Insights into Mechanisms of Damage Recognition and Catalysis by APE1-like Enzymes
Abstract
:1. Introduction
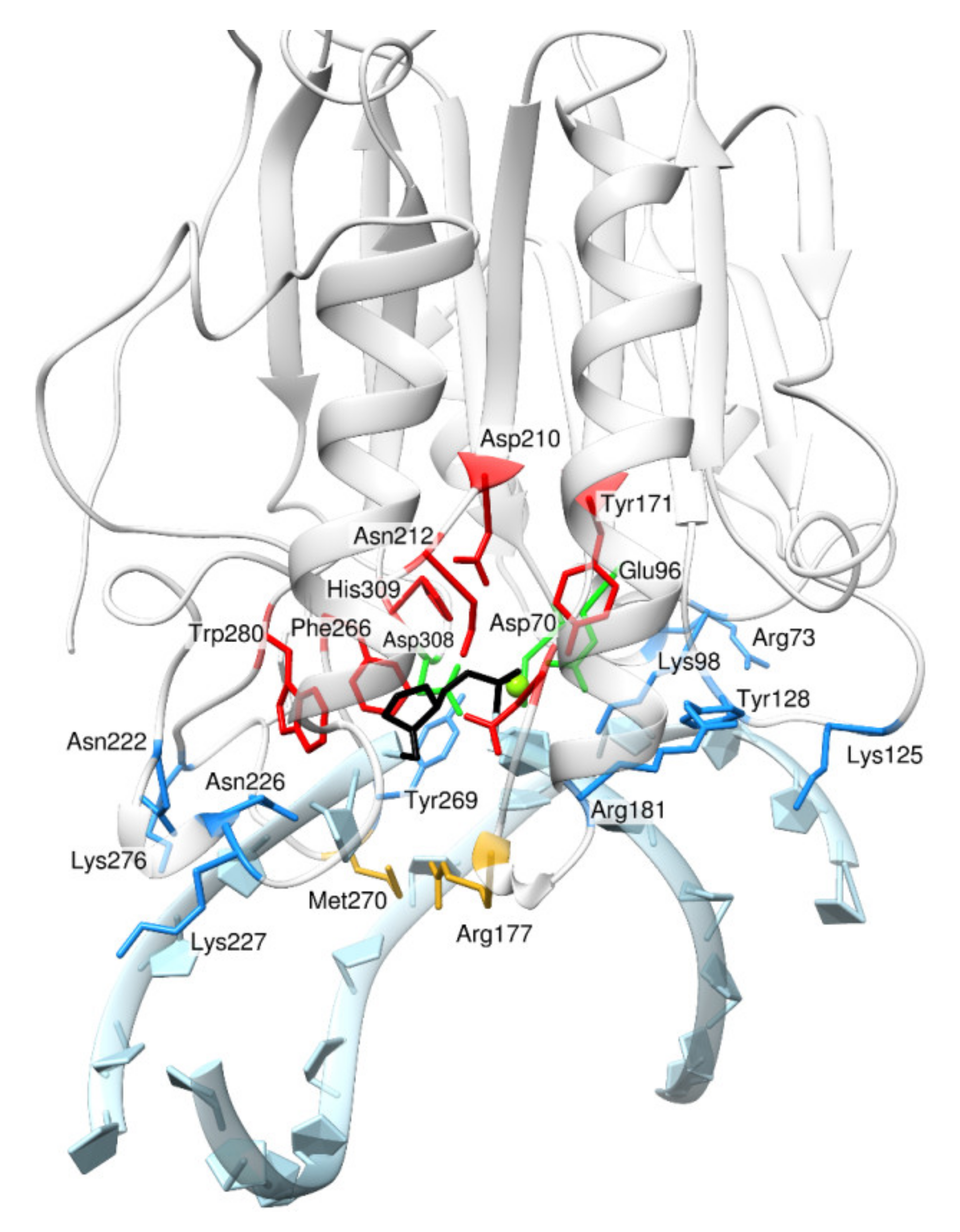
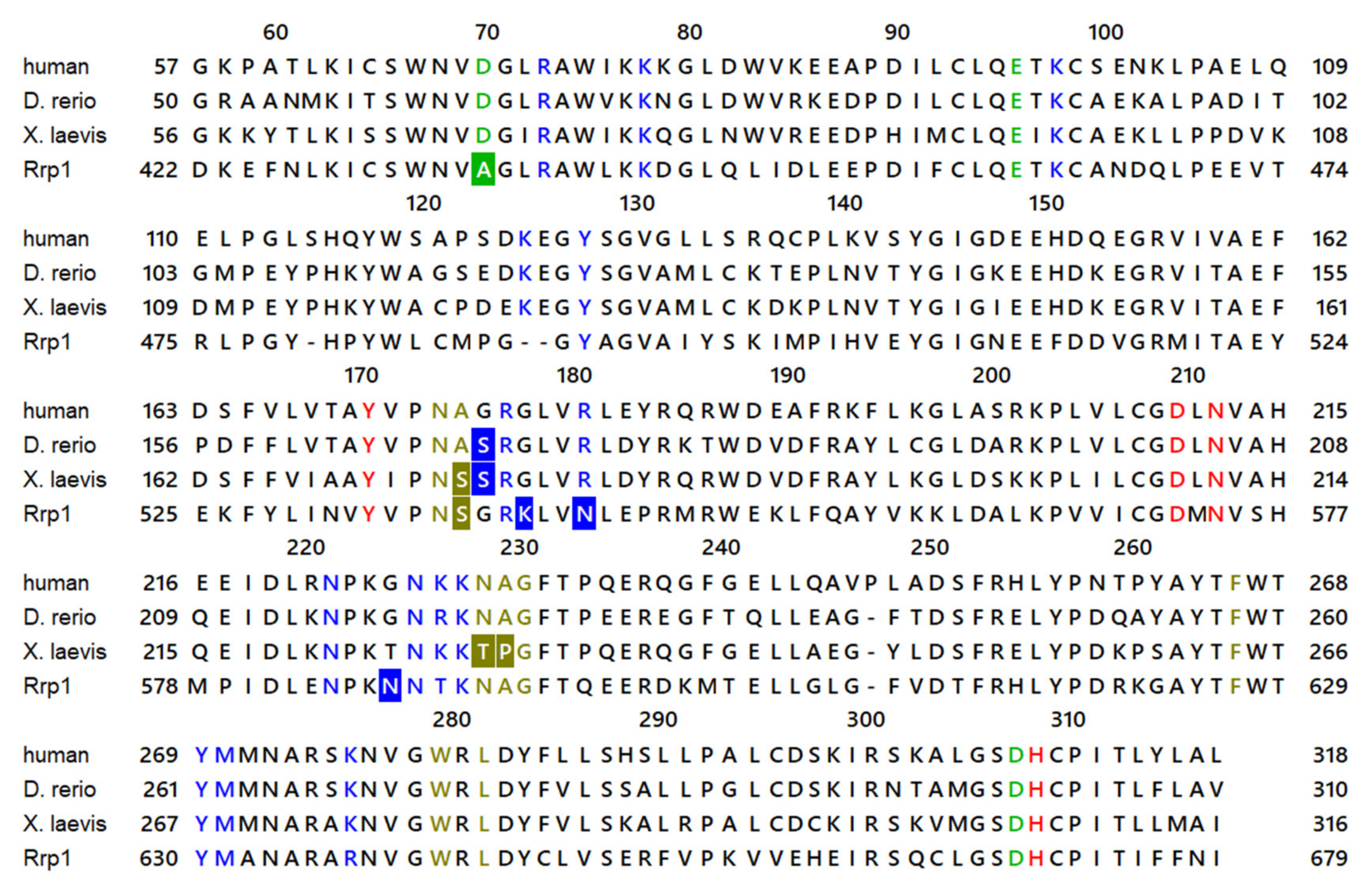
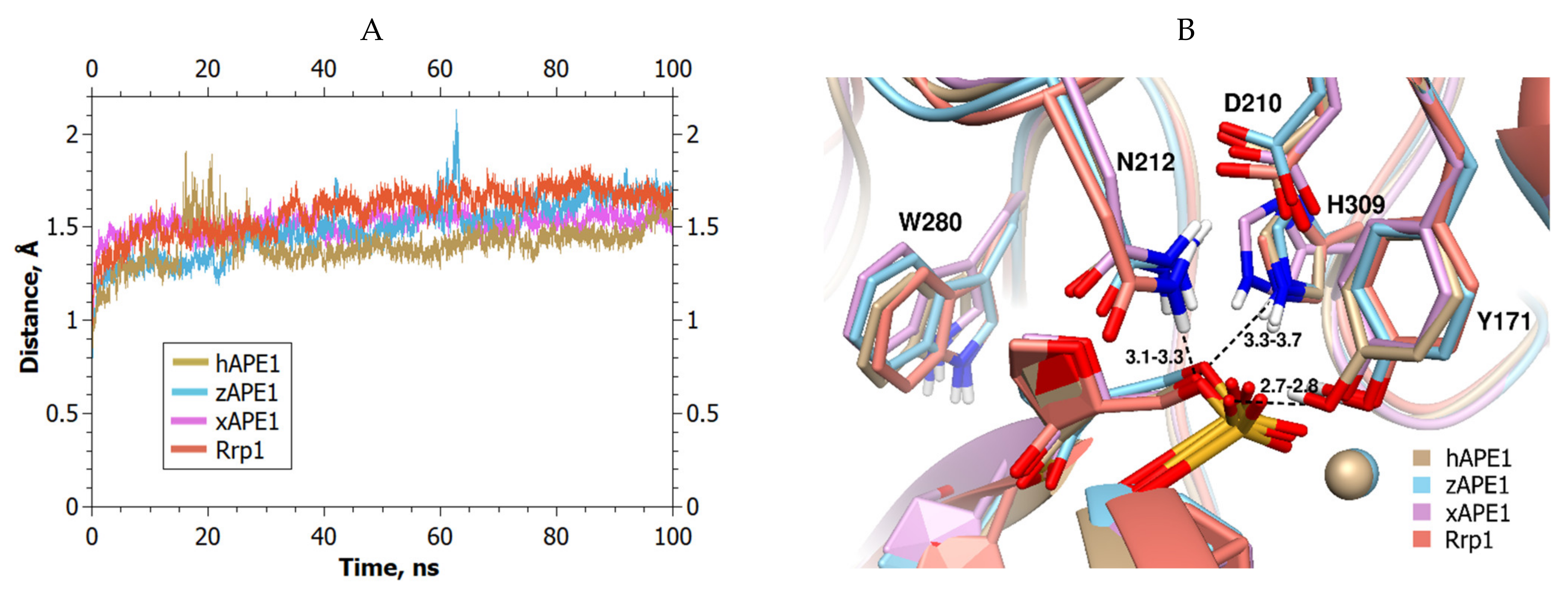
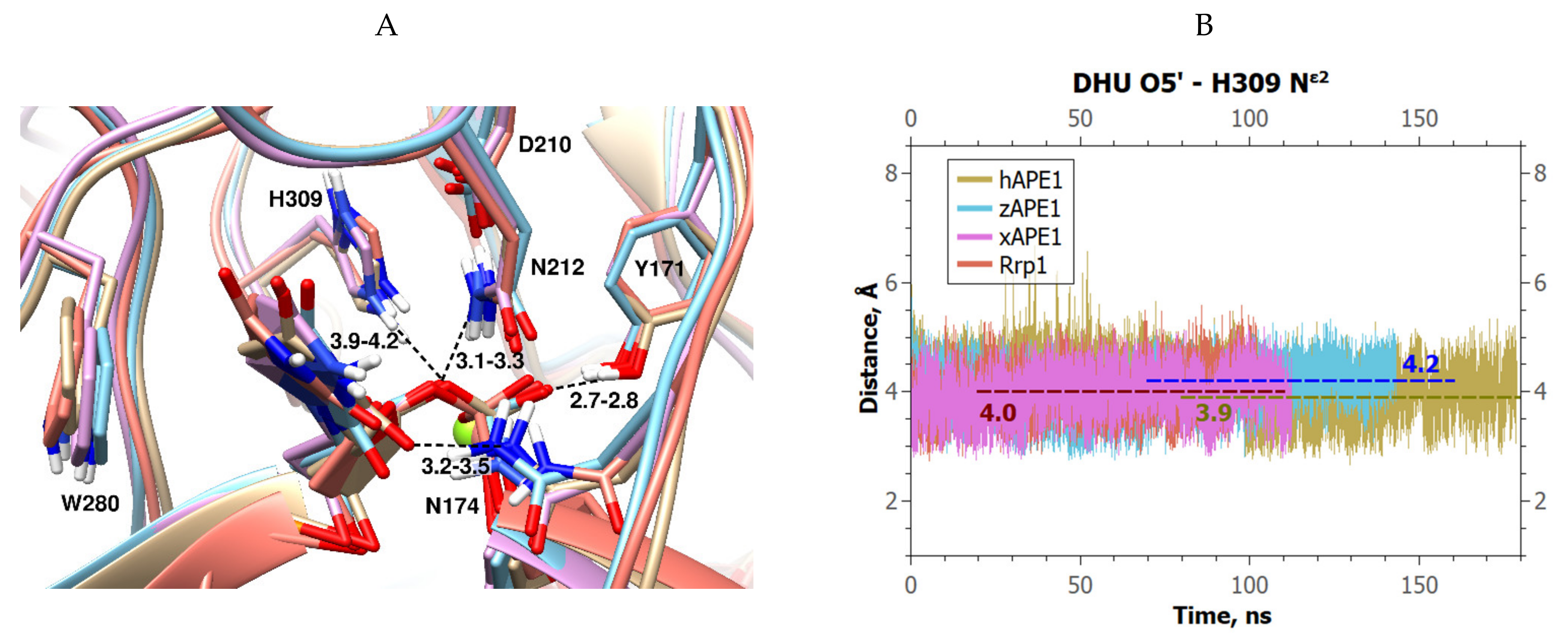
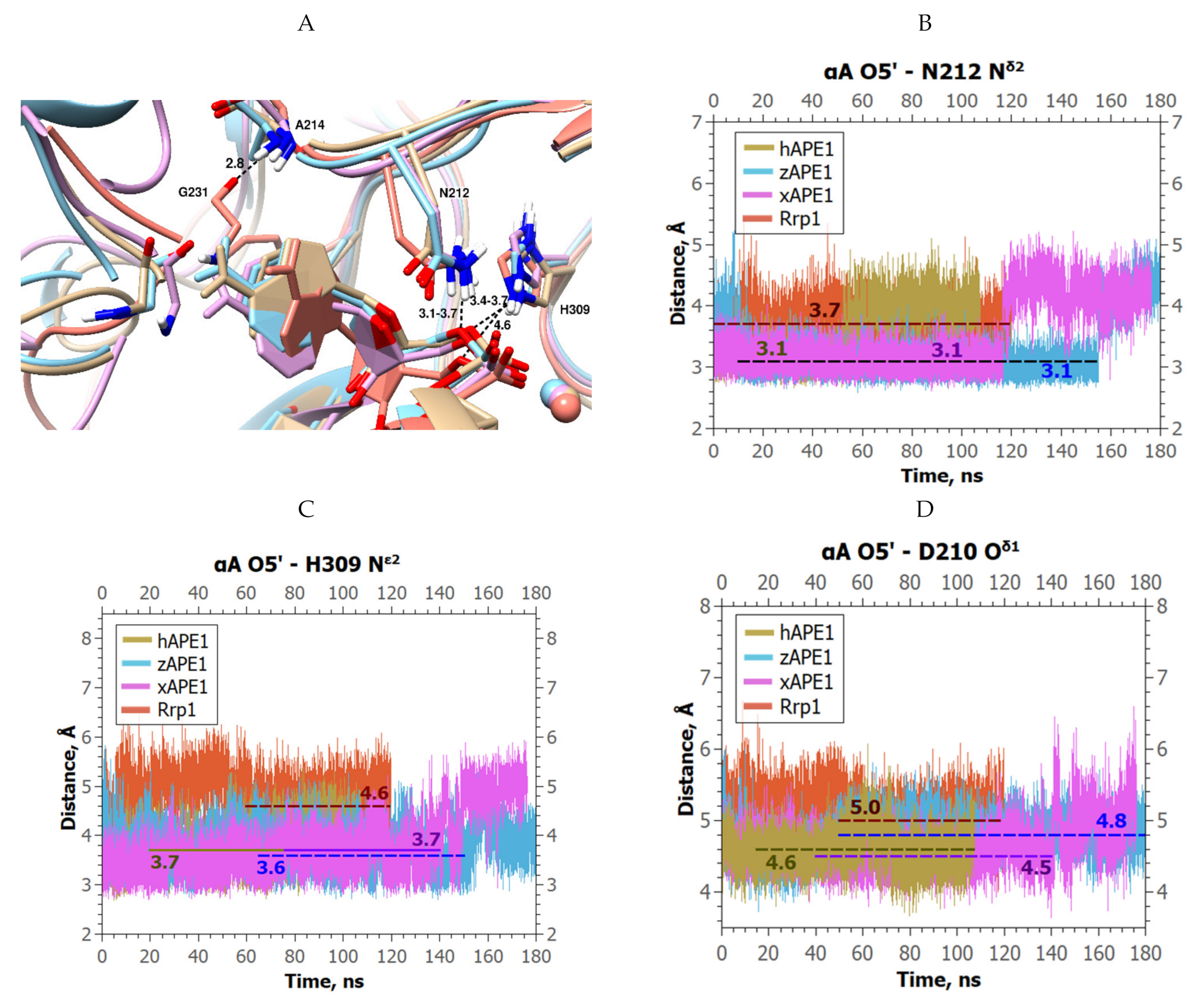
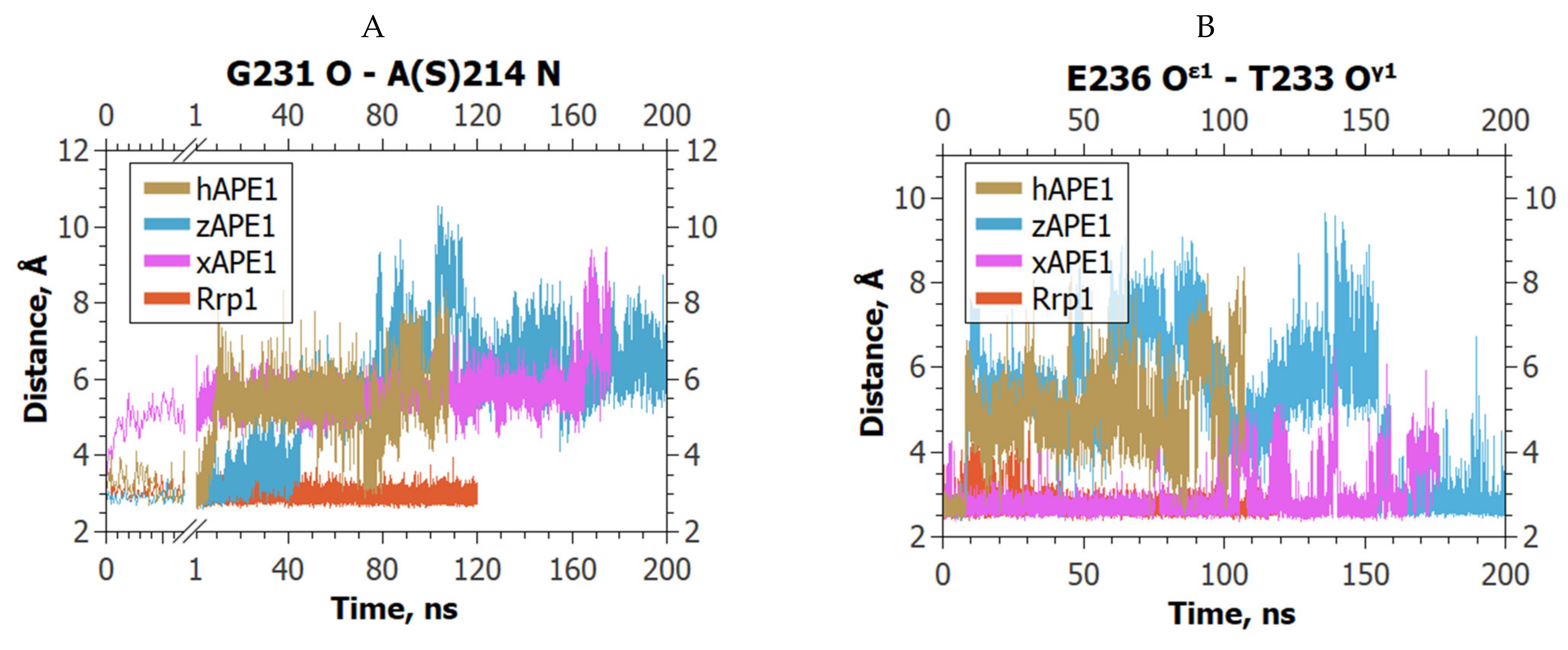
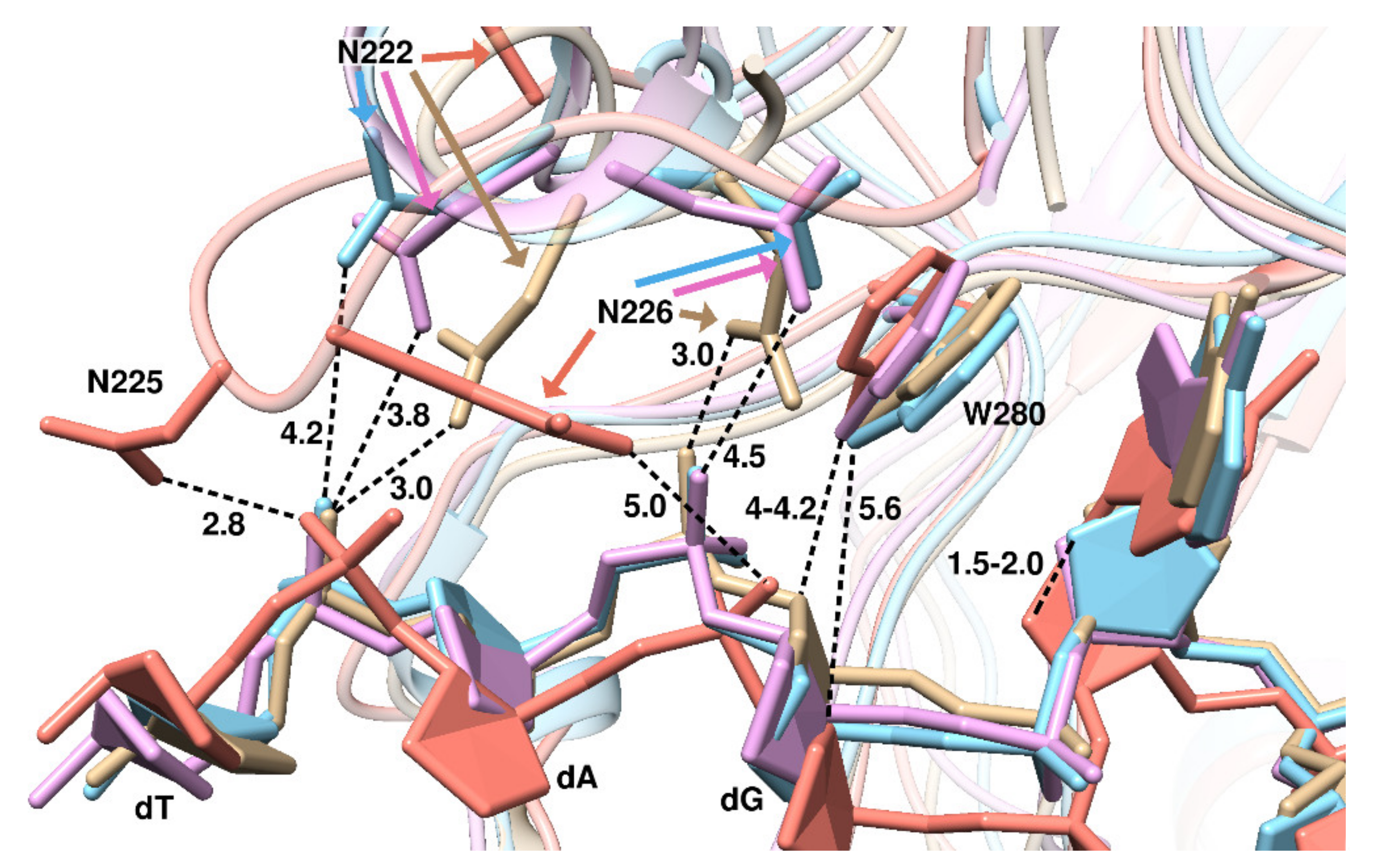
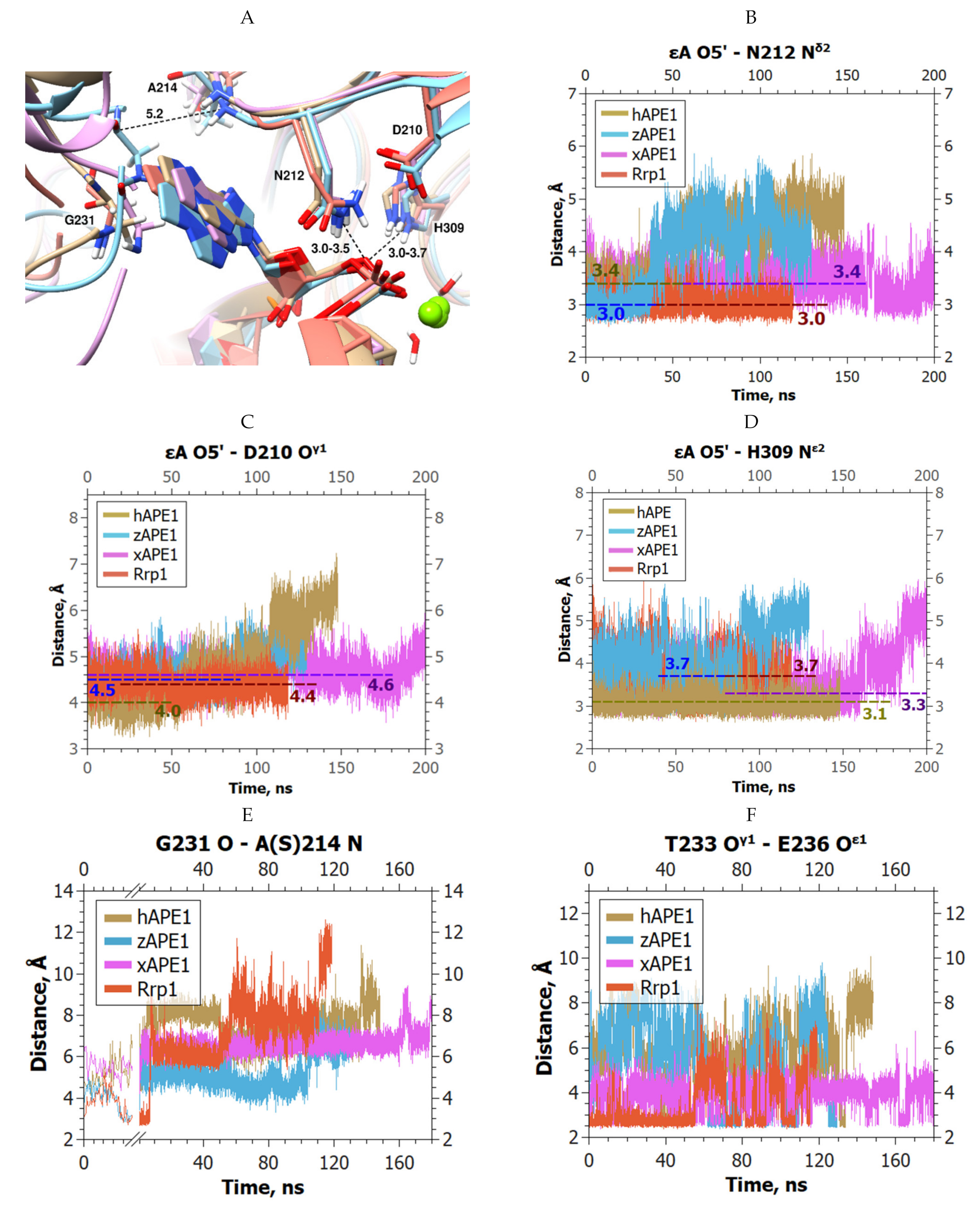
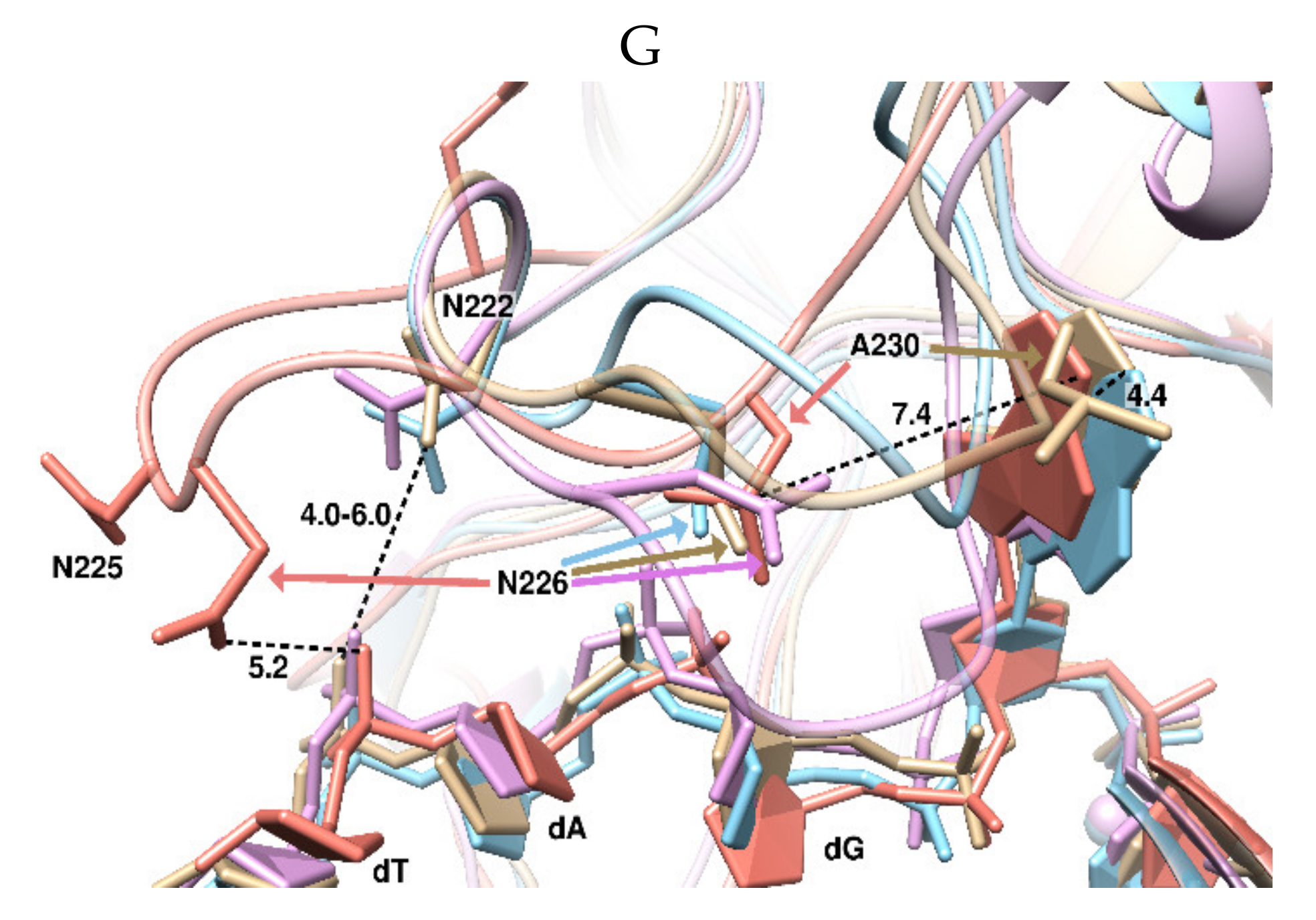
2. Results and Discussion
3. Conclusions
4. Materials and Methods
MD Simulations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D.; Schultz, R.A.; Ellenberger, T. DNA Repair and Mutagenesis; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Gros, L.; Saparbaev, M.K.; Laval, J. Enzymology of the Repair of Free Radicals-Induced DNA Damage. Oncogene 2002, 21, 8905–8925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fromme, J.C.; Banerjee, A.; Verdine, G.L. DNA Glycosylase Recognition and Catalysis. Curr. Opin. Struct. Biol. 2004, 14, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Wilson, D.M. III Overview of Base Excision Repair Biochemistry. Curr. Mol. Pharmacol. 2012, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Daviet, S.; Couve-Privat, S.; Gros, L.; Shinozuka, K.; Ide, H.; Saparbaev, M.; Ishchenko, A.A. Major Oxidative Products of Cytosine Are Substrates for the Nucleotide