Sequencing of the Viral UL111a Gene Directly from Clinical Specimens Reveals Variants of HCMV-Encoded IL-10 That Are Associated with Altered Immune Responses to HCMV
Abstract
1. Introduction
2. Results
2.1. Most Clinical Samples Contain More Than One Variant of HCMV
2.2. Several Polymorphisms Were Group-Specific
2.3. Amino Acid Haplotypes Differ between Samples from Australia and Indonesia
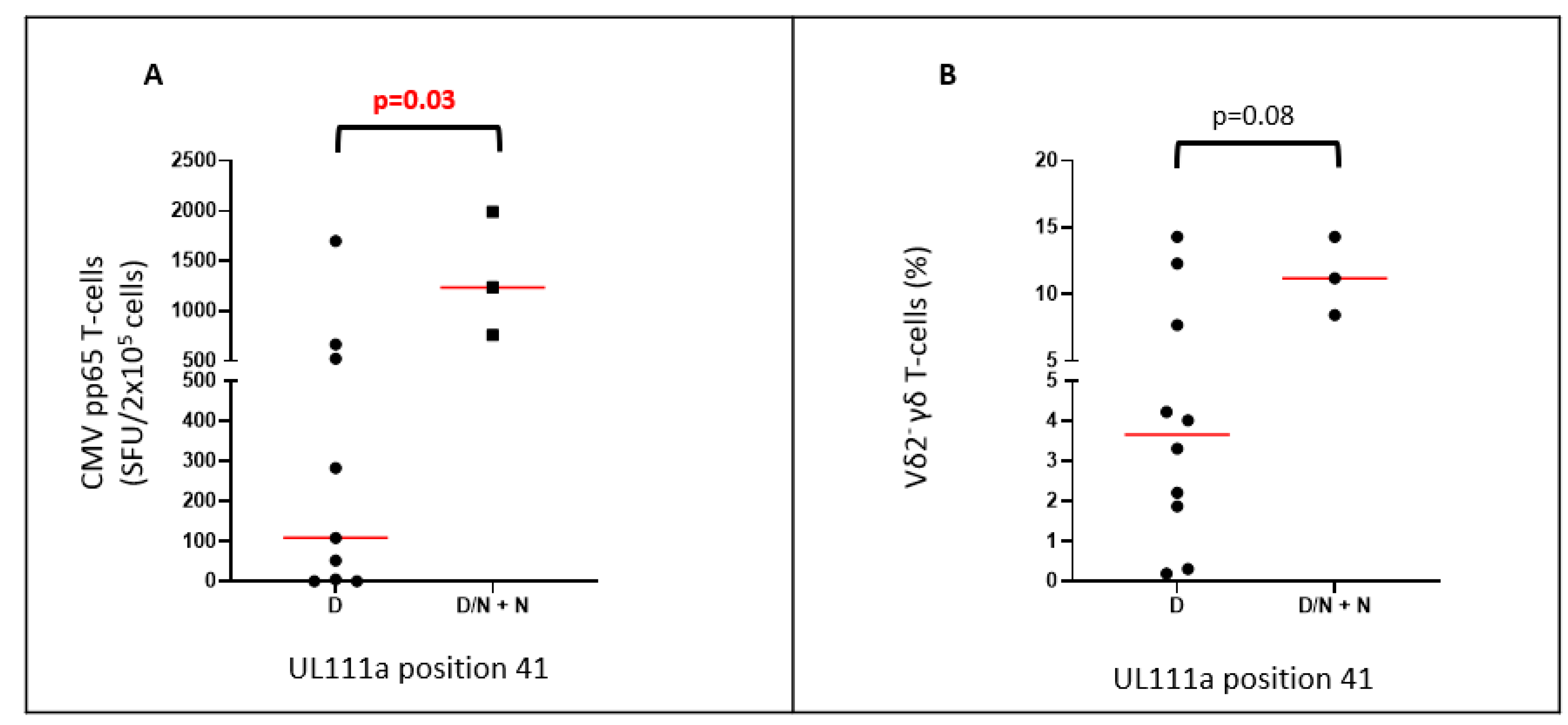
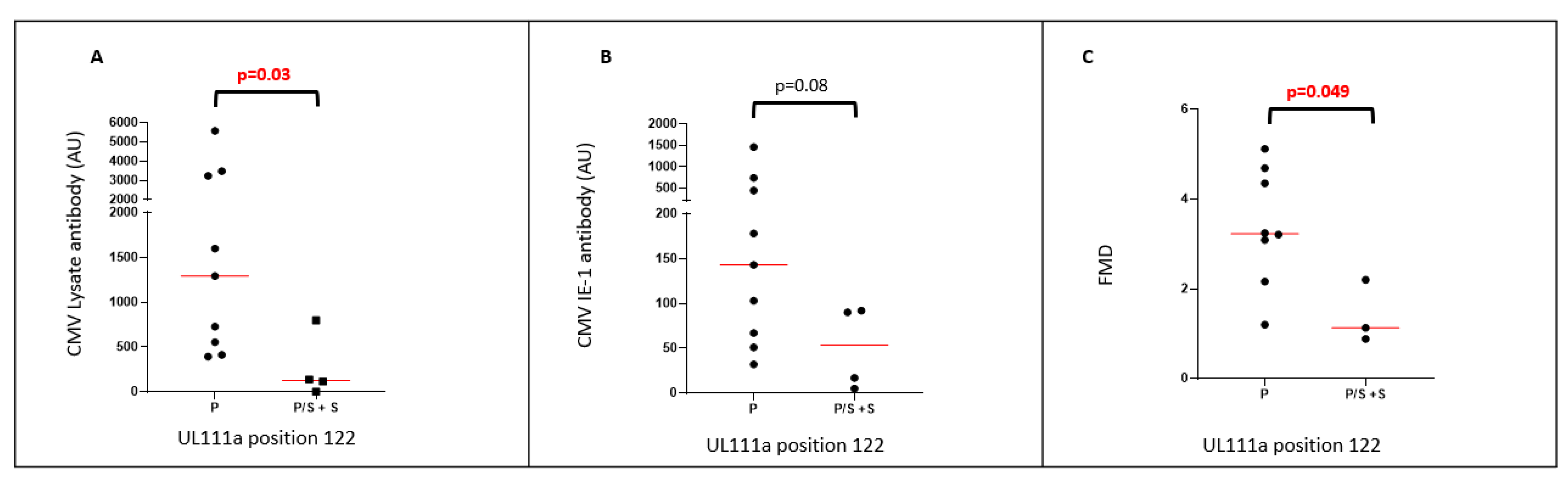
2.4. UL111a Variations Are Associated with Levels of HCMV-Reactive Antibody
3. Discussion
4. Materials and Methods
4.1. RTR and Healthy Controls from Perth, Western Australia
4.2. People with HIV from Jakarta, Indonesia
4.3. Australia Neonates
4.4. Extraction and Detection of HCMV DNA
4.5. Targeted Whole Gene Amplification
4.6. Preparation of Ion Ampliseq™ DNA Libraries
4.7. Libraries Were Sequenced Using an Ion Proton Sequencer
4.8. Immunological Assessments of HCMV
4.9. Assessment of Vascular Pathology
4.10. Data Analysis
4.11. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, G.; Bai, J.; He, B.; Huang, K.; Hu, X.; Liu, D. Cytomegalovirus Infection and Relative Risk of Cardiovascular Disease (Ischemic Heart Disease, Stroke, and Cardiovascular Death): A Meta-Analysis of Prospective Studies Up to 2016. J. Am. Heart Assoc. 2017, 6, e005025. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Brook, E.; Lee, S.; Estiasari, R.; Ariyanto, I.; Price, P. HIV patients, healthy aging and transplant recipients can reveal the hidden footprints of CMV. Clin. Immunol. 2018, 187, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Akpan, U.S.; Pillarisetty, L.S. Congenital Cytomegalovirus Infection; StatPearls publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Cukuranovic, J.; Ugrenovic, S.; Jovanovic, I.; Visnjic, M.; Stefanovic, V. Viral infection in renal transplant recipients. Sci. World J. 2012, 2012, 820621. [Google Scholar] [CrossRef]
- Fischer, S.A.; Avery, R.K.; American Society for Transplantation Infectious Disease Community of Practice. Screening of donor and recipient prior to solid organ transplantation. Am. J. Transplant. 2009, 9 (Suppl. S4), S7–S18. [Google Scholar] [CrossRef]
- Fowotade, A.; Okonko, I.O.; Agbede, O.O.; Suleiman, S.T. High seropositivity of IgG and IgM antibodies against cytomegalovirus (CMV) among HIV-1 seropositive patients in Ilorin, Nigeria. Afr. Health Sci. 2015, 15, 1–9. [Google Scholar] [CrossRef]
