Lewis, A.S.; Rudolph, M.C.; Ruehle, M.D.; Jackman, M.R.; Yun, U.J.; Ilkun, O.; Pereira, R.; Abel, E.D.; Anderson, S.M. Modulation of Glucose Transporter 1 (GLUT1) Expression Levels Alters Mouse Mammary Tumor Cell Growth in Vitro and in Vivo. PLoS ONE 2011, 6, e23205. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a Versatile Platform for Intracellular Delivery of Macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef]
- Hoffman, M.L.; Peck, K.N.; Wegrzyn, J.L.; Reed, S.A.; Zinn, S.A.; Govoni, K.E. Poor Maternal Nutrition during Gestation Alters the Expression of Genes Involved in Muscle Development and Metabolism in Lambs. J. Anim. Sci. 2016, 94, 3093–3099. [Google Scholar] [CrossRef] [PubMed]
- Furlow, J.D.; Watson, M.L.; Waddell, D.S.; Neff, E.S.; Baehr, L.M.; Ross, A.P.; Bodine, S.C. Altered Gene Expression Patterns in Muscle Ring Finger 1 Null Mice during Denervation- and Dexamethasone-Induced Muscle Atrophy. Physiol. Genom. 2013, 45, 1168–1185. [Google Scholar] [CrossRef]
- Gordon, B.S.; Rossetti, M.L.; Eroshkin, A.M. Arrdc2 and Arrdc3 Elicit Divergent Changes in Gene Expression in Skeletal Muscle Following Anabolic and Catabolic Stimuli. Physiol. Genom. 2019, 51, 208–217. [Google Scholar] [CrossRef]
- Parikh, H.; Carlsson, E.; Chutkow, W.A.; Johansson, L.E.; Storgaard, H.; Poulsen, P.; Saxena, R.; Ladd, C.; Schulze, P.C.; Mazzini, M.J.; et al. TXNIP Regulates Peripheral Glucose Metabolism in Humans. PLoS Med. 2007, 4, e158. [Google Scholar] [CrossRef] [PubMed]
- Chutkow, W.A.; Birkenfeld, A.L.; Brown, J.D.; Lee, H.-Y.; Frederick, D.W.; Yoshioka, J.; Patwari, P.; Kursawe, R.; Cushman, S.W.; Plutzky, J.; et al. Deletion of the Alpha-Arrestin Protein Txnip in Mice Promotes Adiposity and Adipogenesis While Preserving Insulin Sensitivity. Diabetes 2010, 59, 1424–1434. [Google Scholar] [CrossRef] [PubMed]
- Alhawiti, N.M.; Al Mahri, S.; Aziz, M.A.; Malik, S.S.; Mohammad, S. TXNIP in Metabolic Regulation: Physiological Role and Therapeutic Outlook. Curr. Drug Targets 2017, 18, 1095–1103. [Google Scholar] [CrossRef]
- Ahn, B.; Soundarapandian, M.M.; Sessions, H.; Peddibhotla, S.; Roth, G.P.; Li, J.-L.; Sugarman, E.; Koo, A.; Malany, S.; Wang, M.; et al. MondoA Coordinately Regulates Skeletal Myocyte Lipid Homeostasis and Insulin Signaling. J. Clin. Investig. 2016, 126, 3567–3579. [Google Scholar] [CrossRef]
- Richards, P.; Rachdi, L.; Oshima, M.; Marchetti, P.; Bugliani, M.; Armanet, M.; Postic, C.; Guilmeau, S.; Scharfmann, R. MondoA Is an Essential Glucose-Responsive Transcription Factor in Human Pancreatic β-Cells. Diabetes 2018, 67, 461–472. [Google Scholar] [CrossRef]
- Richards, P.; Ourabah, S.; Montagne, J.; Burnol, A.-F.; Postic, C.; Guilmeau, S. MondoA/ChREBP: The Usual Suspects of Transcriptional Glucose Sensing; Implication in Pathophysiology. Metabolism 2017, 70, 133–151. [Google Scholar] [CrossRef]