Incision Repair Pathway. DNA Repair 2007, 6, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Vrouwe, M.G.; Pines, A.; Overmeer, R.M.; Hanada, K.; Mullenders, L.H. UV-Induced Photolesions Elicit ATR-Kinase-Dependent Signaling in Non-Cycling Cells through Nucleotide Excision Repair-Dependent and-Independent Pathways. J. Cell Sci. 2011, 124, 435–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guliaev, A.B.; Hang, B.; Singer, B. Structural Insights by Molecular Dynamics Simulations into Specificity of the Major Human AP Endonuclease toward the Benzene-Derived DNA Adduct, PBQ-C. Nucleic Acids Res. 2004, 32, 2844–2852. [Google Scholar] [CrossRef] [PubMed]
- Prorok, P.; Saint-Pierre, C.; Gasparutto, D.; Fedorova, O.S.; Ishchenko, A.A.; Leh, H.; Buckle, M.; Tudek, B.; Saparbaev, M. Highly Mutagenic Exocyclic DNA Adducts Are Substrates for the Human Nucleotide Incision Repair Pathway. PLoS ONE 2012, 7, e51776. [Google Scholar] [CrossRef]
- Christov, P.P.; Banerjee, S.; Stone, M.P.; Rizzo, C.J. Selective Incision of the Alpha-N-Methyl-Formamidopyrimidine Anomer by Escherichia Coli Endonuclease IV. J. Nucleic Acids 2010, 2010, 850234. [Google Scholar] [CrossRef] [Green Version]