- Varo, R.; Buck, W.C.; Kazembe, P.N.; Phiri, S.; Andrianarimanana, D.; Weigel, R. Seroprevalence of CMV, HSV-2 and HBV among HIV-Infected Malawian Children: A Cross-sectional Survey. J. Trop. Pediatr. 2016, 62, 220–226. [Google Scholar] [CrossRef][Green Version]
- Lim, R.B.; Tan, M.T.; Young, B.; Lee, C.C.; Leo, Y.S.; Chua, A.; Ng, O.T. Risk factors and time-trends of cytomegalovirus (CMV), syphilis, toxoplasmosis and viral hepatitis infection and seroprevalence in human immunodeficiency virus (HIV) infected patients. Ann. Acad. Med. Singap. 2013, 42, 667–673. [Google Scholar]
- Deayton, J.R.; Wilson, P.; Sabin, C.A.; Davey, C.C.; Johnson, M.A.; Emery, V.C.; Griffiths, P.D. Changes in the natural history of cytomegalovirus retinitis following the introduction of highly active antiretroviral therapy. AIDS 2000, 14, 1163–1170. [Google Scholar] [CrossRef]
- Lee, S.; Saraswati, H.; Yunihastuti, E.; Gani, R.; Price, P. Patients co-infected with hepatitis C virus (HCV) and human immunodeficiency virus recover genotype cross-reactive neutralising antibodies to HCV during antiretroviral therapy. Clin. Immunol. 2014, 155, 149–159. [Google Scholar] [CrossRef]
- Lichtner, M.; Cicconi, P.; Vita, S.; Cozzi-Lepri, A.; Galli, M.; Caputo, S.L.; Saracino, A.; de Luca, A.; Moioli, M.; Maggiolo, F.; et al. Cytomegalovirus coinfection is associated with an increased risk of severe non-AIDS-defining events in a large cohort of HIV-infected patients. J. Infect. Dis. 2015, 211, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Sijmons, S.; Thys, K.; Ngwese, M.M.; van Damme, E.; Dvorak, J.; van Loock, M.; Li, G.; Tachezy, R.; Busson, L.; Aerssens, J.; et al. High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination. J. Virol. 2015, 89, 7673–7695. [Google Scholar] [CrossRef] [PubMed]
- Shenk, T.E.; Stinski, M.F. Human cytomegalovirus. Preface. Curr. Top Microbiol. Immunol. 2008, 325, 336–339. [Google Scholar]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.; van Loock, M. Functional annotation of human cytomegalovirus gene products: An update. Front. Microbiol. 2014, 5, 218. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Silva, M.C.; Shenk, T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 12396–12401. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Abendroth, A.; Slobedman, B. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 2004, 78, 1440–1447. [Google Scholar] [CrossRef]