- Rauch, S.; Martin-Serrano, J. Multiple Interactions between the ESCRT Machinery and Arrestin-Related Proteins: Implications for PPXY-Dependent Budding. J. Virol. 2011, 85, 3546–3556. [Google Scholar] [CrossRef]
- Gardinassi, L.G. A Cross-Study Biomarker Signature of Human Bronchial Epithelial Cells Infected with Respiratory Syncytial Virus. Adv. Virol. 2016, 2016, 3605302. [Google Scholar] [CrossRef] [PubMed]
- Saçar Demirci, M.D.; Toprak, M.; Allmer, J. A Machine Learning Approach for MicroRNA Precursor Prediction in Retro-Transcribing Virus Genomes. J. Integr. Bioinform. 2016, 13, 303. [Google Scholar] [CrossRef] [PubMed]
- Johanning, G.L.; Malouf, G.G.; Zheng, X.; Esteva, F.J.; Weinstein, J.N.; Wang-Johanning, F.; Su, X. Expression of Human Endogenous Retrovirus-K Is Strongly Associated with the Basal-like Breast Cancer Phenotype. Sci. Rep. 2017, 7, 41960. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; Kukimoto, I.; Li, Z.; Li, S.; Li, N.; Hu, Z.; Takahashi, A.; Inoue, S.; Yokoi, S.; Chen, J.; et al. Genome-Wide Association Study of Cervical Cancer Suggests a Role for ARRDC3 Gene in Human Papillomavirus Infection. Hum. Mol. Genet. 2019, 28, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Tardivel, A.; Thorens, B.; Choi, I.; Tschopp, J. Thioredoxin-Interacting Protein Links Oxidative Stress to Inflammasome Activation. Nat. Immunol. 2010, 11, 136–140. [Google Scholar] [CrossRef]
- Masters, S.L.; Dunne, A.; Subramanian, S.L.; Hull, R.L.; Tannahill, G.M.; Sharp, F.A.; Becker, C.; Franchi, L.; Yoshihara, E.; Chen, Z.; et al. Activation of the NLRP3 Inflammasome by Islet Amyloid Polypeptide Provides a Mechanism for Enhanced IL-1β in Type 2 Diabetes. Nat. Immunol. 2010, 11, 897–904. [Google Scholar] [CrossRef]
- Muri, J.; Thut, H.; Feng, Q.; Kopf, M. Thioredoxin-1 Distinctly Promotes NF-ΚB Target DNA Binding and NLRP3 Inflammasome Activation Independently of Txnip. Elife 2020, 9, e53627. [Google Scholar] [CrossRef]
- Meng, J.; Yao, Z.; He, Y.; Zhang, R.; Zhang, Y.; Yao, X.; Yang, H.; Chen, L.; Zhang, Z.; Zhang, H.; et al. ARRDC4 Regulates Enterovirus 71-Induced Innate Immune Response by Promoting K63 Polyubiquitination of MDA5 through TRIM65. Cell Death Dis. 2017, 8, e2866. [Google Scholar] [CrossRef]
- Kim, H.-R.; Mun, Y.; Lee, K.-S.; Park, Y.-J.; Park, J.-S.; Park, J.-H.; Jeon, B.-N.; Kim, C.-H.; Jun, Y.; Hyun, Y.-M.; et al. T Cell Microvilli Constitute Immunological Synaptosomes That Carry Messages to Antigen-Presenting Cells. Nat. Commun. 2018, 9, 3630. [Google Scholar] [CrossRef]