- Gros, L.; Ishchenko, A.A.; Ide, H.; Elder, R.H.; Saparbaev, M.K. The Major Human AP Endonuclease (Ape1) Is Involved in the Nucleotide Incision Repair Pathway. Nucleic Acids Res. 2004, 32, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Prorok, P.; Alili, D.; Saint-Pierre, C.; Gasparutto, D.; Zharkov, D.O.; Ishchenko, A.A.; Tudek, B.; Saparbaev, M.K. Uracil in Duplex DNA Is a Substrate for the Nucleotide Incision Repair Pathway in Human Cells. Proc. Natl. Acad. Sci. USA 2013, 110, E3695–E3703. [Google Scholar] [CrossRef] [Green Version]
- Gorman, M.A.; Morera, S.; Rothwell, D.G.; de La Fortelle, E.; Mol, C.D.; Tainer, J.A.; Hickson, I.D.; Freemont, P.S. The Crystal Structure of the Human DNA Repair Endonuclease HAP1 Suggests the Recognition of Extra-Helical Deoxyribose at DNA Abasic Sites. EMBO J. 1997, 16, 6548–6558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beernink, P.T.; Segelke, B.W.; Hadi, M.Z.; Erzberger, J.P.; Wilson, D.M., 3rd; Rupp, B. Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism. J. Mol. Biol. 2001, 307, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Manvilla, B.A.; Pozharski, E.; Toth, E.A.; Drohat, A.C. Structure of Human Apurinic/Apyrimidinic Endonuclease 1 with the Essential Mg2+ Cofactor. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2555–2562. [Google Scholar] [CrossRef] [Green Version]
- Mol, C.D.; Izumi, T.; Mitra, S.; Tainer, J.A. DNA-Bound Structures and Mutants Reveal Abasic DNA Binding by APE1 and DNA Repair Coordination. Nature 2000, 403, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Mol, C.D.; Hosfield, D.J.; Tainer, J.A. Abasic Site Recognition by Two Apurinic/Apyrimidinic Endonuclease Families in DNA Base Excision Repair: The 3′ Ends Justify the Means. Mutat. Res. 2000, 460, 211–229. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Beard, W.A.; Cuneo, M.J.; Dyrkheeva, N.S.; Wilson, S.H. Capturing Snapshots of APE1 Processing DNA Damage. Nat. Struct. Mol. Biol. 2015, 22, 924–931. [Google Scholar] [CrossRef] [Green Version]
- Tsutakawa, S.E.; Shin, D.S.; Mol, C.D.; Izumi, T.; Arvai, A.S.; Mantha, A.K.; Szczesny, B.; Ivanov, I.N.; Hosfield, D.J.; Maiti, B.; et al. Conserved Structural Chemistry for Incision Activity in Structurally Non-Homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes. J. Biol. Chem. 2013, 288, 8445–8455. [Google Scholar] [CrossRef] [Green Version]
- Timofeyeva, N.A.; Koval, V.V.; Knorre, D.G.; Zharkov, D.O.; Saparbaev, M.K.; Ishchenko, A.A.; Fedorova, O.S. Conformational Dynamics of Human AP Endonuclease in Base Excision and Nucleotide Incision Repair Pathways. J. Biomol. Struct. Dyn. 2009, 26, 637–652. [Google Scholar] [CrossRef]
- Timofeyeva, N.A.; Koval, V.V.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Kinetic Mechanism of Human Apurinic/Apyrimidinic Endonuclease Action in Nucleotide Incision Repair. Biochemistry 2011, 76, 273–281. [Google Scholar] [CrossRef]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Kuznetsov, N.A.; Fedorova, O.S. Thermodynamics