Genome-Wide Identification of the NAC Gene Family in Zanthoxylum bungeanum and Their Transcriptional Responses to Drought Stress
Abstract
:1. Introduction
2. Results
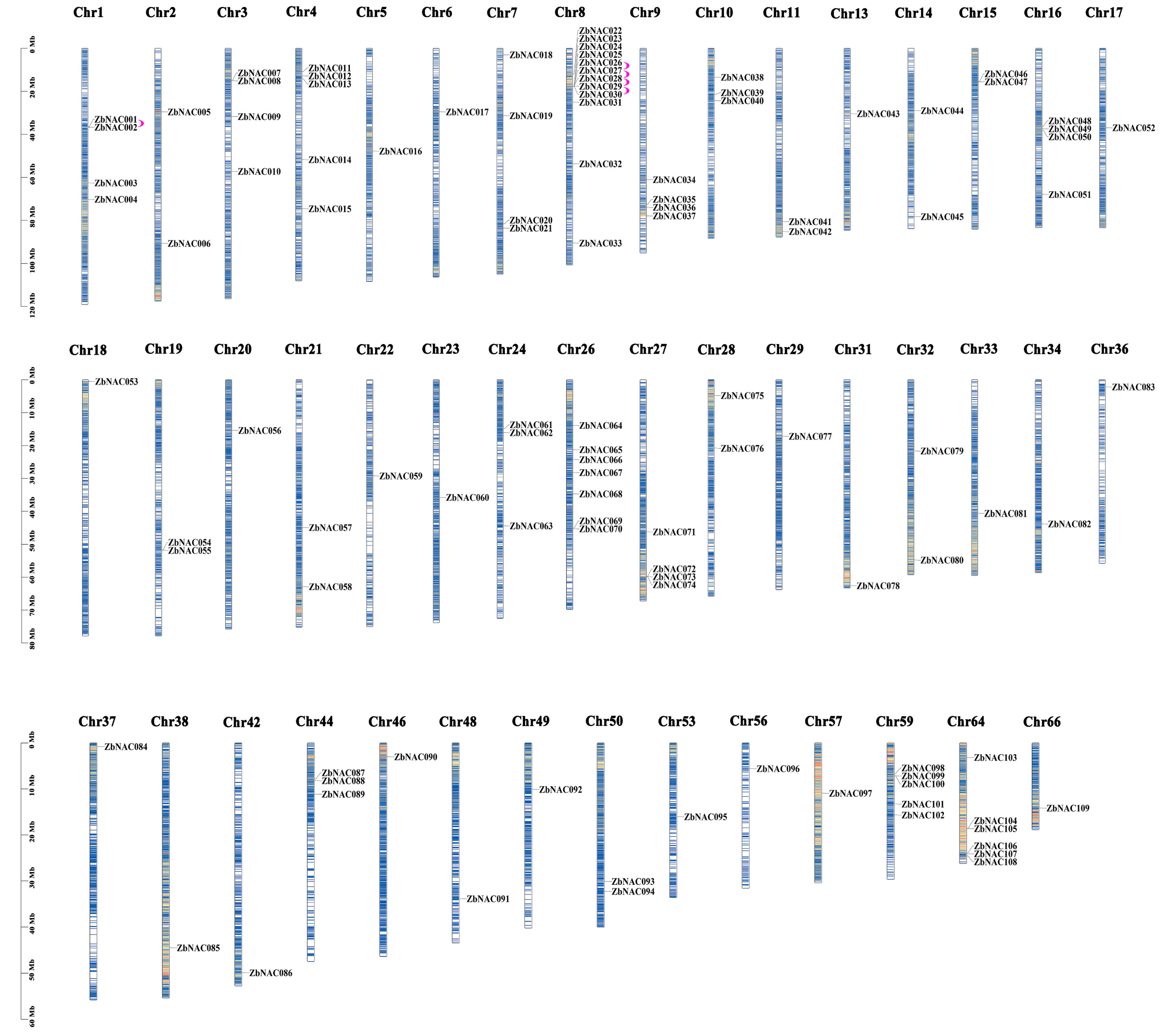
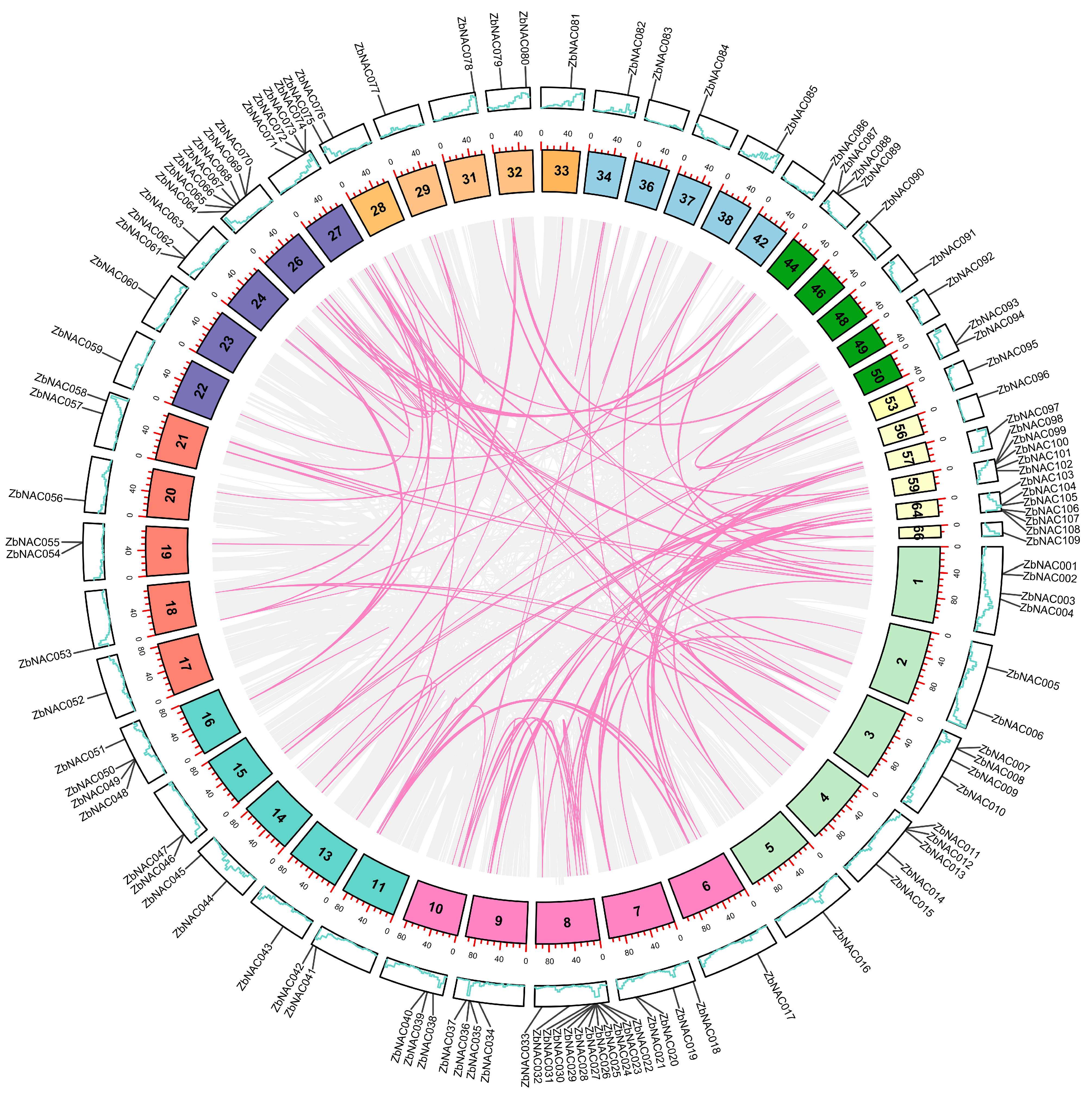
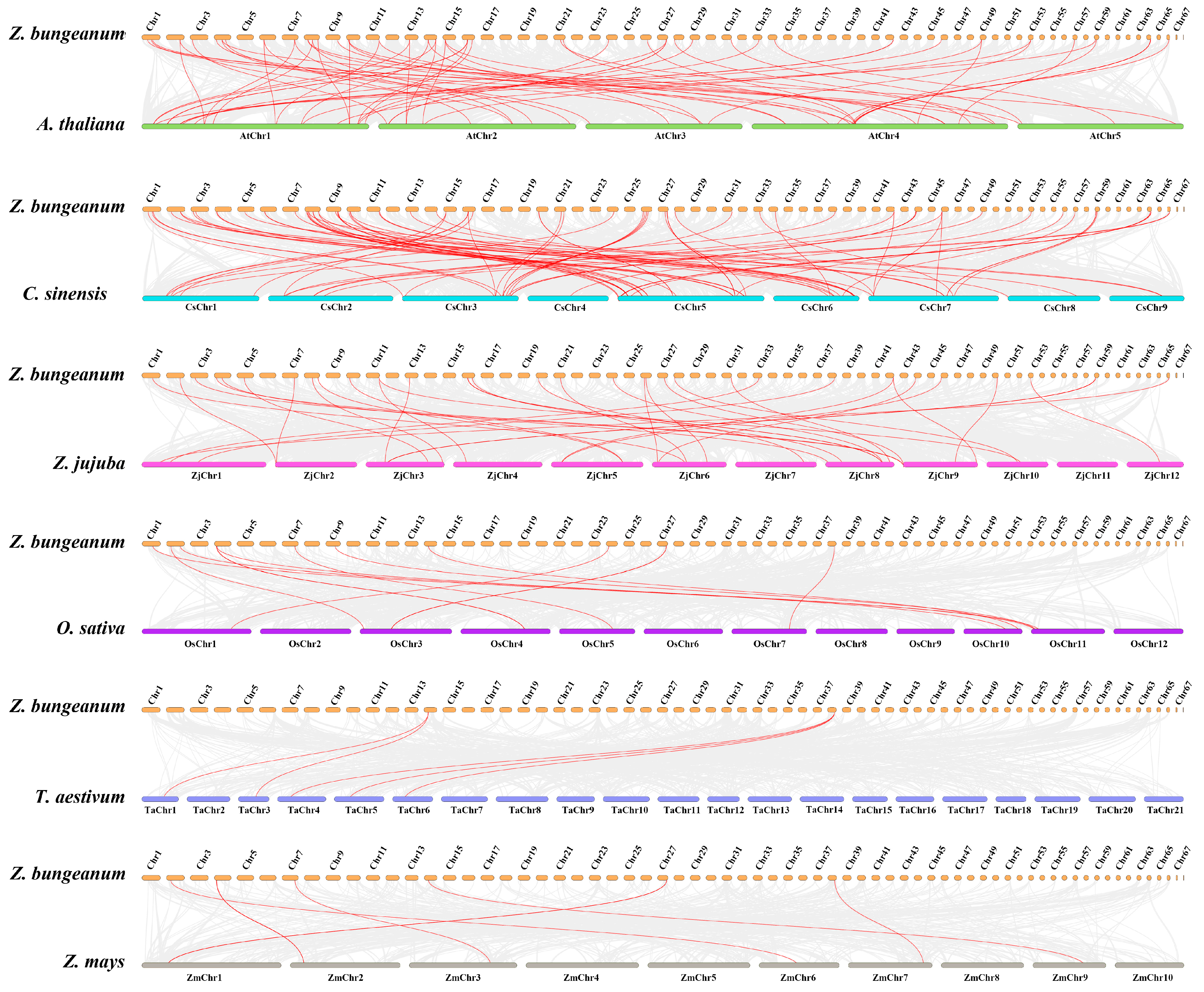
2.1. Genome-Wide Identification, Chromosomal Distribution, Synteny Analysis, and Gene Duplication of Z. bungeanum NAC Genes
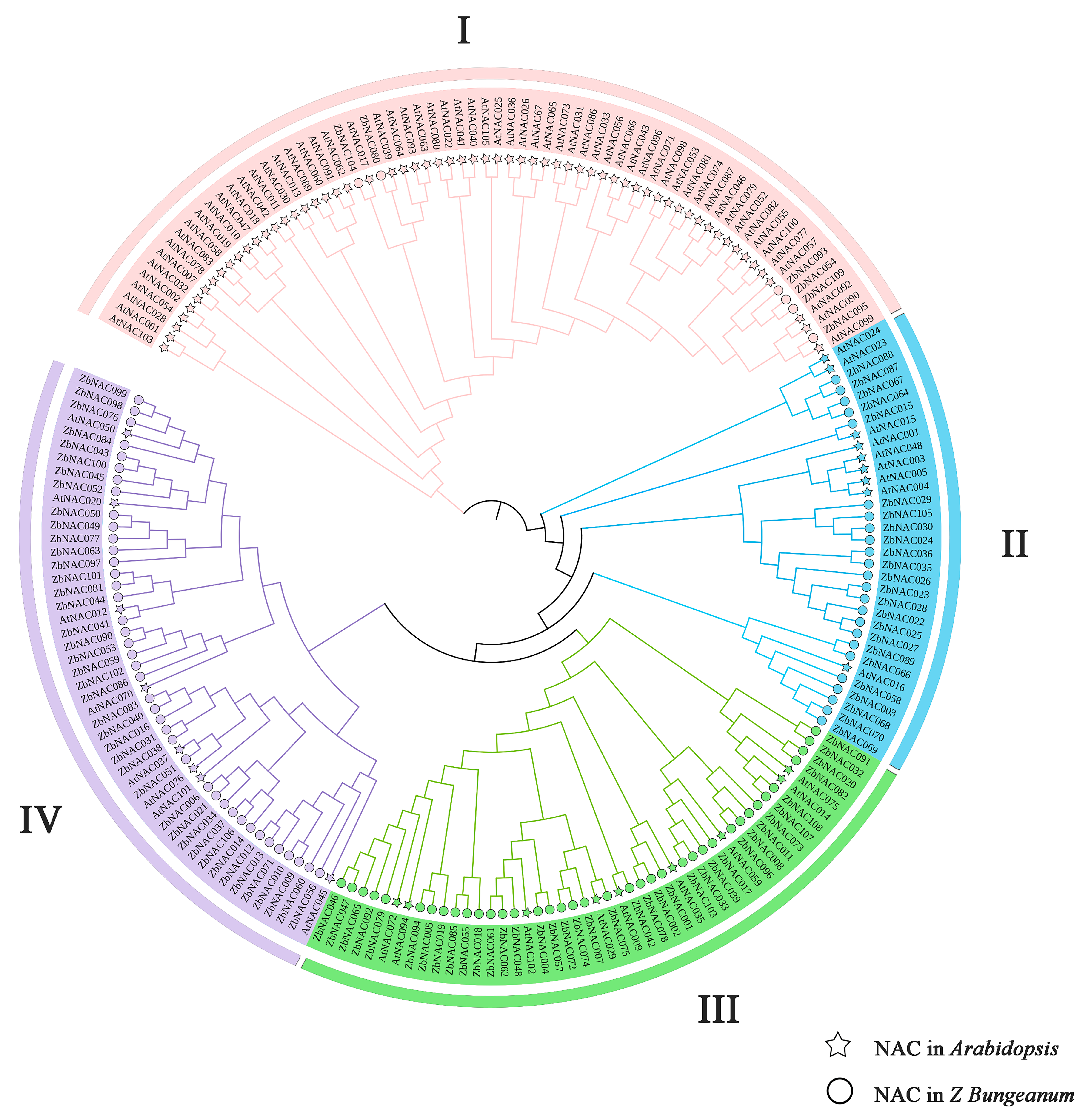
2.2. Phylogenetic Analysis of ZbNAC Proteins
2.3. Gene Structures and Conserved Motifs in ZbNAC Proteins
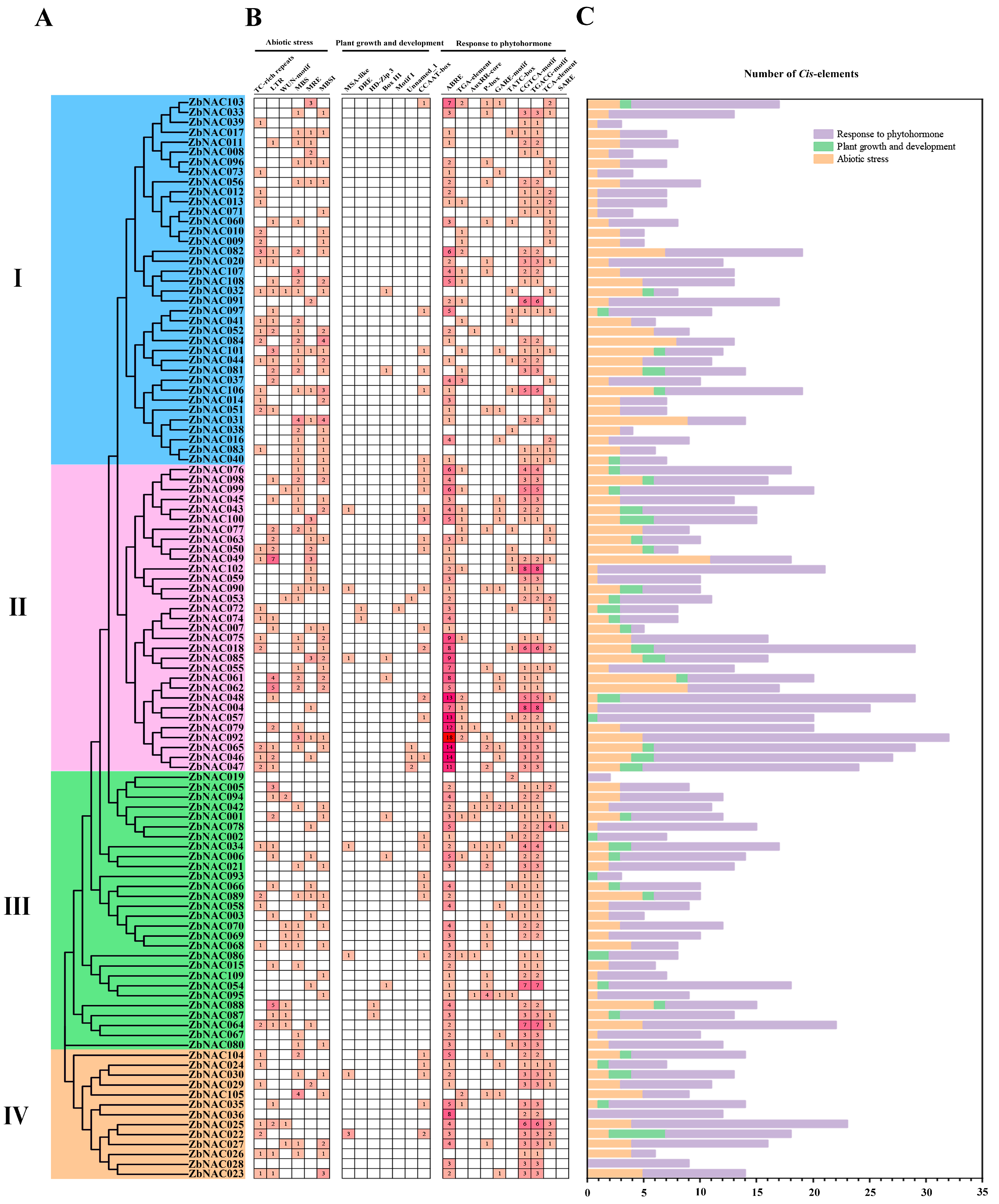
2.4. Cis-element Analysis of ZbNAC Genes
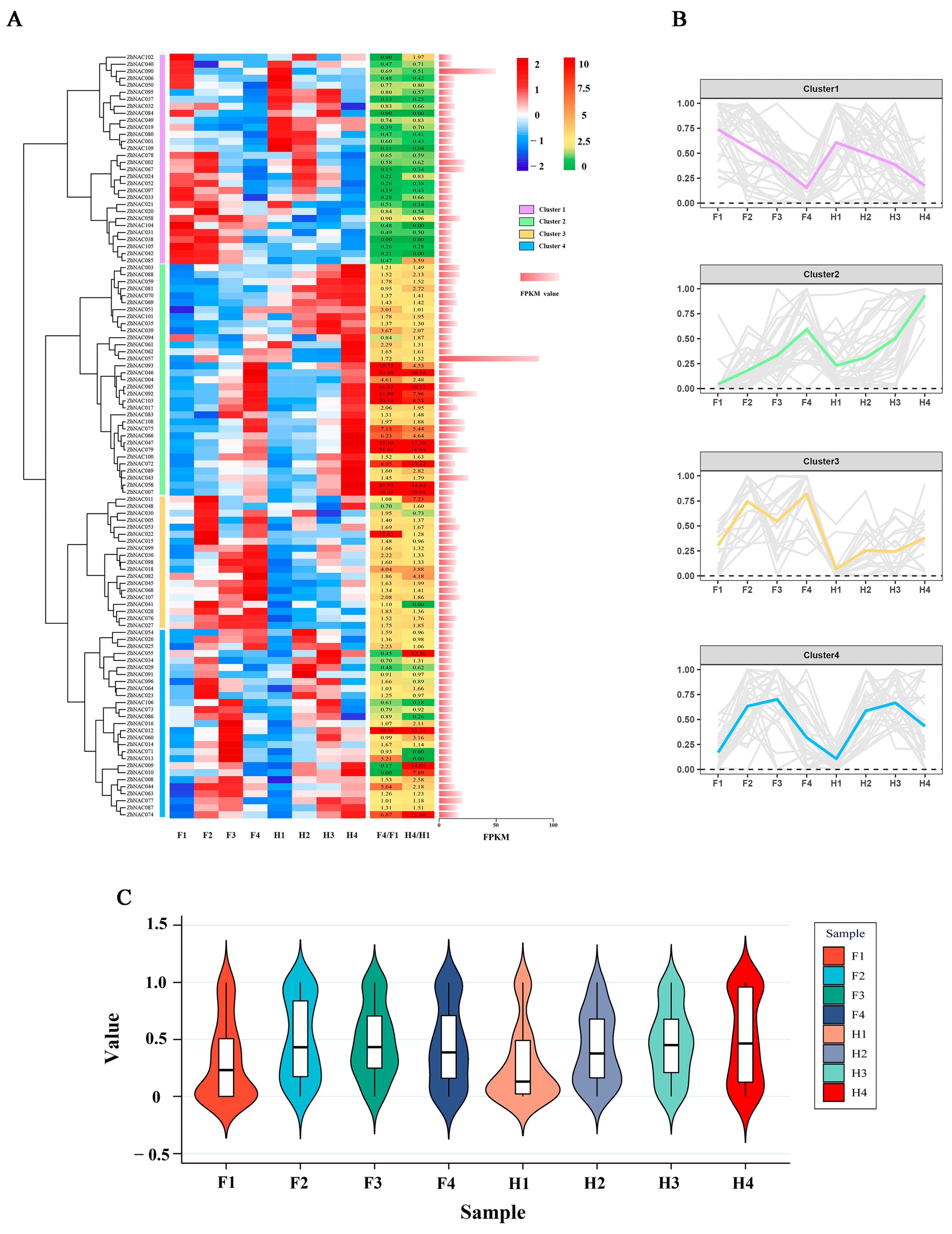
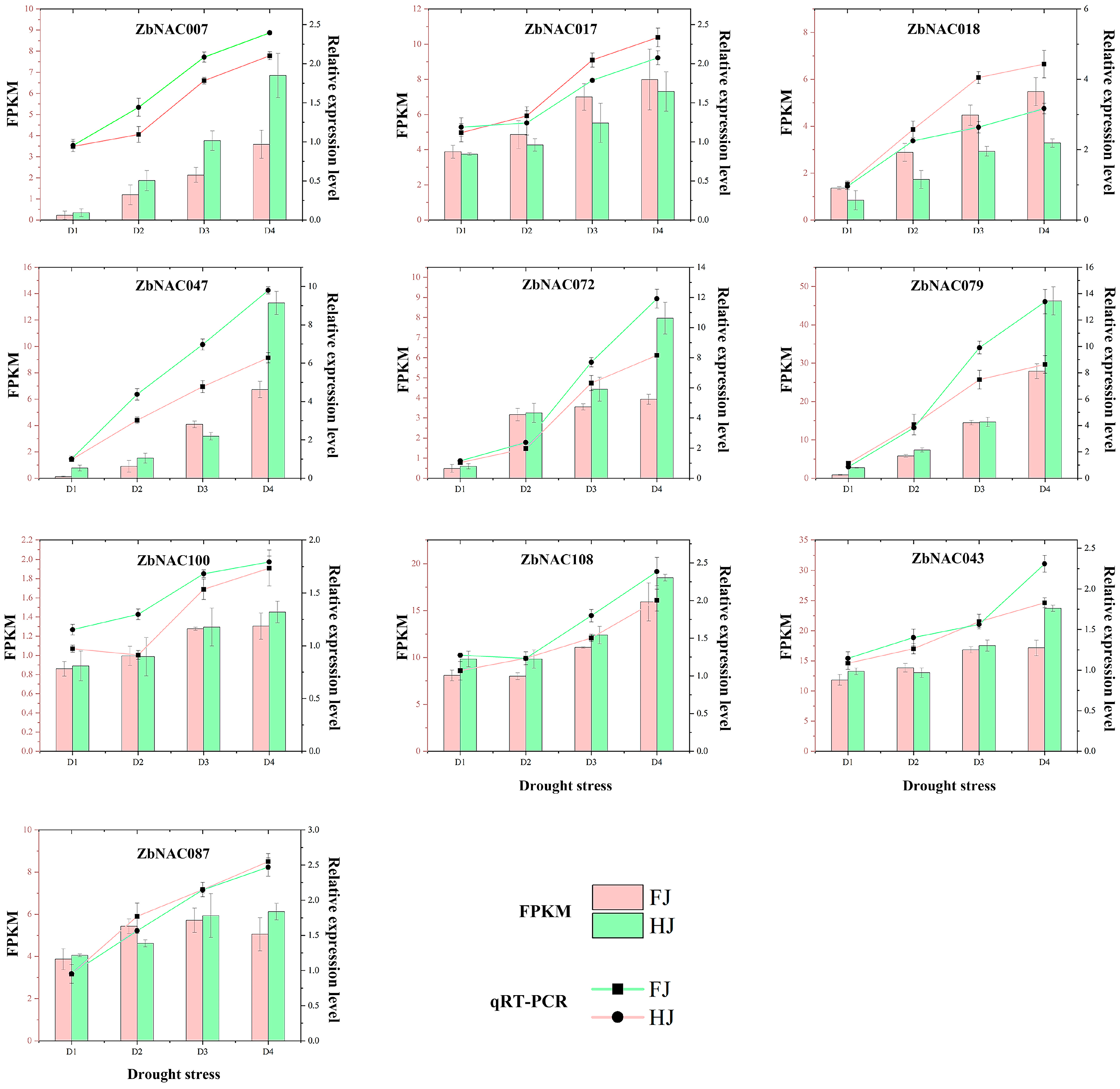
2.5. RNA-seq Analysis of 109 ZbNAC Genes under Drought Stress
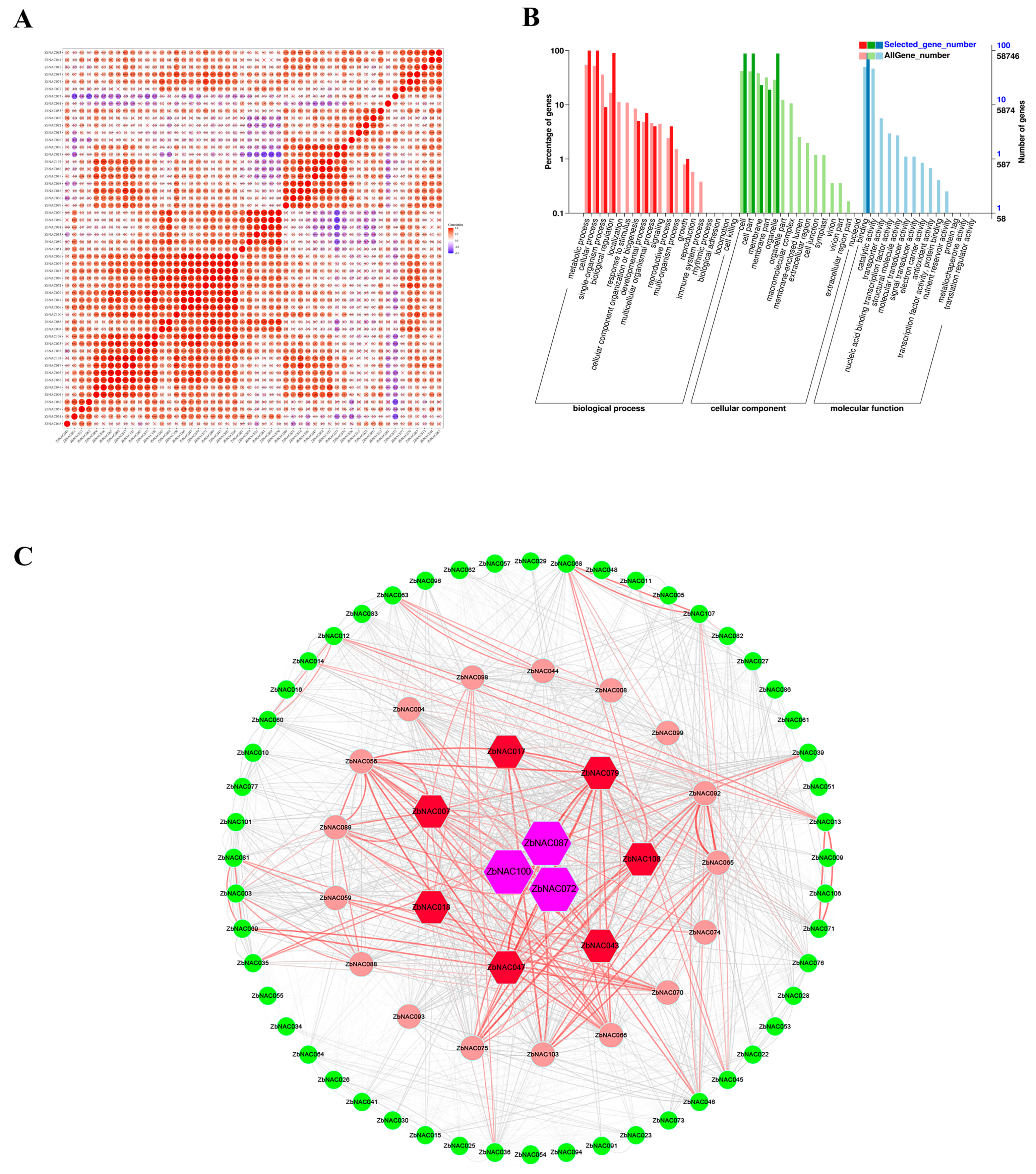
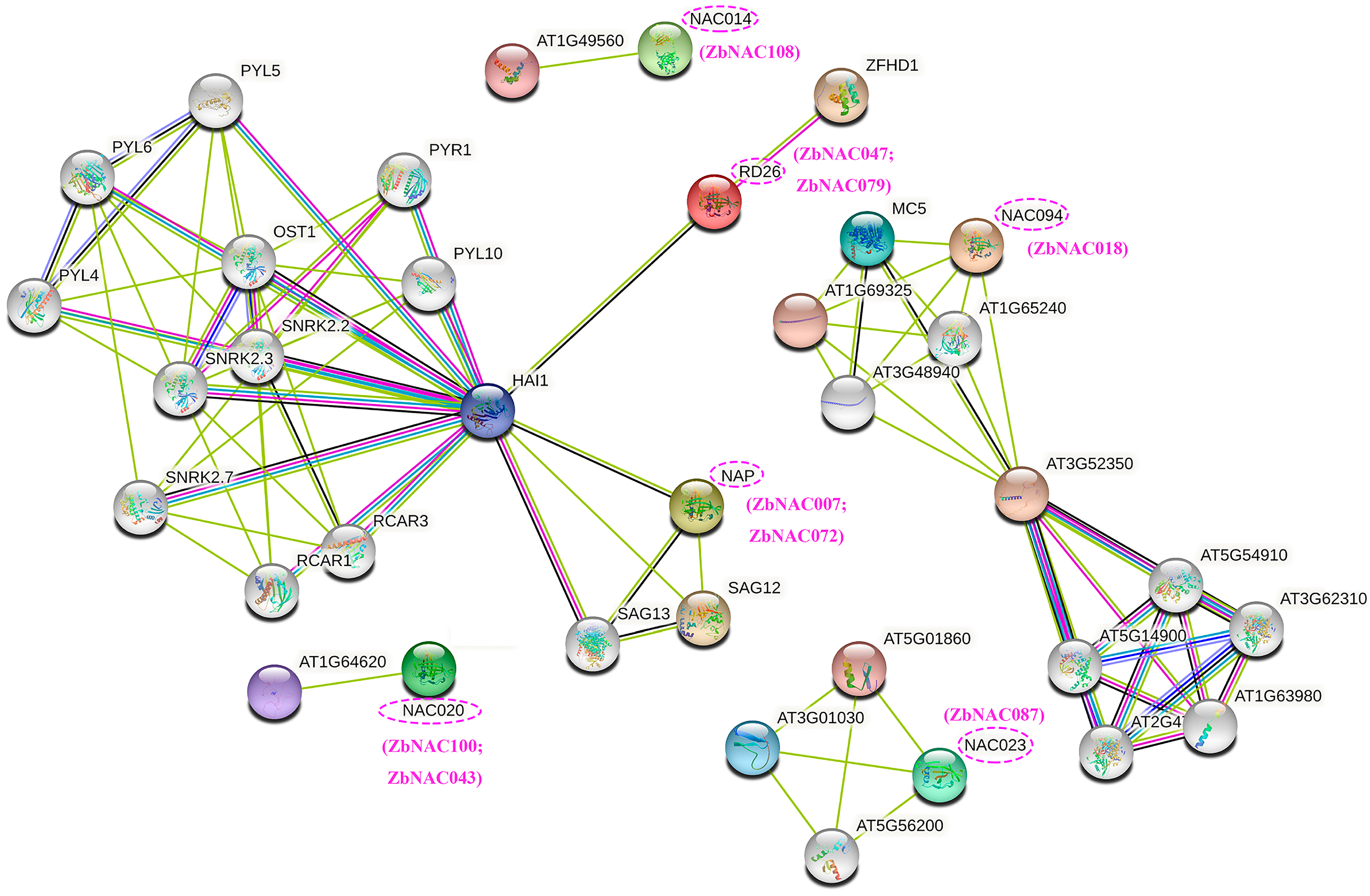
2.6. Comprehensive Analysis of Drought-Related ZbNAC Genes
3. Discussion
4. Materials and Methods
4.1. Identification of NAC Genes in Z. bungeanum
4.2. Chromosomal Distribution and Gene Duplication
4.3. Sequence Alignment and Phylogenetic Analysis of ZbNAC TFs
4.4. Gene Structure, Conserved Motif, and Cis-element Analyses of the ZbNAC TFs
4.5. Plant Materials and RNA-seq
4.6. RNA Isolation, cDNA Synthesis, and qRT-PCR
4.7. Co-expression Network and Protein–Protein Interaction Network Analysis of ZbNAC Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yao, L.; Zhang, N.; Yang, J.; Zhu, X.; Tang, X.; Calderón-Urrea, A.; Si, H. Lateral Root Development in Potato Is Mediated by Stu-mi164 Regulation of NAC Transcription Factor. Front. Plant Sci. 2018, 9, 383. [Google Scholar] [CrossRef] [PubMed]
- Kucukoglu, M. A novel NAC domain transcription factor XVP controls the balance of xylem formation and cambial cell divisions. New Phytol. 2020, 226, 5–7. [Google Scholar] [CrossRef] [Green Version]