- Parruti, G.; Peracchia, F.; Sallese, M.; Ambrosini, G.; Masini, M.; Rotilio, D.; De Blasi, A. Molecular Analysis of Human Beta-Arrestin-1: Cloning, Tissue Distribution, and Regulation of Expression. Identification of Two Isoforms Generated by Alternative Splicing. J. Biol. Chem. 1993, 268, 9753–9761. [Google Scholar] [CrossRef]
- Saitoh, T.; Tanaka, S.; Koike, T. Rapid Induction and Ca(2+) Influx-Mediated Suppression of Vitamin D3 up-Regulated Protein 1 (VDUP1) MRNA in Cerebellar Granule Neurons Undergoing Apoptosis. J. Neurochem. 2001, 78, 1267–1276. [Google Scholar] [CrossRef]
- Regard, J.B.; Sato, I.T.; Coughlin, S.R. Anatomical Profiling of G Protein-Coupled Receptor Expression. Cell 2008, 135, 561–571. [Google Scholar] [CrossRef]
- Sharma, P.; Srivastava, P.; Seth, A.; Tripathi, P.N.; Banerjee, A.G.; Shrivastava, S.K. Comprehensive Review of Mechanisms of Pathogenesis Involved in Alzheimer’s Disease and Potential Therapeutic Strategies. Prog. Neurobiol. 2019, 174, 53–89. [Google Scholar] [CrossRef]
- Ismael, S.; Nasoohi, S.; Li, L.; Aslam, K.S.; Khan, M.M.; El-Remessy, A.B.; McDonald, M.P.; Liao, F.-F.; Ishrat, T. Thioredoxin Interacting Protein Regulates Age-Associated Neuroinflammation. Neurobiol. Dis. 2021, 156, 105399. [Google Scholar] [CrossRef]
- Jia, J.; Zeng, X.; Xu, G.; Wang, Z. The Potential Roles of Redox Enzymes in Alzheimer’s Disease: Focus on Thioredoxin. ASN Neuro 2021, 13, 1759091421994351. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, G.; Shao, N.; Qin, Y.; Chen, Q.; Wang, Y.; Zhou, P.; Cai, B. Thioredoxin-Interacting Protein (TXNIP) as a Target for Alzheimer’s Disease: Flavonoids and Phenols. Inflammopharmacology 2021, 29, 1317–1329. [Google Scholar] [CrossRef]
- Pan, Q.; Guo, K.; Xue, M.; Tu, Q. Estrogen Protects Neuroblastoma Cell from Amyloid-β 42 (Aβ42)-Induced Apoptosis via TXNIP/TRX Axis and AMPK Signaling. Neurochem. Int. 2020, 135, 104685. [Google Scholar] [CrossRef]
- Melone, M.A.B.; Dato, C.; Paladino, S.; Coppola, C.; Trebini, C.; Giordana, M.T.; Perrone, L. Verapamil Inhibits Ser202/Thr205 Phosphorylation of Tau by Blocking TXNIP/ROS/P38 MAPK Pathway. Pharm. Res. 2018, 35, 44. [Google Scholar] [CrossRef]
- Xu, L.; Lin, X.; Guan, M.; Zeng, Y.; Liu, Y. Verapamil Attenuated Prediabetic Neuropathy in High-Fat Diet-Fed Mice through Inhibiting TXNIP-Mediated Apoptosis and Inflammation. Oxidative Med. Cell. Longev. 2019, 2019, 1896041. [Google Scholar] [CrossRef]
- Liu, H.; Guo, W.; Guo, H.; Zhao, L.; Yue, L.; Li, X.; Feng, D.; Luo, J.; Wu, X.; Cui, W.; et al. Bakuchiol Attenuates Oxidative Stress and Neuron Damage by Regulating Trx1/TXNIP and the Phosphorylation of AMPK After Subarachnoid Hemorrhage in Mice. Front. Pharm. 2020, 11, 712. [Google Scholar] [CrossRef]