of Damaged DNA Binding and Catalysis by Human AP Endonuclease 1. Acta Nat. 2016, 8, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Alekseeva, I.V.; Bakman, A.S.; Vorobjev, Y.N.; Fedorova, O.S.; Kuznetsov, N.A. Role of Ionizing Amino Acid Residues in the Process of DNA Binding by Human AP Endonuclease 1 and in Its Catalysis. J. Phys. Chem. B 2019, 123, 9546–9556. [Google Scholar] [CrossRef] [PubMed]
- Davletgildeeva, A.T.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Activity of Human Apurinic/Apyrimidinic Endonuclease APE1 Toward Damaged DNA and Native RNA With Non-Canonical Structures. Front. Cell Dev. Biol. 2020, 8, 590848. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.C.; Loeb, L.A. Mutations in the A8 Loop of Human APE1 Alter Binding and Cleavage of DNA Containing an Abasic Site. J. Biol. Chem. 2003, 278, 46994–47001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelin, A.; Redrejo-Rodríguez, M.; Laval, J.; Fedorova, O.S.; Saparbaev, M.; Ishchenko, A.A. Genetic and Biochemical Characterization of Human AP Endonuclease 1 Mutants Deficient in Nucleotide Incision Repair Activity. PLoS ONE 2010, 5, e12241. [Google Scholar] [CrossRef] [Green Version]