- El Mannai, Y.; Akabane, K.; Hiratsu, K.; Satoh-Nagasawa, N.; Wabiko, H. The NAC Transcription Factor Gene OsY37 (ONAC011) Promotes Leaf Senescence and Accelerates Heading Time in Rice. Int. J. Mol. Sci. 2017, 18, 2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.-S.; Li, H.-L.; Grierson, D.; Fu, D.-Q. NAC Transcription Factor Family Regulation of Fruit Ripening and Quality: A Review. Cells 2022, 11, 525. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Zhang, J.; Ren, Y.; Li, M.; Tian, S.; Yu, Y.; Zuo, Y.; Gong, G.; Zhang, H.; et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 2021, 8, 214. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef]
- Wang, Z.; Ni, L.; Liu, D.; Fu, Z.; Hua, J.; Lu, Z.; Liu, L.; Yin, Y.; Li, H.; Gu, C. Genome-Wide Identification and Characterization of NAC Family in Hibiscus hamabo Sieb. et Zucc. under Various Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 3055. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, J.; Zhang, N.; Xin, M.; Peng, H.; Hu, Z.; Ni, Z.; Du, J. The Wheat NAC Transcription Factor TaNAC2L Is Regulated at the Transcriptional and Post-Translational Levels and Promotes Heat Stress Tolerance in Transgenic Arabidopsis. PLoS ONE 2015, 10, e0135667. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Ma, G.; da Silva, J.A.T.; Qiu, L.; Xu, J.; Zhou, H.; Wei, M.; Xiong, J.; Li, M.; Zhou, S.; et al. Genome-Wide Identification and Analysis of NAC Transcription Factor Family in Two Diploid Wild Relatives of Cultivated Sweet Potato Uncovers Potential NAC Genes Related to Drought Tolerance. Front. Genet. 2021, 12, 744220. [Google Scholar] [CrossRef] [PubMed]
- Corso, M.; Vannozzi, A.; Maza, E.; Vitulo, N.; Meggio, F.; Pitacco, A.; Telatin, A.; D’Angelo, M.; Feltrin, E.; Negri, A.S.; et al. Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. J. Exp. Bot. 2015, 66, 5739–5752. [Google Scholar] [CrossRef]
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2021, 15, 276–292. [Google Scholar] [CrossRef]
- Yu, M.; Liu, J.; Du, B.; Zhang, M.; Wang, A.; Zhang, L. NAC Transcription Factor PwNAC11 Activates ERD1 by Interaction with ABF3 and DREB2A to Enhance Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6952. [Google Scholar] [CrossRef]
- Srivastava, R.; Kobayashi, Y.; Koyama, H.; Sahoo, L. Overexpression of cowpea NAC transcription factors promoted growth and stress tolerance by boosting photosynthetic activity in Arabidopsis. Plant Sci. 2022, 319, 111251. [Google Scholar] [CrossRef]
