Identification of Key Factors for Anoxic Survival of B. cenocepacia H111
Abstract
1. Introduction
2. Results
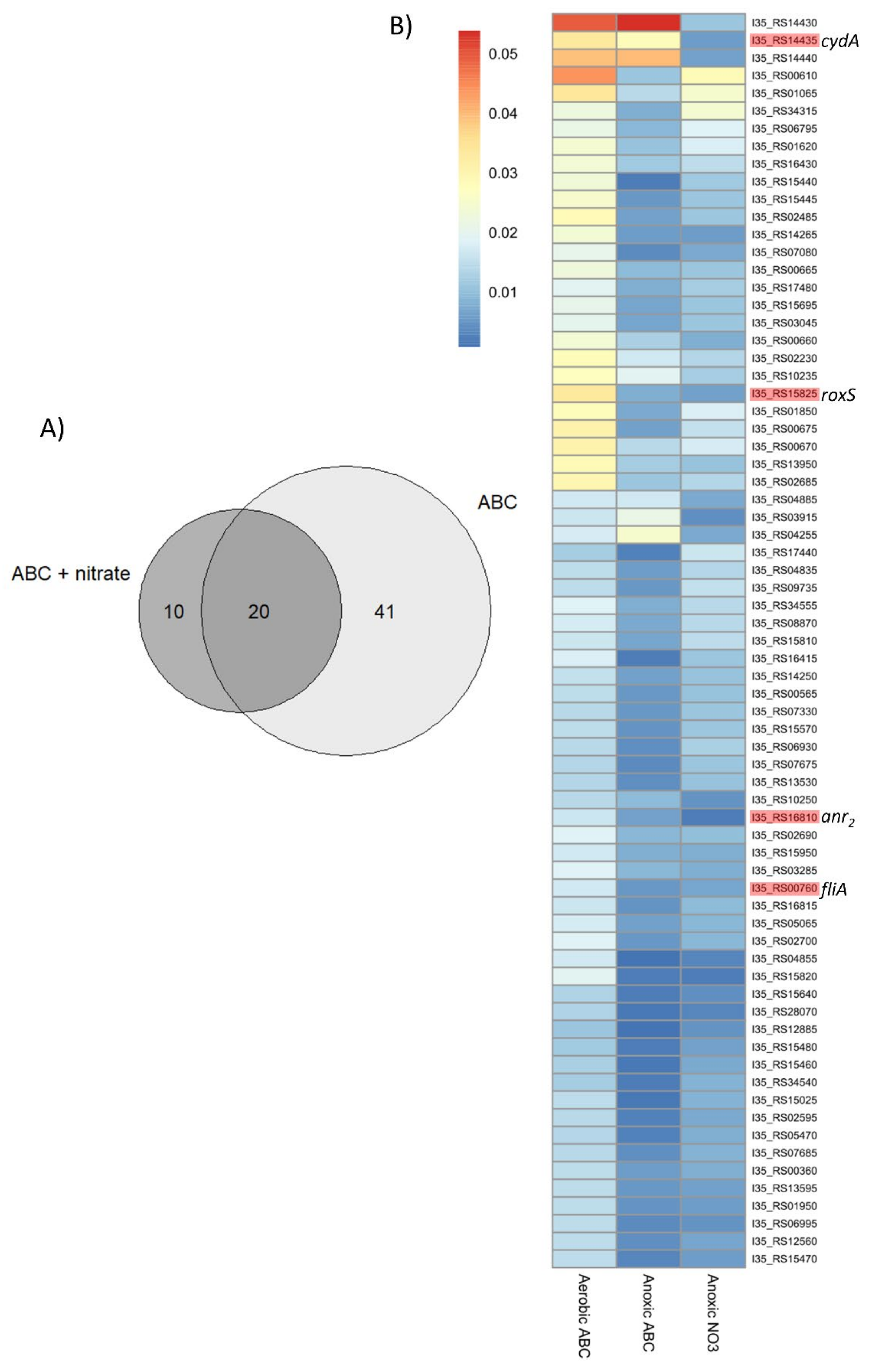
2.1. Identification of Potential Fitness Determinants for the Survival of B. cenocepacia H111 in Anoxic Condition
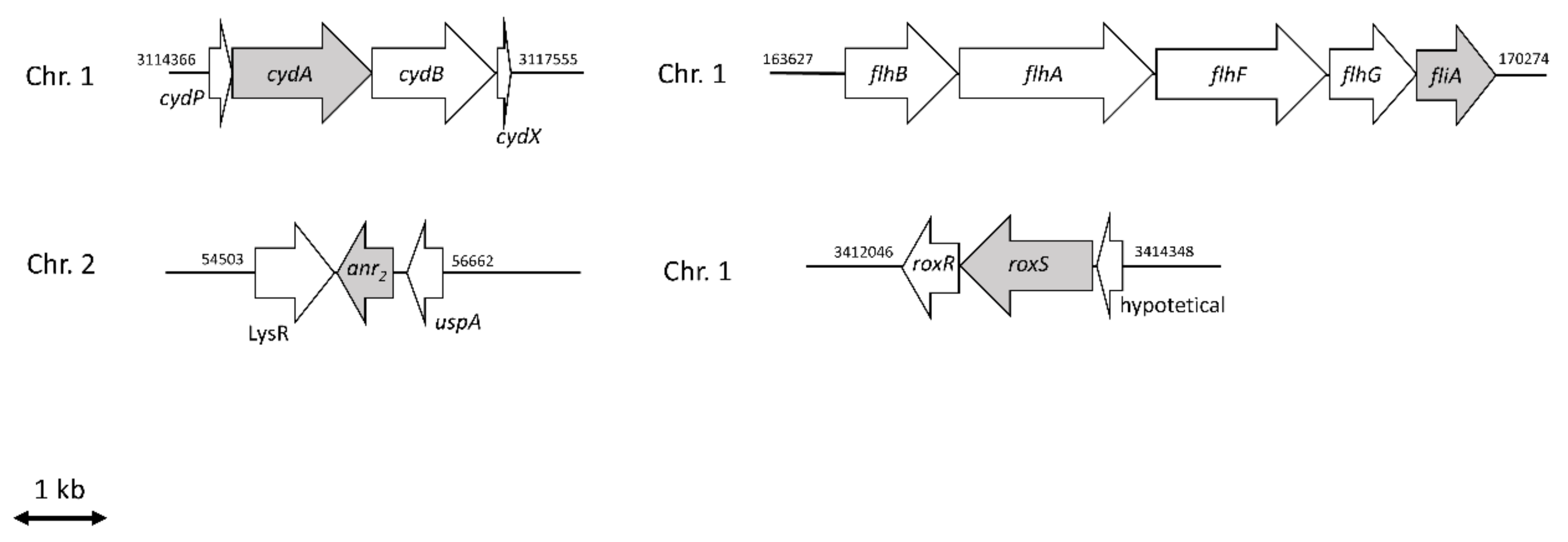
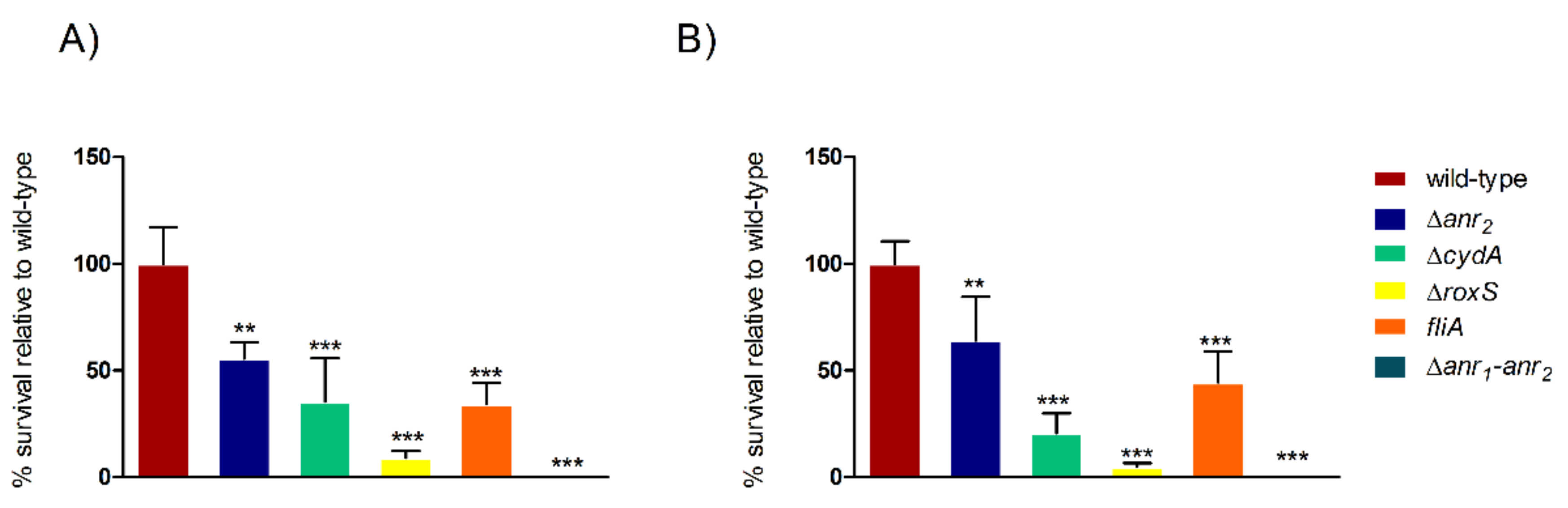
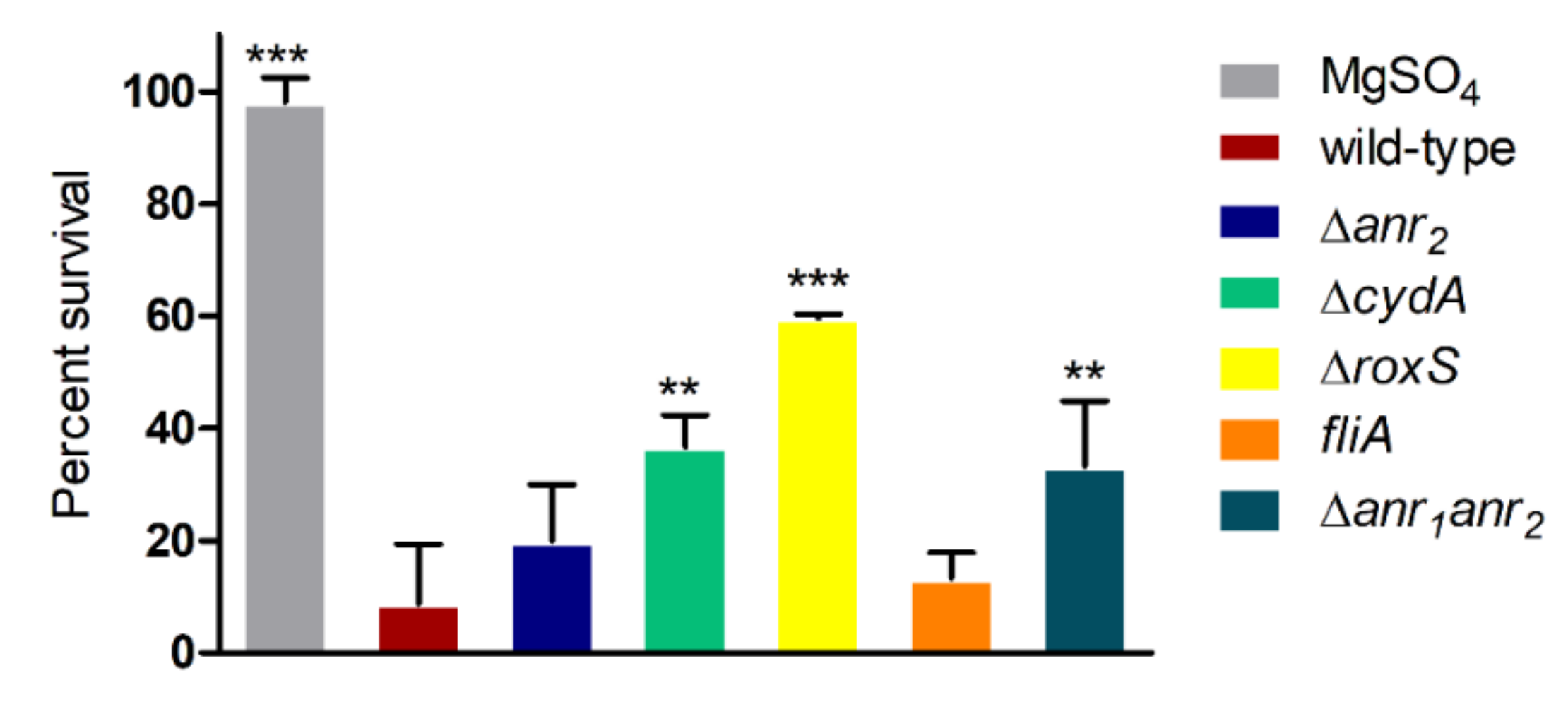
2.2. Anr Is Essential for the Survival of B. cenocepacia H111 under Oxygen Depletion
2.3. RoxS Is Required for Pathogenicity in a Galleria Mellonella Infection Model
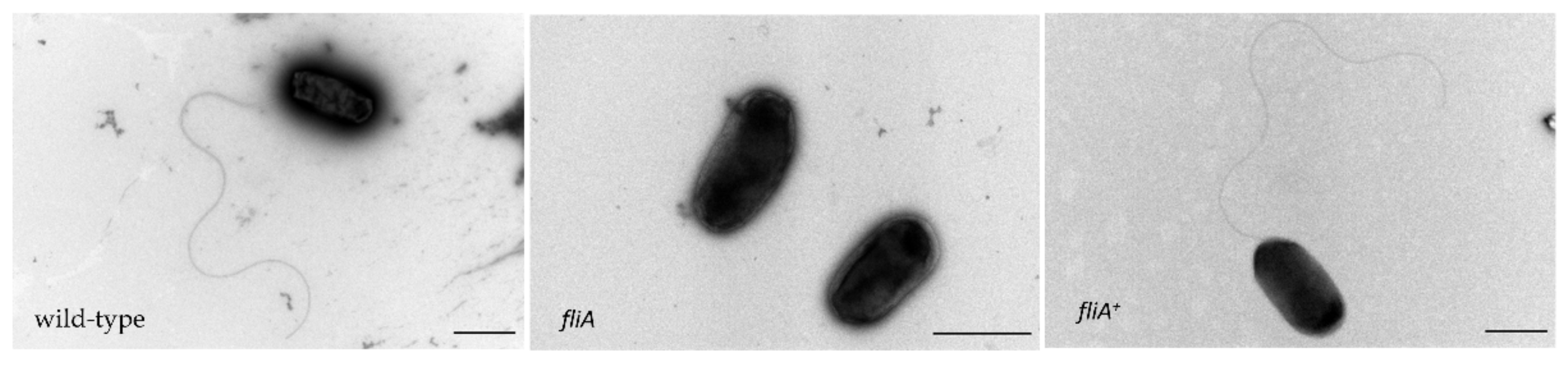
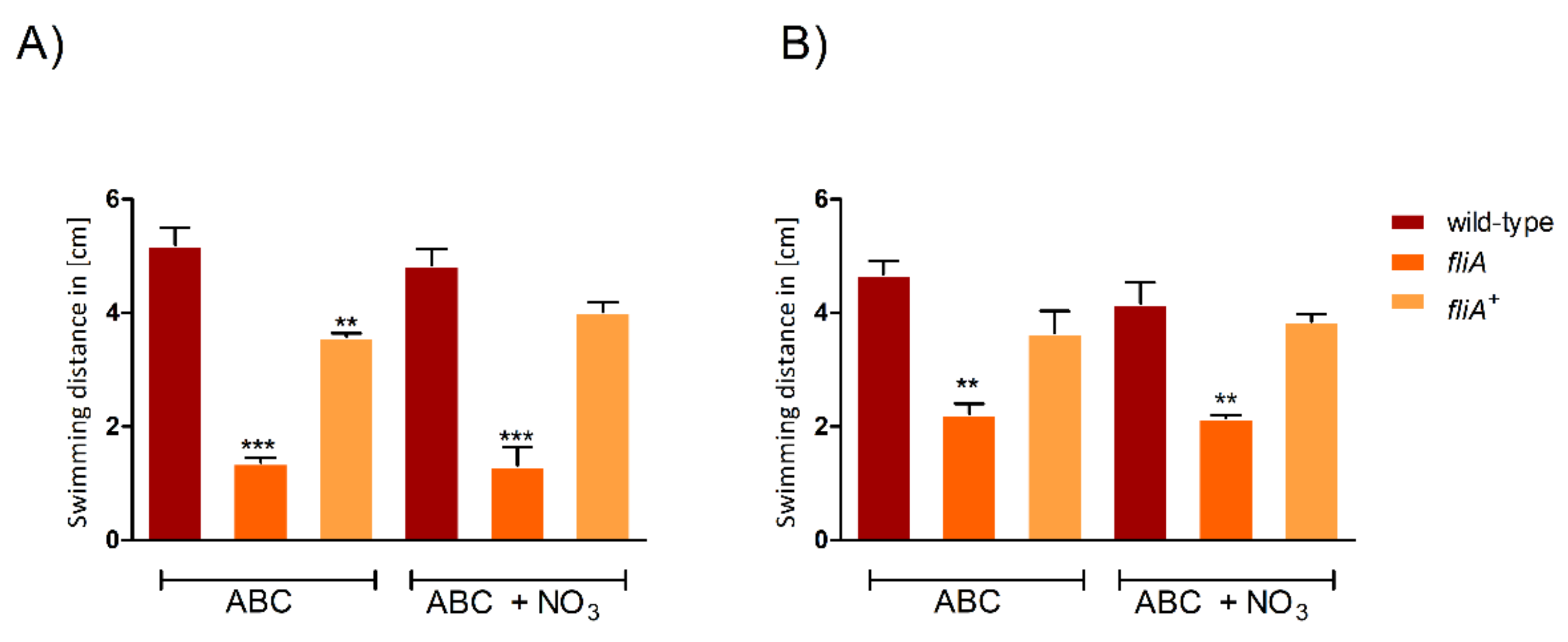
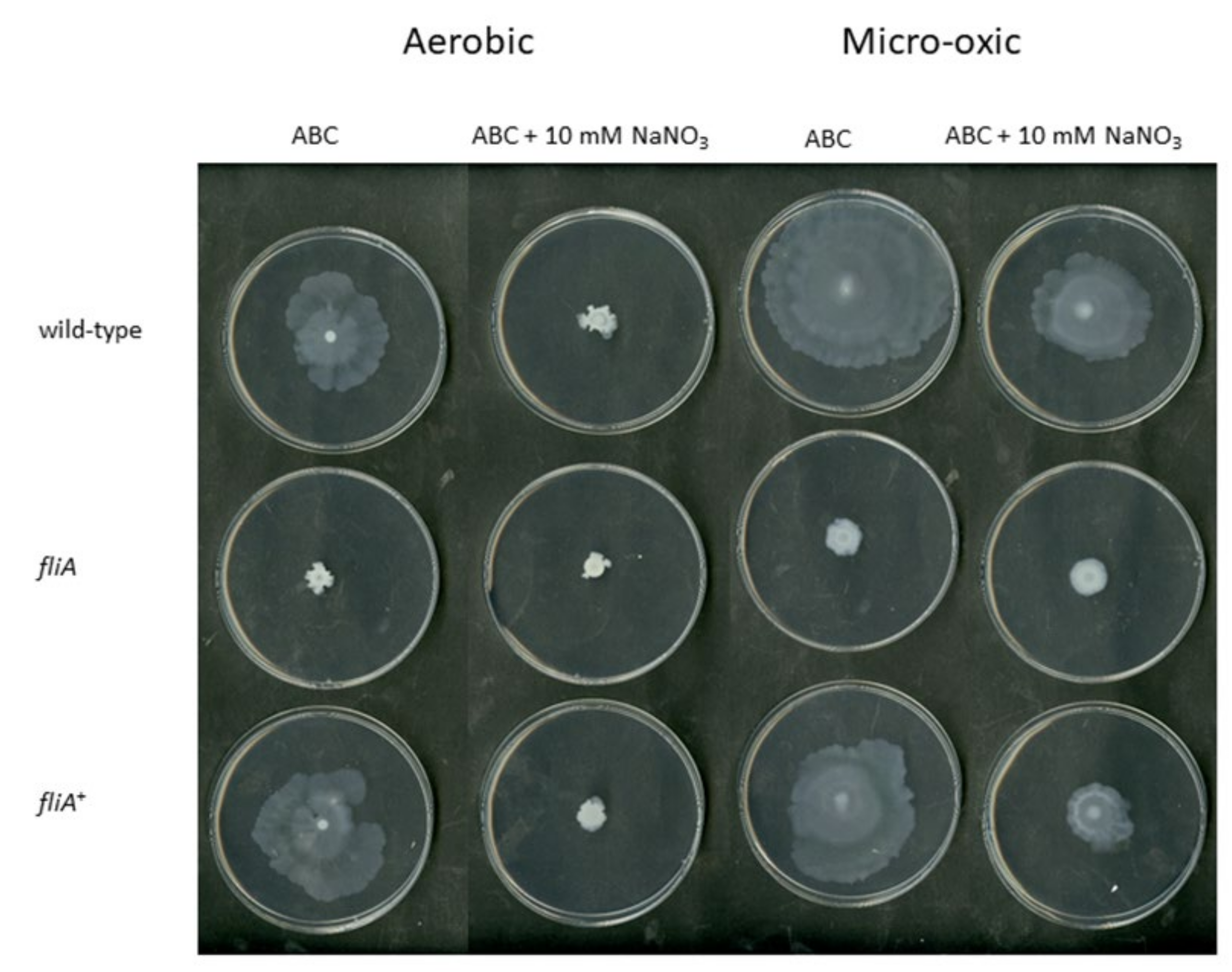
2.4. The Importance of FliA for Swarming and Swimming Motilities in B. cenocepacia H111
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media and Growth Conditions
4.2. Tn-seq Technology
4.3. Construction of B. cenocepacia H111 Mutant Strains
4.4. Phenotypical Analyses
4.5. Pathogenicity Experiments Using Galleria mellonella
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turcios, N.L. Cystic fibrosis lung disease: An overview. Respir. Care 2020, 65, 233–251. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; White, R.; Tobin, P. Keep them breathing: Cystic fibrosis pathophysiology, diagnosis, and treatment. J. Am. Acad. Physician Assist. 2017, 30, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Stutts, M.J.; Canessa, C.M.; Olsen, J.C.; Hamrick, M.; Cohn, J.A.; Rossier, B.C.; Boucher, R.C. CFTR as a cAMP-dependent regulator of sodium channels. Science 1995, 269, 847–850. [Google Scholar] [CrossRef]
- Kh, R.; Bhatt, J.; Kh, R.; Bhatt, J. Eradication therapy for Burkholderia cepacia complex in people with cystic fibrosis. Cochrane Database Syst. Rev. 2019, 11, CD009876. [Google Scholar] [CrossRef]
- Rossi, E.; La Rosa, R.; Bartell, J.A.; Marvig, R.L.; Haagensen, J.A.J.; Sommer, L.M.; Molin, S.; Johansen, H.K. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 2021, 19, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Thornton, C.S.; Surette, M.G. Potential contributions of anaerobes in cystic fibrosis airways. J. Clin. Microbiol. 2021, 59, e01813-19. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Goldberg, J.B. Pseudomonas aeruginosa in cystic fibrosis: A chronic cheater. Proc. Natl. Acad. Sci. USA 2019, 116, 6525–6527. [Google Scholar] [CrossRef] [PubMed]