- Poole, E.; Neves, T.C.; Oliveira, M.T.; Sinclair, J.; da Silva, M.C.C. Human Cytomegalovirus Interleukin 10 Homologs: Facing the Immune System. Front. Cell Infect. Microbiol. 2020, 10, 245. [Google Scholar] [CrossRef]
- Poole, E.; Sinclair, J. Sleepless latency of human cytomegalovirus. Med. Microbiol. Immunol. 2015, 204, 421–429. [Google Scholar] [CrossRef]
- Kotenko, S.V.; Saccani, S.; Izotova, L.S.; Mirochnitchenko, O.V.; Pestka, S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 2000, 97, 1695–1700. [Google Scholar] [CrossRef]
- Jenkins, C.; Garcia, W.; Godwin, M.J.; Spencer, J.V.; Stern, J.L.; Abendroth, A.; Slobedman, B. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J. Virol. 2008, 82, 3736–3750. [Google Scholar] [CrossRef] [PubMed]
- Dargan, D.J.; Douglas, E.; Cunningham, C.; Jamieson, F.; Stanton, R.J.; Baluchova, K.; McSharry, B.P.; Tomasec, P.; Emery, V.C.; Percivalle, E.; et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 2010, 91 Pt 6, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Yu, D.; Grimwood, J.; Schmutz, J.; Dickson, M.; Jarvis, M.A.; Hahn, G.; Nelson, J.A.; Myers, R.M.; Shenk, T.E. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2003, 100, 14976–14981. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Agostino, M.; Lee, S.; Ariyanto, I.; Kresoje, N.; Leary, S.; Munyard, K.; Gaudieri, S.; Gaff, J.; Irish, A.; et al. Sequencing Directly from Clinical Specimens Reveals Genetic Variations in HCMV-Encoded Chemokine Receptor US28 That May Influence Antibody Levels and Interactions with Human Chemokines. Microbiol. Spectr. 2021, 9, e0002021. [Google Scholar] [CrossRef]
- Quinnan, G.V., Jr.; Delery, M.; Rook, A.H.; Frederick, W.R.; Epstein, J.S.; Manischewitz, J.F.; Jackson, L.; Ramsey, K.M.; Mittal, K.; Plotkin, S.A.; et al. Comparative virulence and immunogenicity of the Towne strain and a nonattenuated strain of cytomegalovirus. Ann. Intern. Med. 1984, 101, 478–483. [Google Scholar] [CrossRef]
- Price, P.; Lee, S.; Affandi, J.; Parsons, R.; Naylor, L.H.; Watts, G.F.; Irish, A. Cytomegalovirus antibody and vascular pathology in renal transplant recipients. J. Med. Virol. 2017, 89, 177–181. [Google Scholar] [CrossRef]