- Alekseeva, I.V.; Kuznetsova, A.A.; Bakman, A.S.; Fedorova, O.S.; Kuznetsov, N.A. The Role of Active-Site Amino Acid Residues in the Cleavage of DNA and RNA Substrates by Human Apurinic/Apyrimidinic Endonuclease APE1. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129718. [Google Scholar] [CrossRef]
- Alekseeva, I.V.; Davletgildeeva, A.T.; Arkova, O.V.; Kuznetsov, N.A.; Fedorova, O.S. The Impact of Single-Nucleotide Polymorphisms of Human Apurinic/Apyrimidinic Endonuclease 1 on Specific DNA Binding and Catalysis. Biochimie 2019, 163, 73–83. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Gavrilova, A.A.; Novopashina, D.S.; Fedorova, O.S.; Kuznetsov, N.A. Mutational and Kinetic Analysis of APE1 Endoribonuclease Activity. Mol. Biol. 2021, 55, 211–224. [Google Scholar] [CrossRef]
- Bulygin, A.A.; Kuznetsova, A.A.; Vorobjev, Y.N.; Fedorova, O.S.; AKuznetsov, N. The Role of Active-Site Plasticity in Damaged-Nucleotide Recognition by Human Apurinic/Apyrimidinic Endonuclease APE1. Molecules 2020, 25, 3940. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Senchurova, S.I.; Ishchenko, A.A.; Saparbaev, M.; Fedorova, O.S.; Kuznetsov, N.A. Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases. Int. J. Mol. Sci. 2021, 22, 8874. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Matveeva, A.G.; Milov, A.D.; Vorobjev, Y.N.; Dzuba, S.A.; Fedorova, O.S.; Kuznetsov, N.A. Substrate Specificity of Human Apurinic/Apyrimidinic Endonuclease APE1 in the Nucleotide Incision Repair Pathway. Nucleic Acids Res. 2018, 46, 11454–11465. [Google Scholar] [CrossRef] [Green Version]