- Strohm, T.O.; Griffin, B.; Zumft, W.G.; Schink, B. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 2007, 73, 1420–1424. [Google Scholar] [CrossRef]
- Arai, H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2011, 2, 103. [Google Scholar] [CrossRef]
- Eschbach, M.; Schreiber, K.; Trunk, K.; Buer, J.; Jahn, D.; Schobert, M. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 2004, 186, 4596–4604. [Google Scholar] [CrossRef]
- Jo, J.; Price-whelan, A.; Cornell, C. Interdependency of respiratory metabolism and phenazine associated physiology in Pseudomonas aeruginosa PA14. J. Bacteriol. 2020, 202, e00700-19. [Google Scholar] [CrossRef]
- Ciemniecki, J.A.; Newman, D.K. The potential for redox-active metabolites to enhance or unlock anaerobic survival metabolisms in aerobes. J. Bacteriol. 2020, 202, 1–14. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Chiarelli, L.R.; Trespidi, G.; Mentasti, M.; Riccardi, G.; Buroni, S. Burkholderia cenocepacia infections in cystic fibrosis patients: Drug resistance and therapeutic approaches. Front. Microbiol. 2017, 8, 1592. [Google Scholar] [CrossRef]
- De Volder, A.L.; Teves, S.; Isasmendi, A.; Pinheiro, J.L.; Ibarra, L.; Breglia, N.; Herrera, T.; Vazquez, M.; Hernandez, C.; Degrossi, J. Distribution of Burkholderia cepacia complex species isolated from industrial processes and contaminated products in Argentina. Int. Microbiol. 2021, 24, 157–167. [Google Scholar] [CrossRef]
- Sauty, A.A.; Manuel, O.; Koutsokera, A.; Tullis, E. Burkholderia cepacia complex. Unisante 2019, 1–7. [Google Scholar]
- Mannaa, M.; Park, I.; Seo, Y.S. Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated Burkholderia species. Int. J. Mol. Sci. 2019, 20, 121. [Google Scholar] [CrossRef]
- Vial, L.; Chapalain, A.; Groleau, M.C.; Déziel, E. The various lifestyles of the Burkholderia cepacia complex species: A tribute to adaptation. Environ. Microbiol. 2011, 13, 1–12. [Google Scholar] [CrossRef]
- Pessi, G.; Braunwalder, R.; Grunau, A.; Omasits, U.; Ahrens, C.H.; Eberl, L. Response of Burkholderia cenocepacia H111 to micro-oxia. PLoS ONE 2013, 8, e72939. [Google Scholar] [CrossRef]
- Pitcher, R.S.; Watmough, N.J. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta Bioenerg. 2004, 1655, 388–399. [Google Scholar] [CrossRef]