- Karim, B.; Wijaya, I.P.; Rahmaniyah, R.; Ariyanto, I.; Waters, S.; Estiasari, R.; Price, P. Factors affecting affect cardiovascular health in Indonesian HIV patients beginning ART. AIDS Res. Ther. 2017, 14, 52. [Google Scholar] [CrossRef][Green Version]
- Lee, S.; Affandi, J.S.; Irish, A.B.; Price, P. Cytomegalovirus infection alters phenotypes of different gammadelta T-cell subsets in renal transplant recipients with long-term stable graft function. J. Med. Virol. 2017, 89, 1442–1452. [Google Scholar] [CrossRef]
- Suarez, N.M.; Wilkie, G.S.; Hage, E.; Camiolo, S.; Holton, M.; Hughes, J.; Maabar, M.; Vattipally, S.B.; Dhingra, A.; Gompels, U.A.; et al. Human Cytomegalovirus Genomes Sequenced Directly from Clinical Material: Variation, Multiple-Strain Infection, Recombination, and Gene Loss. J. Infect. Dis. 2019, 220, 781–791. [Google Scholar] [CrossRef]
- Waters, S.; Lee, S.; Irish, A.; Price, P. Challenging the Conventional Interpretation of HCMV Seronegativity. Microorganisms 2021, 9, 2382. [Google Scholar] [CrossRef]
- Murthy, S.; Hayward, G.S.; Wheelan, S.; Forman, M.S.; Ahn, J.H.; Pass, R.F.; Arav-Boger, R. Detection of a single identical cytomegalovirus (CMV) strain in recently seroconverted young women. PLoS ONE 2011, 6, e15949. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.A.; Pati, P.; Jensen, T.L.; Goll, J.B.; Gelber, C.E.; Singh, A.; McNeal, M.; Boppana, S.B.; Bernstein, D.I. Cytomegalovirus Genetic Diversity Following Primary Infection. J. Infect. Dis. 2020, 221, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Manuel, O.; Asberg, A.; Pang, X.; Rollag, H.; Emery, V.C.; Preiksaitis, J.K.; Kumar, D.; Pescovitz, M.D.; Bignamini, A.A.; Hartmann, A.; et al. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin. Infect. Dis. 2009, 49, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Coaquette, A.; Bourgeois, A.; Dirand, C.; Varin, A.; Chen, W.; Herbein, G. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 2004, 39, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Garcia, W.; Abendroth, A.; Slobedman, B. Expression of a human cytomegalovirus latency-associated homolog of interleukin-10 during the productive phase of infection. Virology 2008, 370, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.V.; Lockridge, K.M.; Barry, P.A.; Lin, G.; Tsang, M.; Penfold, M.E.; Schall, T.J. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 2002, 76, 1285–1292. [Google Scholar] [CrossRef]