- Feng, S.; Liu, Z.; Hu, Y.; Tian, J.; Yang, T.; Wei, A. Genomic analysis reveals the genetic diversity, population structure, evolutionary history and relationships of Chinese pepper. Hortic. Res. 2020, 7, 158. [Google Scholar] [CrossRef]
- Ma, Y.; Li, X.; Hou, L.-X.; Wei, A.-Z. Extraction solvent affects the antioxidant, antimicrobial, cholinesterase and HepG2 human hepatocellular carcinoma cell inhibitory activities of Zanthoxylum bungeanum pericarps and the major chemical components. Ind. Crop. Prod. 2019, 142, 111872. [Google Scholar] [CrossRef]
- Li, M.; Hou, L.; Liu, S.; Zhang, C.; Yang, W.; Pang, X.; Li, Y. Genome-wide identification and expression analysis of NAC transcription factors in Ziziphus jujuba Mill. reveal their putative regulatory effects on tissue senescence and abiotic stress responses. Ind. Crop. Prod. 2021, 173, 114093. [Google Scholar] [CrossRef]
- Luoni, S.A.B.; Cenci, A.; Moschen, S.; Nicosia, S.; Radonic, L.M.; García, J.V.S.Y.; Langlade, N.B.; Vile, D.; Rovere, C.V.; Fernandez, P. Genome-wide and comparative phylogenetic analysis of senescence-associated NAC transcription factors in sunflower (Helianthus annuus). BMC Genom. 2021, 22, 893. [Google Scholar] [CrossRef]
- Li, X.; Cai, K.; Pei, X.; Li, Y.; Hu, Y.; Meng, F.; Song, X.; Tigabu, M.; Ding, C.; Zhao, X. Genome-Wide Identification of NAC Transcription Factor Family in Juglans mandshurica and Their Expression Analysis during the Fruit Development and Ripening. Int. J. Mol. Sci. 2021, 22, 12414. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Peng, Z.; Xu, P.; Tang, G.; Ma, C.; Zhu, J.; Shan, L.; Wan, S. Genome-Wide Identification of NAC Transcription Factors and Their Functional Prediction of Abiotic Stress Response in Peanut. Front. Genet. 2021, 12, 630292. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Liu, Z.; Cheng, J.; Li, Z.; Tian, L.; Liu, M.; Yang, T.; Liu, Y.; Liu, Y.; Dai, H.; et al. Zanthoxylum-specific whole genome duplication and recent activity of transposable elements in the highly repetitive paleotetraploid Z. bungeanum genome. Hortic. Res. 2021, 8, 205. [Google Scholar] [CrossRef]