- Timofeyeva, N.A.; Fedorova, O.S. A Kinetic Mechanism of Repair of DNA Containing Alpha-Anomeric Deoxyadenosine by Human Apurinic/Apyrimidinic Endonuclease 1. Mol. Biosyst. 2016, 12, 3435–3446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davletgildeeva, A.T.; Ishchenko, A.A.; Saparbaev, M.; Fedorova, O.S.; Kuznetsov, N.A. The Enigma of Substrate Recognition and Catalytic Efficiency of APE1-Like Enzymes. Front. Cell Dev. Biol. 2021, 9, 617161. [Google Scholar] [CrossRef] [PubMed]
- Erzberger, J.P.; Wilson, D.M., 3rd. The Role of Mg2+ and Specific Amino Acid Residues in the Catalytic Reaction of the Major Human Abasic Endonuclease: New Insights from EDTA-Resistant Incision of Acyclic Abasic Site Analogs and Site-Directed Mutagenesis. J. Mol. Biol. 1999, 290, 447–457. [Google Scholar] [CrossRef]
- Georgiadis, M.M.; Luo, M.; Gaur, R.K.; Delaplane, S.; Li, X.; Kelley, M.R. Evolution of the Redox Function in Mammalian Apurinic/Apyrimidinic Endonuclease. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2008, 643, 54–63. [Google Scholar] [CrossRef] [Green Version]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Aduri, R.; Psciuk, B.T.; Saro, P.; Taniga, H.; Schlegel, H.B.; SantaLucia, J. AMBER Force Field Parameters for the Naturally Occurring Modified Nucleosides in RNA. J. Chem. Theory Comput. 