- Spencer, J.V.; Cadaoas, J.; Castillo, P.R.; Saini, V.; Slobedman, B. Stimulation of B lymphocytes by cmvIL-10 but not LAcmvIL-10. Virology 2008, 374, 164–169. [Google Scholar] [CrossRef]
- Wijaya, I.P.; Karim, B.; Azizi, M.S.; Ariyanto, I.; Mansjoer, A.; Yunihastuti, E.; Harimurti, K.; Alwi, I.; Lee, S.; Price, P. Cytomegalovirus may influence vascular endothelial health in Indonesian HIV-infected patients after 5 years on ART. AIDS Res. Ther. 2021, 18, 83. [Google Scholar] [CrossRef]
- Lee, S.; Brook, E.; Affandi, J.; Howson, P.; Tanudjaja, S.A.; Dhaliwal, S.; Irish, A.; Price, P. A high burden of cytomegalovirus marks poor vascular health in transplant recipients more clearly than in the general population. Clin. Transl. Immunol. 2019, 8, e1043. [Google Scholar] [CrossRef]
- Waters, S.; Lee, S.; Lloyd, M.; Irish, A.; Price, P. The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients. Int. J. Mol. Sci. 2019, 20, 5230. [Google Scholar] [CrossRef]
- Currenti, J.; Chopra, A.; John, M.; Leary, S.; McKinnon, E.; Alves, E.; Pilkinton, M.; Smith, R.; Barnett, L.; McDonnell, W.J.; et al. Deep sequence analysis of HIV adaptation following vertical transmission reveals the impact of immune pressure on the evolution of HIV. PLoS Pathog. 2019, 15, e1008177. [Google Scholar] [CrossRef] [PubMed]
- Estiasari, R.; Aryanto, I.; Lee, S.; Pramana, S.; Djauzi, S.; Price, P. Determinants of cognitive health in Indonesian HIV patients beginning antiretroviral therapy. J. Neurovirol. 2020, 26, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Affandi, J.S.; Lee, S.; Chih, H.; Brook, E.; Waters, S.; Howson, P.; Reid, C.M.; Irish, A.; Price, P. Cytomegalovirus burden improves a predictive model identifying measures of vascular risk in renal transplant recipients and healthy adults. J. Med. Virol. 2020, 92, 3650–3657. [Google Scholar] [CrossRef] [PubMed]
- Scheet, P.; Stephens, M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006, 78, 629–644. [Google Scholar] [CrossRef]


| Residue Position | Toledo Reference | Neonates n = 4 | Adults n = 55 | Australian n = 28 | Indonesian n = 27 | Buffy Coat n = 31 | Saliva n = 24 |
|---|---|---|---|---|---|---|---|
| 9 | S | S | S/T | S/T | S/T | S/T | S/T |
| 16 | F | F | F/L | F/L | F | F/L | F/L |
| 30 | I | I/T/M | I/T/M | I/T/M | I | I/T/M | I/T/M |