- Eshao, H.-B.; Ewang, H.; Etang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Nie, G.; Feng, G.; Han, J.; Huang, L.; Zhang, X. Genome-wide identification, characterization, and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata L.). BMC Genom. 2021, 22, 178. [Google Scholar] [CrossRef]
- Wei, S.; Gao, L.; Zhang, Y.; Zhang, F.; Yang, X.; Huang, D. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Rep. 2016, 35, 1827–1839. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wei, Y.; Xia, Z.; Yan, Y.; Hou, X.; Zou, M.; Lu, C.; Wang, W.; Peng, M. Genome-Wide Identification and Expression Analysis of the NAC Transcription Factor Family in Cassava. PLoS ONE 2015, 10, e0136993. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Shiriga, K.; Sharma, R.; Kumar, K.; Yadav, S.K.; Hossain, F.; Thirunavukkarasu, N. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2014, 2, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-Wide Survey and Expression Analysis of the Plant-Specific NAC Transcription Factor Family in Soybean During Development and Dehydration Stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowers, J.; Chapman, B.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A.; Tabassum, B.; Khan, A.L.; Abd_Allah, E.F.; Al-Harrasi, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Shan, Z.; Jiang, Y.; Li, H.; Guo, J.; Dong, M.; Zhang, J.; Liu, G. Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genom. 2020, 21, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Fan, R.; Yang, Q.; Hu, C.; Sheng, O.; Deng, G.; Dong, T.; Li, C.; Peng, X.; Bi, F.; et al. Genome-Wide Identification and Characterization of the NAC Transcription Factor Family in Musa Acuminata and Expression Analysis during Fruit Ripening. Int. J. Mol. Sci. 2020, 21, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Kandul, N.P.; Noor, M.A. Large introns in relation to alternative splicing and gene evolution: A case study of Drosophila bruno-3. BMC Genet. 2009, 10, 67. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Kudapa, H.; Garg, V.; Varshney, R.K. Comprehensive analysis and identification of drought-responsive candidate NAC genes in three semi-arid tropics (SAT) legume crops. BMC Genom. 2021, 22, 289. [Google Scholar] [CrossRef]