- Su, C.-J.; Shen, Z.; Cui, R.-X.; Huang, Y.; Xu, D.-L.; Zhao, F.-L.; Pan, J.; Shi, A.-M.; Liu, T.; Yu, Y.-L. Thioredoxin-Interacting Protein (TXNIP) Regulates Parkin/PINK1-Mediated Mitophagy in Dopaminergic Neurons under High-Glucose Conditions: Implications for Molecular Links Between Parkinson’s Disease and Diabetes. Neurosci. Bull. 2020, 36, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Ou, W.; Chen, C.; Liu, Y.; Li, H.; Zhang, X.; Chai, H.; Ding, X.; Wang, Q. Endoplasmic Reticulum Stress and Oxidative Stress Contribute to Neuronal Pyroptosis Caused by Cerebral Venous Sinus Thrombosis in Rats: Involvement of TXNIP/Peroxynitrite-NLRP3 Inflammasome Activation. Neurochem. Int. 2020, 141, 104856. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-J.; Feng, Y.; Liu, T.-T.; Liu, X.; Bao, J.-J.; Shi, A.-M.; Hu, D.-M.; Liu, T.; Yu, Y.-L. Thioredoxin-Interacting Protein Induced α-Synuclein Accumulation via Inhibition of Autophagic Flux: Implications for Parkinson’s Disease. CNS Neurosci. 2017, 23, 717–723. [Google Scholar] [CrossRef]
- Ables, J.L.; Breunig, J.J.; Eisch, A.J.; Rakic, P. Not(Ch) Just Development: Notch Signalling in the Adult Brain. Nat. Rev. Neurosci. 2011, 12, 269–283. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, Q. Plasma Membrane-Derived Extracellular Microvesicles Mediate Non-Canonical Intercellular NOTCH Signaling. Nat. Commun. 2017, 8, 709. [Google Scholar] [CrossRef]
- Nichols, C.D.; Sanders-Bush, E. Molecular Genetic Responses to Lysergic Acid Diethylamide Include Transcriptional Activation of MAP Kinase Phosphatase-1, C/EBP-Beta and ILAD-1, a Novel Gene with Homology to Arrestins. J. Neurochem. 2004, 90, 576–584. [Google Scholar] [CrossRef]
- Jefsen, O.H.; Elfving, B.; Wegener, G.; Müller, H.K. Transcriptional Regulation in the Rat Prefrontal Cortex and Hippocampus after a Single Administration of Psilocybin. J. Psychopharm. 2021, 35, 483–493. [Google Scholar] [CrossRef]
- Ly, C.; Greb, A.C.; Cameron, L.P.; Wong, J.M.; Barragan, E.V.; Wilson, P.C.; Burbach, K.F.; Soltanzadeh Zarandi, S.; Sood, A.; Paddy, M.R.; et al. Psychedelics Promote Structural and Functional Neural Plasticity. Cell Rep. 2018, 23, 3170–3182. [Google Scholar] [CrossRef]
- Frahm, K.A.; Peffer, M.E.; Zhang, J.Y.; Luthra, S.; Chakka, A.B.; Couger, M.B.; Chandran, U.R.; Monaghan, A.P.; DeFranco, D.B. Research Resource: The Dexamethasone Transcriptome in Hypothalamic Embryonic Neural Stem Cells. Mol. Endocrinol. 2016, 30, 144–154. [Google Scholar] [CrossRef]
- Murani, E.; Trakooljul, N.; Hadlich, F.; Ponsuksili, S.; Wimmers, K. Brain Transcriptome Responses to Dexamethasone Depending on Dose and Sex Reveal Factors Contributing to Sex-Specific Vulnerability to Stress-Induced Disorders. Neuroendocrinology 2022, 112, 235–251. [Google Scholar] [CrossRef]
- Sapolsky, R.M. Do Glucocorticoid Concentrations Rise with Age in the Rat? Neurobiol. Aging 1992, 13, 171–174. [Google Scholar] [CrossRef]