- Comolli, J.C.; Donohue, T.J. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol. Microbiol. 2002, 45, 755–768. [Google Scholar] [CrossRef]
- Vitale, A.; Paszti, S.; Takahashi, K.; Toyofuku, M.; Pessi, G.; Eberl, L. Mapping of the denitrification pathway in Burkholderia thailandensis by genome-wide mutant profiling. J. Bacteriol. 2020, 202, e00304-20. [Google Scholar] [CrossRef] [PubMed]
- Sass, A.M.; Schmerk, C.; Agnoli, K.; Norville, P.J.; Eberl, L.; Valvano, M.A.; Mahenthiralingam, E. The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. ISME J. 2013, 7, 1568–1581. [Google Scholar] [CrossRef] [PubMed]
- Carlier, A.; Agnoli, K.; Pessi, G.; Suppiger, A.; Jenul, C.; Schmid, N.; Tümmler, B.; Pinto-carbo, M. Genome Sequence of Burkholderia cenocepacia H111, a cystic fibrosis airway isolate. ASM 2014, 2, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Higgins, S.; Sanchez-Contreras, M.; Gualdi, S.; Pinto-Carbó, M.; Carlier, A.; Eberl, L. The essential genome of Burkholderia cenocepacia H111. J. Bacteriol. 2017, 199, e00260-17. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Wang, S.; Fleming, R.T.; Westbrook, E.M.; Matsumura, P.; McKay, D.B. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 2006, 355, 798–808. [Google Scholar] [CrossRef]
- Winsor, G.L.; Khaira, B.; Van Rossum, T.; Lo, R.; Whiteside, M.D.; Brinkman, F.S.L. The Burkholderia Genome Database: Facilitating flexible queries and comparative analyses. Bioinformatics 2008, 24, 2803–2804. [Google Scholar] [CrossRef]
- Nagy, P.L.; Marolewski, A.; Benkovic, S.J.; Zalkin, H. Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J. Bacteriol. 1995, 177, 1292–1298. [Google Scholar] [CrossRef]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Vanorsdel, C.E.; Bhatt, S.; Allen, R.J.; Brenner, E.P.; Hobson, J.J.; Jamil, A.; Haynes, B.M.; Genson, A.M.; Hemma, M.R. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J. Bacteriol. 2013, 195, 3640–3650. [Google Scholar] [CrossRef] [PubMed]