- Li, W.; Zeng, Y.; Yin, F.; Wei, R.; Mao, X. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in sunflower during salt and drought stress. Sci. Rep. 2021, 11, 19865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, R.; Gao, Z.; Chen, C.; Jiang, Z.; Shu, H. A genome-wide analysis of the expansin genes in Malus × Domestica. Mol. Genet. Genom. 2013, 289, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Yang, Z.; He, J.; Liu, A.; Chen, J.; Wang, S.; Wang, X.; Feng, G.; Li, D.; Peng, Y.; et al. Genome-Wide Investigation of the NAC Transcription Factor Family in Miscanthus sinensis and Expression Analysis Under Various Abiotic Stresses. Front. Plant Sci. 2021, 12, 766550. [Google Scholar] [CrossRef]
- Wu, R.; Duan, L.; Pruneda-Paz, J.L.; Oh, D.-H.; Pound, M.; Kay, S.; Dinneny, J.R. The 6xABRE Synthetic Promoter Enables the Spatiotemporal Analysis of ABA-Mediated Transcriptional Regulation. Plant Physiol. 2018, 177, 1650–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehong, Y.; Ezhang, H.; Ehuang, L.; Eli, D.; Esong, F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Han Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Jiang, Y.; Jin, Y.; Wang, C.; Yang, J.; Qi, H. Drought-induced ABA, H2O2 and JA positively regulate CmCAD genes and lignin synthesis in melon stems. BMC Plant Biol. 2021, 21, 83. [Google Scholar] [CrossRef]
- Aroca, R.; Alguacil, M.D.M.; Vernieri, P.; Ruiz-Lozano, J.M. Plant Responses to Drought Stress and Exogenous ABA Application are Modulated Differently by Mycorrhization in Tomato and an ABA-deficient Mutant (Sitiens). Microb. Ecol. 2008, 56, 704–719. [Google Scholar] [CrossRef]
- Jiang, Y.; Ye, J.; Li, S. Niinemets, Ülo Methyl jasmonate-induced emission of biogenic volatiles is biphasic in cucumber: A high-resolution analysis of dose dependence. J. Exp. Bot. 2017, 68, 4679–4694. [Google Scholar] [CrossRef]