2007, 3, 1464–1475. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A Flexible Algorithm for Calculating Pair Interactions on SIMD Architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Thakur, A.R. 20ns Molecular Dynamics Simulation of the Antennapedia Homeodomain-Dna Complex: Water Interaction and Dna Structure Analysis. J. Biomol. Struct. Dyn. 2010, 27, 443–455. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulygin, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Insights into Mechanisms of Damage Recognition and Catalysis by APE1-like Enzymes. Int. J. Mol. Sci. 2022, 23, 4361. https://doi.org/10.3390/ijms23084361
Bulygin AA, Fedorova OS, Kuznetsov NA. Insights into Mechanisms of Damage Recognition and Catalysis by APE1-like Enzymes. International Journal of Molecular Sciences. 2022; 23(8):4361. https://doi.org/10.3390/ijms23084361
Chicago/Turabian StyleBulygin, Anatoly A., Olga S. Fedorova, and Nikita A. Kuznetsov. 2022. "Insights into Mechanisms of Damage Recognition and Catalysis by APE1-like Enzymes" International Journal of Molecular Sciences 23, no. 8: 4361. https://doi.org/10.3390/ijms23084361
APA StyleBulygin, A. A., Fedorova, O. S., & Kuznetsov, N. A. (2022). Insights into Mechanisms of Damage Recognition and Catalysis by APE1-like Enzymes. International Journal of Molecular Sciences, 23(8), 4361. https://doi.org/10.3390/ijms23084361