| 41 | D | D | D/N | D/N | D | D/N | D/N |
| 73 | S | S/Y | S/Y | S/Y | S/Y | S/Y | S/Y |
| 83 | P | P | P/L | P/L | P/L | P/L | P/L |
| 99 | G | G | G/V | G/V | G/V | G/V | G/V |
| 109 | H | H | H/Y | H/Y | H/Y | H/Y | H/Y |
| 114 | K | E | K/E | E/K | E/K | K/E | K/E |
| 122 | P | P/S | P/S | P/S | P/S | P/S | P/S |
| 123 | R | R | R/C | R/C | R/C | R/C | R/C |
| 126 | P | P | P/T/S | P/T/S | P/T | P/T/S | P/S/T |
| 127 | R | R | R/G | R/G | R/G | R/G | R/G |
| 128 | L | L | L/Q/V | L/Q/V | L/Q/V | L/Q/V | L/Q/V |
| 129 | S | S | S/F/A/C | S/F/A/C | S/F/A/C | S/F/A/C | S/F/A/C |
| 130 | R | R | R/L/G | R/L/G | R/L/G | R/L/G | R/L/G |
| 131 | T | T | T/A/S/P | T/A/S/P | T/A/S/P | T/A/S/P | T/A/S/P |
| 132 | Q | Q/* | Q/* | Q/* | Q/* | Q/* | Q/* |
| 147 | Q | Q | Q/* | Q/* | Q/* | Q/* | Q/* |
| 153 | R | R | R/C | R/C | R/C | R/C | R/C |
| 163 | A | A | A/T | A/T | A/T | A/T | A/T |
| 170 | * | * | */Q | */Q | */Q | */Q | */Q |
| 174 | L | L/F | L/F | L/F/W | L/F/W | L/F/W | L/F/W |
| 180 | Q | Q | Q/H/L/R | Q/H/R | Q/H/L/R | Q/H/L/R | Q/H/R |
| 181 | P | P | P/L/T | P/L/T | P/L/T | P/L/T | P/L/T |
| 182 | L | L | L/V/Q | L/V/Q | L/V/Q | L/V/Q | L/V/Q |
| 183 | L | L | L/F/S | L/F | L/F/S | L/F/S | L/F |
| 184 | G | G | G/V | G/V | G/V | G/V | G/V |
| 185 | C | C | C/G/S/F | C/S/F | C/G/S/F | C/G/S/F | C/S/F |
| 186 | G | G | G/P/R | G | G/P/R | G/P/R | G |
| 189 | S | S | S/T | S | S/T | S/T | S |
| 214 | D | D | D/N | D | D/N | D/N | D |
| Position Toledo Variant | 9 | 16 | 41 | 73 | 83 | 99 | 109 | 114 | 122 | 123 | 127 | 153 | 170 | 174 | 184 | 189 | 214 | Indo (n = 27) | Aus (n = 28) | p a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | F | D | S | P | G | H | K | P | R | R | R | * | L | G | S | D | ||||
| T | L | N | Y | L | V | Y | E | S | C | G | C | Q | F/W | V | T | N | ||||
| Haplotypes | ||||||||||||||||||||
| UL111a-1 | S | F | D | S | P | G | H | E | P | R | R | R | * | L | G | S | D | 12 | 12 | 0.99 |
| UL111a-2 | S | F | D | S | L | G | H | E | P | R | R | R | * | L | G | S | D | 14 | 5 | 0.01 |
| UL111a-3 | S | F | D | S | P | G | H | E | P | R | R | R | * | F/W | G | S | D | 3 | 3 | 0.99 |
| UL111a-4 | S | F | D | Y | P | G | H | E | P | R | R | R | * | L | G | S | D | 2 | 3 | 0.99 |
| UL111a-5 | S | F | D | S | P | G | H | E | S | R | R | R | * | L | G | S | D | 1 | 0 | 0.49 |