- Liu, T.; Li, C.-X.; Zhong, J.; Shu, D.; Luo, D.; Li, Z.-M.; Zhou, J.-Y.; Yang, J.; Tan, H.; Ma, X.-R. Exogenous 1’,4’-trans-Diol-ABA Induces Stress Tolerance by Affecting the Level of Gene Expression in Tobacco (Nicotiana tabacum L.). Int. J. Mol. Sci. 2021, 22, 2555. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Li, C.; Lu, X.; Zhao, X.; Yan, C.; Wang, J.; Sun, Q.; Shan, S. Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol. 2020, 20, 454. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M.H.S.A. Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network. Int. J. Mol. Sci. 2018, 19, 1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, W.A.; Afzal, M.; Chen, X.; Cui, R.; Lu, X.; Wang, S.; Wang, J.; Mahmood, I.; Ye, W. Systematic analysis and comparison of ABC proteins superfamily confer structural, functional and evolutionary insights into four cotton species. Ind. Crop. Prod. 2021, 177, 114433. [Google Scholar] [CrossRef]
- Hou, Q.; Qiu, Z.; Wen, Z.; Zhang, H.; Li, Z.; Hong, Y.; Qiao, G.; Wen, X. Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L. Int. J. Mol. Sci. 2021, 22, 11968. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Zhou, W.; Xie, L.; Wang, L.; Zhang, Y.; Zhang, Q. Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genom. 2021, 22, 361. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Wong, M.M.; Bhaskara, G.B.; Wen, T.-N.; Lin, W.-D.; Nguyen, T.T.; Chong, G.L.; Verslues, P.E. Phosphoproteomics of Arabidopsis Highly ABA-Induced1 identifies AT-Hook– Like10 phosphorylation required for stress growth regulation. Proc. Natl. Acad. Sci. USA 2019, 116, 2354–2363. [Google Scholar] [CrossRef] [Green Version]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, Y.-S.; Han, S.-H.; Lee, B.-D.; Paek, N.-C. The Arabidopsis Transcription Factor NAC016 Promotes Drought Stress Responses by Repressing AREB1 Transcription through a Trifurcate Feed-Forward Regulatory Loop Involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.; Walley, J.; Yin, Y. GSK 3-like kinase BIN 2 phosphorylates RD 26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, N.; Lam, E. Recent advance in the study of caspase-like proteases and Bax inhibitor-1 in plants: Their possible roles as regulator of programmed cell death. Mol. Plant Pathol. 2004, 5, 65–70. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef] [Green Version]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinform. 