- Schwager, S.; Agnoli, K.; Köthe, M.; Feldmann, F.; Givskov, M.; Carlier, A.; Eberl, L. Identification of Burkholderia cenocepacia strain H111 virulence factors using nonmammalian infection hosts. Infect. Immun. 2013, 81, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Uehlinger, S.; Schwager, S.; Bernier, S.P.; Riedel, K.; Nguyen, D.T.; Sokol, P.A.; Eberl, L. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect. Immun. 2009, 77, 4102–4110. [Google Scholar] [CrossRef] [PubMed]
- Starnbach, M.N.; Lory, S. The filA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 1992, 6, 459–469. [Google Scholar] [CrossRef]
- Lo, Y.L.; Chen, C.L.; Shen, L.; Chen, Y.C.; Wang, Y.H.; Lee, C.C.; Wang, L.C.; Chuang, C.H.; Janapatla, R.P.; Chiu, C.H.; et al. Characterization of the role of global regulator FliA in the pathophysiology of Pseudomonas aeruginosa infection. Res. Microbiol. 2018, 169, 135–144. [Google Scholar] [CrossRef]
- Cullen, L.; O’Connor, A.; McCormack, S.; Owens, R.A.; Holt, G.S.; Collins, C.; Callaghan, M.; Doyle, S.; Smith, D.; Schaffer, K.; et al. The involvement of the low-oxygen-activated locus of Burkholderia cenocepacia in adaptation during cystic fibrosis infection. Sci. Rep. 2018, 8, 13386. [Google Scholar] [CrossRef]
- Lieberman, T.D.; Michel, J.B.; Aingaran, M.; Potter-Bynoe, G.; Roux, D.; Davis, M.R.; Skurnik, D.; Leiby, N.; Lipuma, J.J.; Goldberg, J.B.; et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet. 2011, 43, 1275–1280. [Google Scholar] [CrossRef]
- Schaefers, M.M.; Liao, T.L.; Boisvert, N.M.; Roux, D.; Yoder-Himes, D.; Priebe, G.P. An oxygen-sensing two-component system in the Burkholderia cepacia complex regulates biofilm, intracellular invasion, and pathogenicity. PLoS Pathog. 2017, 13, e1006116. [Google Scholar] [CrossRef]
- Han, H.; Hemp, J.; Pace, L.A.; Ouyang, H.; Ganesan, K.; Roh, J.H.; Daldal, F.; Blanke, S.R.; Gennis, R.B. Adaptation of aerobic respiration to low O2 environments. Proc. Natl. Acad. Sci. USA 2012, 109, 7947. [Google Scholar] [CrossRef]
- Morris, R.L.; Schmidt, T.M. Shallow breathing: Bacterial life at low O2. Nat. Rev. Microbiol. 2013, 11, 205–212. [Google Scholar] [CrossRef]
- Arai, H.; Kawakami, T.; Osamura, T.; Hirai, T.; Sakai, Y.; Ishii, M. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 4206–4215. [Google Scholar] [CrossRef]
- Zufferey, R.; Preisig, O.; Hennecke, H.; Thöny-Meyer, L. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 1996, 271, 9114–9119. [Google Scholar] [CrossRef]