- Sharman, E.H.; Bondy, S.C.; Sharman, K.G.; Lahiri, D.; Cotman, C.W.; Perreau, V.M. Effects of Melatonin and Age on Gene Expression in Mouse CNS Using Microarray Analysis. Neurochem. Int. 2007, 50, 336–344. [Google Scholar] [CrossRef]
- Davis, O.S.P.; Butcher, L.M.; Docherty, S.J.; Meaburn, E.L.; Curtis, C.J.C.; Simpson, M.A.; Schalkwyk, L.C.; Plomin, R. A Three-Stage Genome-Wide Association Study of General Cognitive Ability: Hunting the Small Effects. Behav. Genet. 2010, 40, 759–767. [Google Scholar] [CrossRef]
- Zhang, L.; Ju, X.; Cheng, Y.; Guo, X.; Wen, T. Identifying Tmem59 Related Gene Regulatory Network of Mouse Neural Stem Cell from a Compendium of Expression Profiles. BMC Syst. Biol. 2011, 5, 152. [Google Scholar] [CrossRef]
- Lindvall, O.; Kokaia, Z.; Martinez-Serrano, A. Stem Cell Therapy for Human Neurodegenerative Disorders-How to Make It Work. Nat. Med. 2004, 10, S42–S50. [Google Scholar] [CrossRef]
- Noh, H.; Park, C.; Park, S.; Lee, Y.S.; Cho, S.Y.; Seo, H. Prediction of MiRNA-MRNA Associations in Alzheimer’s Disease Mice Using Network Topology. BMC Genom. 2014, 15, 644. [Google Scholar] [CrossRef]
- Haouari, S.; Vourc’h, P.; Jeanne, M.; Marouillat, S.; Veyrat-Durebex, C.; Lanznaster, D.; Laumonnier, F.; Corcia, P.; Blasco, H.; Andres, C.R. The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 3882. [Google Scholar] [CrossRef]
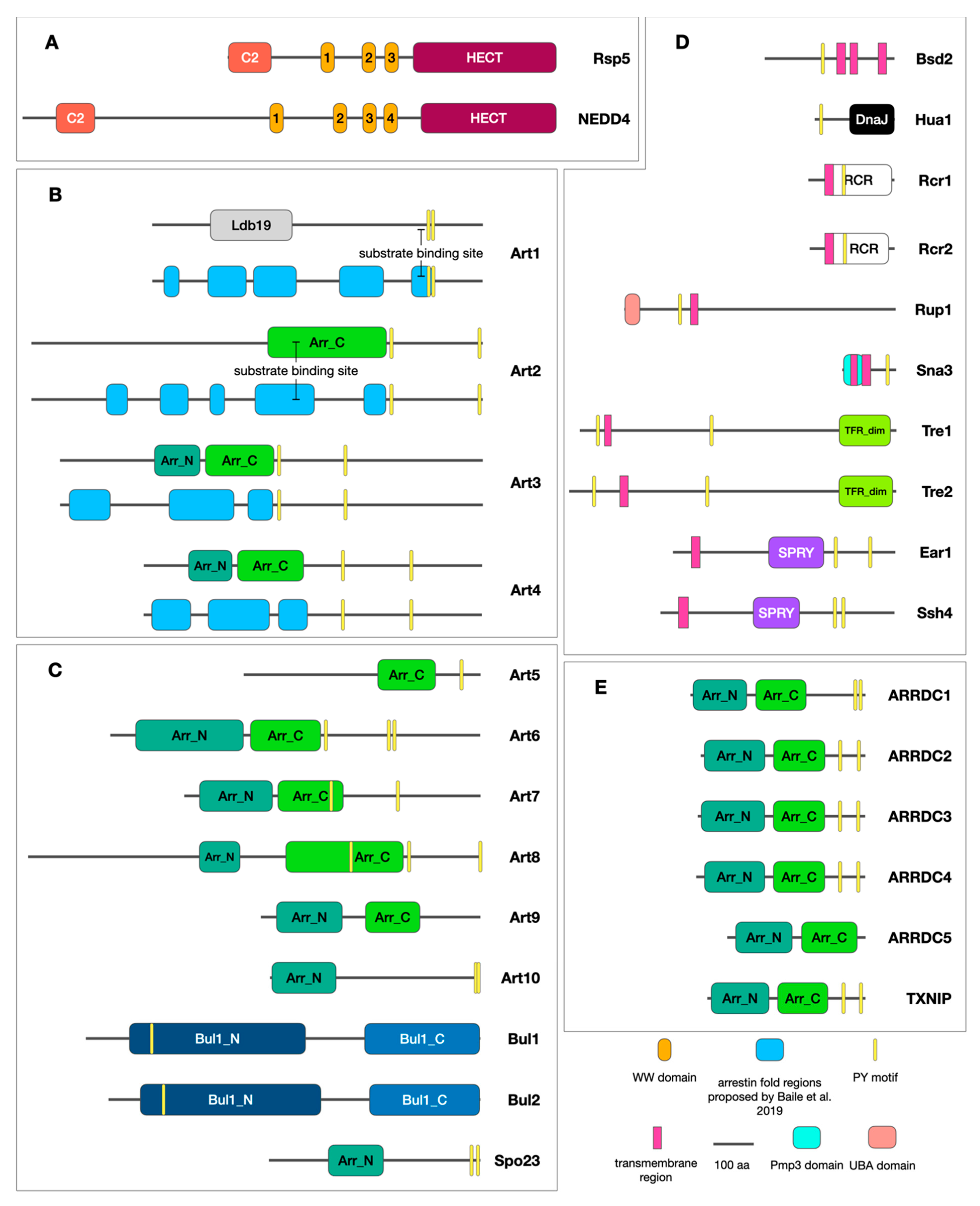
| Name | Systematic Name | Subcellular Localization | Posttranslational Regulation (Experimentally Determined) | Substrates | Identified Substrate Binding Sites |
|---|---|---|---|---|---|
| Art1/ Ldb19 | YOR322C | Cytoplasm, PM, trans-Golgi network (TGN) [11,44], early endosomes [45] | Inactivated through Npr1-dependent [45] and Clg1-dependent [26] phosphorylation; activated through Pho80-dependent phosphorylation [26] and Rsp5-dependent ubiquitination [11] | Mup1 [11,26,46,47,48] Can1 [11,26,45,46], Tat2 [13], Fur4 [13], Lyp1 [11,26], Ste2 [48,49], Ste3 [48] | Basic residues in the C-terminal half of the protein (R653, R660) [46] |
| Art2/ Ecm21 | YBL101C | Cytoplasm [44] | N/D | Mup1 [47], Can1 [47], Lyp1 [47], Ina1 [47], Smf1 [50] Tat2 [13,47], Fur4 [13], Thi7 [51], Thi72 [51], Nrt1 [51], Bap2 [52] | Basic residues in the C-terminal half of the protein (K664D, R665D, R666D, K667D) [47] |
| Art3/ Aly2 | YJL084C | Cytoplasm [44], endosomes, TGN [53] | Monoubiquitinated at K392 (unknown function) [54] | Gap1 [53], Dip5 [55,56], Ste3 [48], Acr3 [57], Git1 [54], Ena1 [58], Put4 [59] | N/D |
| Art4/ Rod1 | YOR018W | PM [60], GA, vacuole [61] | In the absence of glucose negatively regulated by Snf1-dependent phosphorylation; activated through PPI-dependent dephosphorylation and Rsp5-dependent activating ubiquitination [62] | Jen1 [62,63,64,65], Hxt6 [13,26,61,66], Hxt1 [61,67] Hxt3 [47,67] Acr3 [57], Ste2 [48,49], Stl1 [68], Gal2 [69] | The unspecified region at the N-terminal portion (amino acids 1-395) of the protein [61] |