2017, 60, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2006, 35, D883–D887. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Fei, X.; Shi, Q.; Yang, T.; Fei, Z.; Wei, A. Expression Stabilities of Ten Candidate Reference Genes for RT-qPCR in Zanthoxylum bungeanum Maxim. Molecules 2018, 23, 802. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Duplication Type |
|---|---|---|---|---|---|
| ZbNAC001 | ZbNAC002 | 0.10 | 0.43 | 0.24 | tandem |
| ZbNAC001 | ZbNAC078 | 0.15 | 0.40 | 0.37 | segmental |
| ZbNAC003 | ZbNAC069 | 0.09 | 0.27 | 0.31 | segmental |
| ZbNAC003 | ZbNAC058 | 0.11 | 0.30 | 0.35 | segmental |
| ZbNAC004 | ZbNAC057 | 0.07 | 0.36 | 0.20 | segmental |
| ZbNAC006 | ZbNAC021 | 0.06 | 0.30 | 0.21 | segmental |
| ZbNAC009 | ZbNAC010 | 0.01 | 0.03 | 0.18 | segmental |
| ZbNAC009 | ZbNAC013 | 0.09 | 0.29 | 0.32 | segmental |
| ZbNAC010 | ZbNAC012 | 0.09 | 0.26 | 0.35 | segmental |
| ZbNAC024 | ZbNAC035 | 0.46 | 1.06 | 0.43 | segmental |
| ZbNAC024 | ZbNAC028 | 0.47 | 0.73 | 0.64 | segmental |
| ZbNAC025 | ZbNAC026 | 0.20 | 0.27 | 0.74 | segmental |
| ZbNAC026 | ZbNAC027 | 0.21 | 0.26 | 0.82 | tandem |
| ZbNAC026 | ZbNAC036 | 0.47 | 0.65 | 0.72 | segmental |
| ZbNAC027 | ZbNAC028 | 0.20 | 0.17 | 1.22 | tandem |
| ZbNAC028 | ZbNAC029 | 0.52 | 0.84 | 0.61 | tandem |
| ZbNAC029 | ZbNAC030 | 0.36 | 0.53 | 0.68 | tandem |
| ZbNAC035 | ZbNAC036 | 0.02 | 0.04 | 0.64 | segmental |
| ZbNAC038 | ZbNAC031 | 0.11 | 0.35 | 0.30 | segmental |
| ZbNAC039 | ZbNAC033 | 0.07 | 0.22 | 0.34 | segmental |
| ZbNAC041 | ZbNAC097 | 0.11 | 0.33 | 0.32 | segmental |
| ZbNAC043 | ZbNAC045 | 0.07 | 0.22 | 0.31 | segmental |
| ZbNAC044 | ZbNAC101 | 0.06 | 0.18 | 0.33 | segmental |
| ZbNAC046 | ZbNAC092 | 0.08 | 0.36 | 0.23 | segmental |
| ZbNAC049 | ZbNAC063 | 0.09 | 0.19 | 0.47 | segmental |
| ZbNAC049 | ZbNAC050 | 0.01 | 0.01 | 1.13 | segmental |
| ZbNAC050 | ZbNAC063 | 0.10 | 0.21 | 0.49 | segmental |
| ZbNAC053 | ZbNAC090 | 0.04 | 0.19 | 0.20 | segmental |
| ZbNAC058 | ZbNAC069 | 0.10 | 0.27 | 0.38 | segmental |
| ZbNAC059 | ZbNAC090 | 0.04 | 0.17 | 0.24 | segmental |
| ZbNAC064 | ZbNAC088 | 0.17 | 0.27 | 0.62 | segmental |