- Gong, X.; Garcia-Robledo, E.; Lund, M.B.; Lehner, P.; Borisov, S.M.; Klimant, I.; Revsbech, N.P.; Schramm, A. Gene expression of terminal oxidases in two marine bacterial strains exposed to nanomolar oxygen concentrations. FEMS Microbiol. Ecol. 2018, 94, fiy072. [Google Scholar] [CrossRef]
- Keightley, J.A.; Zimmermann, B.H.; Mather, M.W.; Springer, P.; Pastuszyn, A.; Lawrence, D.M.; Fee, J.A. Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J. Biol. Chem. 1995, 270, 20345–20358. [Google Scholar] [CrossRef]
- De Meyer, S.E.; Briscoe, L.; Martínez-Hidalgo, P.; Agapakis, C.M.; De-Los Santos, P.E.; Seshadri, R.; Reeve, W.; Weinstock, G.; O’Hara, G.; Howieson, J.G.; et al. Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol. Plant-Microbe Interact. 2016, 29, 609–619. [Google Scholar] [CrossRef]
- Münch, R.; Hiller, K.; Grote, A.; Scheer, M.; Klein, J.; Schobert, M.; Jahn, D. Virtual Footprint and PRODORIC: An integrative framework for regulon prediction in prokaryotes. Bioinformatics 2005, 21, 4187–4189. [Google Scholar] [CrossRef]
- Elsen, S.; Swem, L.R.; Swem, D.L.; Bauer, C.E. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 2004, 68, 263–279. [Google Scholar] [CrossRef]
- Phenn, J.; Pané-Farré, J.; Meukow, N.; Klein, A.; Troitzsch, A.; Tan, P.; Fuchs, S.; Wagner, G.E.; Lichtenegger, S.; Steinmetz, I.; et al. RegAB Homolog of Burkholderia pseudomallei is the master regulator of redox control and involved in virulence. PLoS Pathog. 2021, 17, e1009604. [Google Scholar] [CrossRef]
- Bernier, S.P.; Son, S.; Surette, M.G. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J. Bacteriol. 2018, 200, e00156-18. [Google Scholar] [CrossRef]
- Wang, Y.; Hay, I.D.; Rehman, Z.U.; Rehm, B.H.A. Membrane-anchored MucR mediates nitrate-dependent regulation of alginate production in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2015, 99, 7253–7265. [Google Scholar] [CrossRef]
- Prüß, B.M.; Campbell, J.W.; Van Dyk, T.K.; Zhu, C.; Kogan, Y.; Matsumura, P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 2003, 185, 534–543. [Google Scholar] [CrossRef]
- Hendry, S.; Steinke, S.; Wittstein, K.; Stadler, M.; Harmrolfs, K.; Adewunmi, Y.; Sahukhal, G.; Elasri, M.; Thomashow, L.; Weller, D.; et al. Functional analysis of phenazine biosynthesis genes in Burkholderia spp. Appl. Environ. Microbiol. 2021, 87, 1–18. [Google Scholar] [CrossRef]