| Art5 | YGR068C | Cytoplasm [70] | N/D | Itr1 [13] | N/D |
| Art6/ Aly1 | YKR021W | Endosomes, TGN [53] | Positively regulated by calcineurin-dependent dephosphorylation (56); monoubiquitinated at K391 (unknown function) [54] | Gap1 [53], Dip5 [55,56], Ste3 [48], Git1 [54] | N/D |
| Art7/ Rog3 | YFR022W | Cytoplasm [70] | N/D | Hxt3 [67], Ste2 [48,49], Hxt6 [66] | N/D |
| Art8/ Crs2 | YPR030W | Cytoplasm, nucleus [71] | Activated through Rsp5-dependent ubiquitination; inactivated through deubiquitination and PKA-dependent phosphorylation [63] | Smf1 [50], Fur4 [13], Tat2 [13], Hxt2 [63], Hxt3 [67,72], Hxt4 [63], Hxt6 [63], Hxt7 [63] | N/D |
| Art9/ Rim8 | YGL045W | PM, cytoplasm, nucleus [73] | Requires Rsp5-dependent monoubiquitination for ESCRT recruitment [73]; CK1-dependent phosphorylation prevents the PM association [74] | Rim21 [73,74] Nrt1 [51], Thi72 [51], Pma1 [75], Ena1 [76] | N/D |
| Art10 | YLR392C | Cytoplasm [44] | N/D | N/D | N/D |
| Bul1 | YMR275C | Cytoplasm [44] | Nitrogen starvation causes inhibition through Npr1-dependent phosphorylation; activated trough Sit4-dependent dephosphorylation and Rsp5-dependent ubiquitination [77] | Jen1 [78,79], Gap1 [53,80], Ptr2 [81,82], Tat1 [83], Tat2 [13], Ctr1 [84], Put4 [82], Dal5 [82], Agp1 [85], Fur4 [13], Can1 [86], Gal2 [69] | N/D |
| Bul2 | YML111W | Cytoplasm [44] | Nitrogen starvation causes inhibition through Npr1-dependent phosphorylation; activated trough Sit4-dependent dephosphorylation and Rsp5-dependent ubiquitination [77] | Fur4 [13], Jen1 [79], Gap1 [53,80], Ptr2 [81,82], Tat1 [83], Tat2 [83], Ctr1 [84], Put4 [82], Dal5 [82], Can1 [86] | N/D |
| Bul3 | YNR069C/YNR068C | N/D | N/D | N/D | N/D |
| Spo23 | YBR250W | N/D | N/D | N/D | N/D |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zbieralski, K.; Wawrzycka, D. α-Arrestins and Their Functions: From Yeast to Human Health. Int. J. Mol. Sci. 2022, 23, 4988. https://doi.org/10.3390/ijms23094988
Zbieralski K, Wawrzycka D. α-Arrestins and Their Functions: From Yeast to Human Health. International Journal of Molecular Sciences. 2022; 23(9):4988. https://doi.org/10.3390/ijms23094988
Chicago/Turabian StyleZbieralski, Kacper, and Donata Wawrzycka. 2022. "α-Arrestins and Their Functions: From Yeast to Human Health" International Journal of Molecular Sciences 23, no. 9: 4988. https://doi.org/10.3390/ijms23094988
APA StyleZbieralski, K., & Wawrzycka, D. (2022). α-Arrestins and Their Functions: From Yeast to Human Health. International Journal of Molecular Sciences, 23(9), 4988. https://doi.org/10.3390/ijms23094988