| ZbNAC064 | ZbNAC087 | 0.08 | 0.12 | 0.71 | segmental |
| ZbNAC066 | ZbNAC089 | 0.09 | 0.16 | 0.54 | segmental |
| ZbNAC068 | ZbNAC100 | 0.07 | 0.25 | 0.26 | segmental |
| ZbNAC068 | ZbNAC069 | 0.00 | 0.01 | 0.35 | segmental |
| ZbNAC071 | ZbNAC012 | 0.07 | 0.28 | 0.26 | segmental |
| ZbNAC071 | ZbNAC009 | 0.08 | 0.29 | 0.26 | segmental |
| ZbNAC071 | ZbNAC013 | 0.07 | 0.28 | 0.26 | segmental |
| ZbNAC072 | ZbNAC074 | 0.01 | 0.03 | 0.27 | segmental |
| ZbNAC072 | ZbNAC007 | 0.09 | 0.29 | 0.31 | segmental |
| ZbNAC073 | ZbNAC011 | 0.14 | 0.32 | 0.45 | segmental |
| ZbNAC074 | ZbNAC007 | 0.09 | 0.27 | 0.33 | segmental |
| ZbNAC076 | ZbNAC099 | 0.00 | 0.04 | 0.11 | segmental |
| ZbNAC105 | ZbNAC028 | 0.52 | 1.18 | 0.44 | segmental |
| ZbNAC105 | ZbNAC024 | 0.24 | 0.39 | 0.60 | segmental |
| ZbNAC106 | ZbNAC037 | 0.06 | 0.34 | 0.16 | segmental |
| ZbNAC107 | ZbNAC108 | 0.04 | 0.05 | 0.74 | segmental |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Ma, L.; Chen, X.; Fei, X.; He, B.; Luo, Y.; Liu, Y.; Wei, A. Genome-Wide Identification of the NAC Gene Family in Zanthoxylum bungeanum and Their Transcriptional Responses to Drought Stress. Int. J. Mol. Sci. 2022, 23, 4769. https://doi.org/10.3390/ijms23094769
Hu H, Ma L, Chen X, Fei X, He B, Luo Y, Liu Y, Wei A. Genome-Wide Identification of the NAC Gene Family in Zanthoxylum bungeanum and Their Transcriptional Responses to Drought Stress. International Journal of Molecular Sciences. 2022; 23(9):4769. https://doi.org/10.3390/ijms23094769
Chicago/Turabian StyleHu, Haichao, Lei Ma, Xin Chen, Xitong Fei, Beibei He, Yingli Luo, Yonghong Liu, and Anzhi Wei. 2022. "Genome-Wide Identification of the NAC Gene Family in Zanthoxylum bungeanum and Their Transcriptional Responses to Drought Stress" International Journal of Molecular Sciences 23, no. 9: 4769. https://doi.org/10.3390/ijms23094769
APA StyleHu, H., Ma, L., Chen, X., Fei, X., He, B., Luo, Y., Liu, Y., & Wei, A. (2022). Genome-Wide Identification of the NAC Gene Family in Zanthoxylum bungeanum and Their Transcriptional Responses to Drought Stress. International Journal of Molecular Sciences, 23(9), 4769. https://doi.org/10.3390/ijms23094769