- Cartwright, D.K.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin and phenazine production by Pseudomonas cepacia strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microb. Biotechnol. 1995, 43, 211–216. [Google Scholar] [CrossRef]
- Ramos, C.G.; Sousa, S.A.; Grilo, A.M.; Eberl, L.; Leitão, J.H. The Burkholderia cenocepacia K56-2 pleiotropic regulator Pbr, is required for stress resistance and virulence. Microb. Pathog. 2010, 48, 168–177. [Google Scholar] [CrossRef]
- Hwang, J.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin production by Burkholderia cepacia and biocontrol of Rhizoctonia stem rot of poinsettia. Biol. Control 2002, 25, 56–63. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Clark, D.J.; Maaløe, O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 1967, 23, 99–112. [Google Scholar] [CrossRef]
- Gotschlich, A.; Huber, B.; Geisenberger, O.; Togl, A.; Steidle, A.; Riedel, K.; Hill, P.; Tümmler, B.; Vandamme, P.; Middleton, B.; et al. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 2001, 24, 1–14. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Ramage, E.; Patrapuvich, R.; Weiss, E.; Brittnacher, M.; Manoil, C. Sequence-defined transposon mutant library of Burkholderia thailandensis. MBio 2013, 4, 1–7. [Google Scholar] [CrossRef]
- Solaimanpour, S.; Sarmiento, F.; Mrázek, J. Tn-seq explorer: A tool for analysis of high-throughput sequencing data of transposon mutant libraries. PLoS ONE 2015, 10, e0126070. [Google Scholar] [CrossRef]
- Flannagan, R.; Linn, T.; Valvano, M.A. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 2008, 10, 1652–1660. [Google Scholar] [CrossRef]
- Eberl, L.; Christlansen, G.; Molin, S.; Givskov, M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 1996, 178, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; de Lorenzo Timmis, K.V. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef]
- Miller, V.L.; Mekalanos, J.J. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1998, 170, 2575–2583. [Google Scholar] [CrossRef]
- Römling, U.; Wingender, J.; Müller, H.; Tümmler, B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 1994, 60, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef]