| UL111a-6 | S | F | D | S | L | V | H | E | P | R | R | R | * | L | G | S | D | 1 | 1 | 0.99 |
| UL111a-7 | S | F | N | Y | P | G | H | E | P | R | R | R | * | L | G | S | D | 0 | 5 | 0.05 |
| UL111a-8 | S | F | D | S | P | G | H | E | S | R | R | C | * | L | G | S | D | 0 | 3 | 0.24 |
| UL111a-9 | S | F | D | S | L | G | H | E | P | R | R | R | Q | L | G | S | D | 2 | 0 | 0.24 |
| UL111a-10 | S | L | D | S | P | G | H | E | S | R | R | C | * | L | G | S | D | 0 | 2 | 0.49 |
| UL111a-11 | S | F | D | S | P | G | Y | E | S | R | R | C | * | L | G | S | D | 1 | 1 | 0.99 |
| UL111a-12 | S | F | D | S | L | G | Y | E | P | R | R | R | * | L | G | S | D | 1 | 1 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waters, S.; Lee, S.; Ariyanto, I.; Kresoje, N.; Leary, S.; Munyard, K.; Gaudieri, S.; Irish, A.; Keil, A.D.; Allcock, R.J.N.; et al. Sequencing of the Viral UL111a Gene Directly from Clinical Specimens Reveals Variants of HCMV-Encoded IL-10 That Are Associated with Altered Immune Responses to HCMV. Int. J. Mol. Sci. 2022, 23, 4644. https://doi.org/10.3390/ijms23094644
Waters S, Lee S, Ariyanto I, Kresoje N, Leary S, Munyard K, Gaudieri S, Irish A, Keil AD, Allcock RJN, et al. Sequencing of the Viral UL111a Gene Directly from Clinical Specimens Reveals Variants of HCMV-Encoded IL-10 That Are Associated with Altered Immune Responses to HCMV. International Journal of Molecular Sciences. 2022; 23(9):4644. https://doi.org/10.3390/ijms23094644
Chicago/Turabian StyleWaters, Shelley, Silvia Lee, Ibnu Ariyanto, Nina Kresoje, Shay Leary, Kylie Munyard, Silvana Gaudieri, Ashley Irish, Anthony D. Keil, Richard J. N. Allcock, and et al. 2022. "Sequencing of the Viral UL111a Gene Directly from Clinical Specimens Reveals Variants of HCMV-Encoded IL-10 That Are Associated with Altered Immune Responses to HCMV" International Journal of Molecular Sciences 23, no. 9: 4644. https://doi.org/10.3390/ijms23094644
APA StyleWaters, S., Lee, S., Ariyanto, I., Kresoje, N., Leary, S., Munyard, K., Gaudieri, S., Irish, A., Keil, A. D., Allcock, R. J. N., & Price, P. (2022). Sequencing of the Viral UL111a Gene Directly from Clinical Specimens Reveals Variants of HCMV-Encoded IL-10 That Are Associated with Altered Immune Responses to HCMV. International Journal of Molecular Sciences, 23(9), 4644. https://doi.org/10.3390/ijms23094644