- Phadnis, S.H.; Berg, D.E. Identification of base pairs in the outside end of insertion sequence IS50 that are needed for IS50 and Tn5 transposition. Proc. Natl. Acad. Sci. USA 1987, 84, 9118–9122. [Google Scholar] [CrossRef]
- Shastri, S.; Spiewak, H.L.; Sofoluwe, A.; Eidsvaag, V.A.; Asghar, A.H.; Pereira, T.; Bull, E.H.; Butt, A.T.; Thomas, M.S. An efficient system for the generation of marked genetic mutants in members of the genus Burkholderia. Plasmid 2017, 89, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., II; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Carlier, A.; Burbank, L.; von Bodman, S.B. Identification and characterization of three novel EsaI/EsaR quorum-sensing controlled stewartan exopolysaccharide biosynthetic genes in Pantoea stewartii ssp. stewartii. Mol. Microbiol. 2009, 74, 903–913. [Google Scholar] [CrossRef]
- Agnoli, K.; Schwager, S.; Uehlinger, S.; Vergunst, A.; Viteri, D.F.; Nguyen, D.T.; Sokol, P.A.; Carlier, A.; Eberl, L. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol. Microbiol. 2012, 83, 362–378. [Google Scholar] [CrossRef]
- Wanner, B.L.; Datsenko, K.A. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Lardi, M.; Liu, Y.; Purtschert, G.; Bolzan de Campos, S.; Pessi, G. Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes 2017, 8, 389. [Google Scholar] [CrossRef]
- Choi, K.H.; Schweizer, H.P. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005, 5, 1–11. [Google Scholar] [CrossRef]
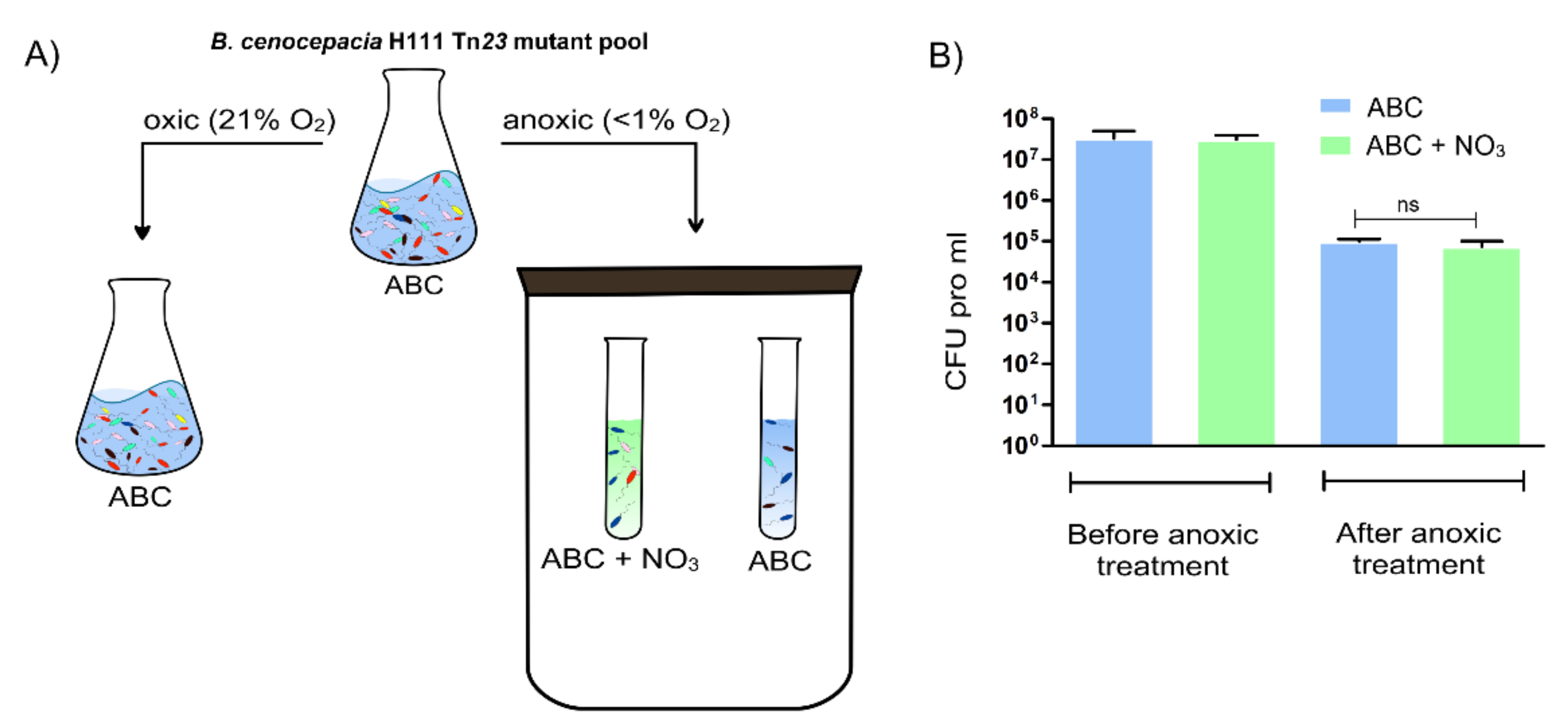
| Condition | Medium | Number of UIC | Number of Potential Fitness Determinants |
|---|---|---|---|
| Aerobic | ABC | 1,106,798 | NA |
| Anoxic | ABC | 987,541 | 61 |
| Anoxic | ABC + 10 mM NaNO3 | 841,867 | 30 |
| Locus_tag | Orthologs 1 | Description | Gene Name 4 | COG 2 | FC 3 (ABC) | FC 3 (ABC + 10 mM NaNO3) |
|---|---|---|---|---|---|---|
| I35_RS00665 | BCAL0125 | transcriptional regulator | flhC | K | −1.27 | −1.02 |
| I35_RS00675 | BCAL0127 | flagellar motor protein | motB | N | −2.32 | −1.01 |
| I35_RS00760 | BCAL0144 | RNA polymerase sigma factor | fliA | K | −1.71 | −1.33 |
| I35_RS01950 | BCAL0387 | GTP-binding protein | - | J | −1.7 | −1.33 |
| I35_RS02485 | BCAL3398 | competence damage-inducible protein A | - | S | −2.19 | −1.35 |
| I35_RS02685 | BCAL3358 | ABC transporter | gltI | P | −1.43 | −1.12 |
| I35_RS02700 | BCAL3354 | arginine ABC transporter ATP-binding protein | - | E | −1.88 | −1.04 |
| I35_RS03285 | - | tRNA-Lys | - | - | −1.03 | −1.28 |
| I35_RS04855 | - | nicotinate phosphoribosyltransferase | - | H | −4.48 | −2.37 |
| I35_RS06995 | BCAL1509 | tRNA pseudouridine synthase B | truB | J | −1.9 | −1.6 |
| I35_RS07080 | BCAL1526 | pilus assembly protein | tadE | U | −2.41 | −1.52 |
| I35_RS13950 | BCAL0883 | TetR family transcriptional regulator | - | K | −1.22 | −1.45 |
| I35_RS14265 | BCAL0820 | adenine phosphoribosyltransferase | - | F | −2.01 | −2.02 |
| I35_RS15445 | BCAL0576 | flagellar hook protein | flgK | N | −2.26 | −1.18 |
| I35_RS15470 | BCAL0569 | flagellar basal body rod protein | flgG | N | −2.23 | −1.35 |
| I35_RS15820 | BCAL0499 | chemotaxis protein | roxR | T | −3.26 | −3.17 |
| I35_RS15825 | BCAL0497 | two-component system sensor histidine kinase | roxS | T | −2.08 | −2.43 |
| I35_RS15950 | BCAL0473 | 7-cyano-7-deazaguanine synthase | - | S | −1.14 | −1.1 |
| I35_RS16810 | BCAM0049 | Crp-Fnr family transcriptional regulator | anr2 | K | −1.32 | −2.86 |
| I35_RS28070 | BCAM2434 | YsiA protein | - | K | −2.84 | −1.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paszti, S.; Vitale, A.; Liu, Y.; Braunwalder, R.; Kalawong, R.; Biner, O.; Pessi, G.; Eberl, L. Identification of Key Factors for Anoxic Survival of B. cenocepacia H111. Int. J. Mol. Sci. 2022, 23, 4560. https://doi.org/10.3390/ijms23094560
Paszti S, Vitale A, Liu Y, Braunwalder R, Kalawong R, Biner O, Pessi G, Eberl L. Identification of Key Factors for Anoxic Survival of B. cenocepacia H111. International Journal of Molecular Sciences. 2022; 23(9):4560. https://doi.org/10.3390/ijms23094560
Chicago/Turabian StylePaszti, Sarah, Alessandra Vitale, Yilei Liu, Rubina Braunwalder, Ratchara Kalawong, Olivier Biner, Gabriella Pessi, and Leo Eberl. 2022. "Identification of Key Factors for Anoxic Survival of B. cenocepacia H111" International Journal of Molecular Sciences 23, no. 9: 4560. https://doi.org/10.3390/ijms23094560
APA StylePaszti, S., Vitale, A., Liu, Y., Braunwalder, R., Kalawong, R., Biner, O., Pessi, G., & Eberl, L. (2022). Identification of Key Factors for Anoxic Survival of B. cenocepacia H111. International Journal of Molecular Sciences, 23(9), 4560. https://doi.org/10.3390/ijms23094560







