PcWRKY11, an II-d WRKY Transcription Factor from Polygonum cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. Identification and Sequence Analysis of PcWRKY11
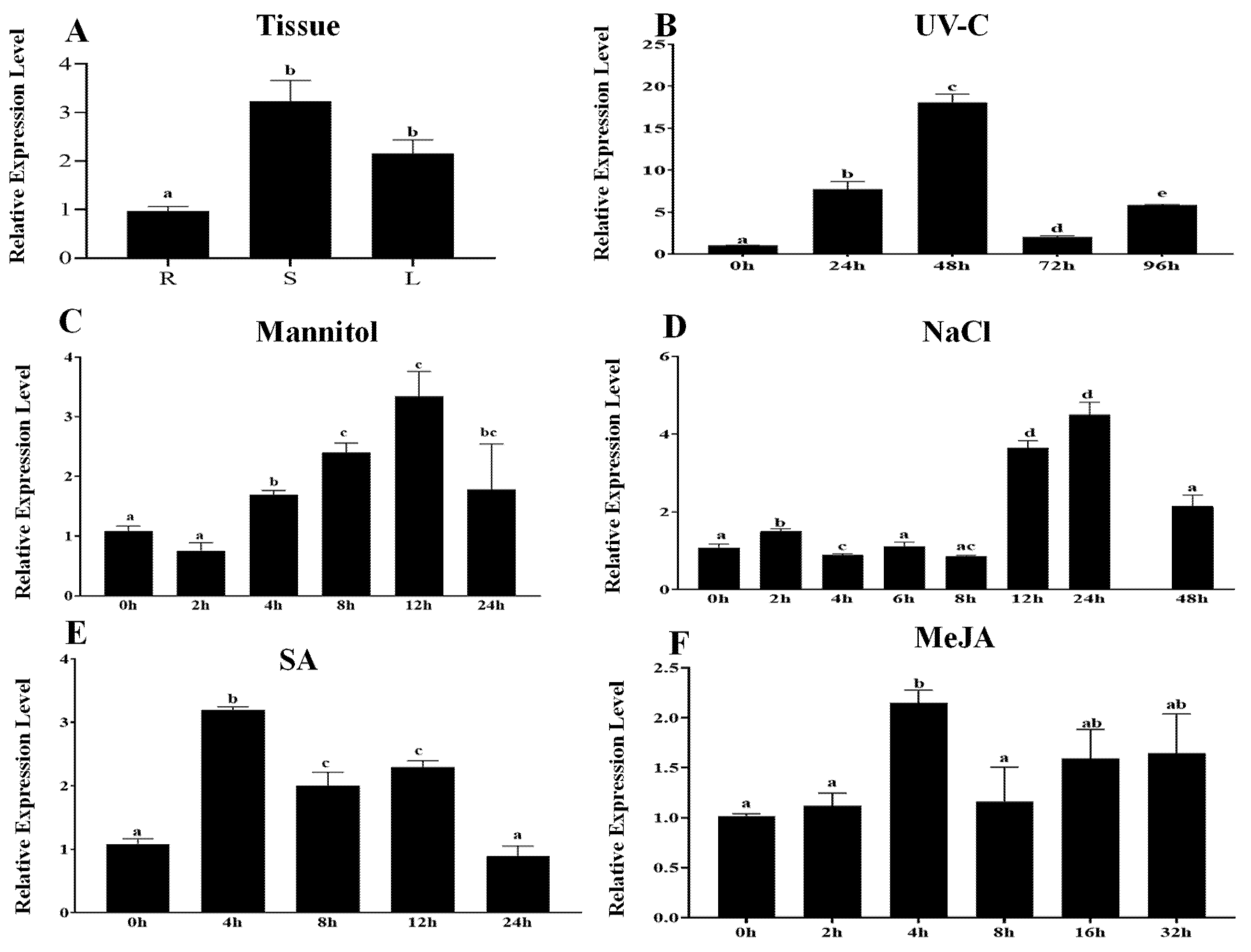
2.2. Expression Analysis of PcWRKY11
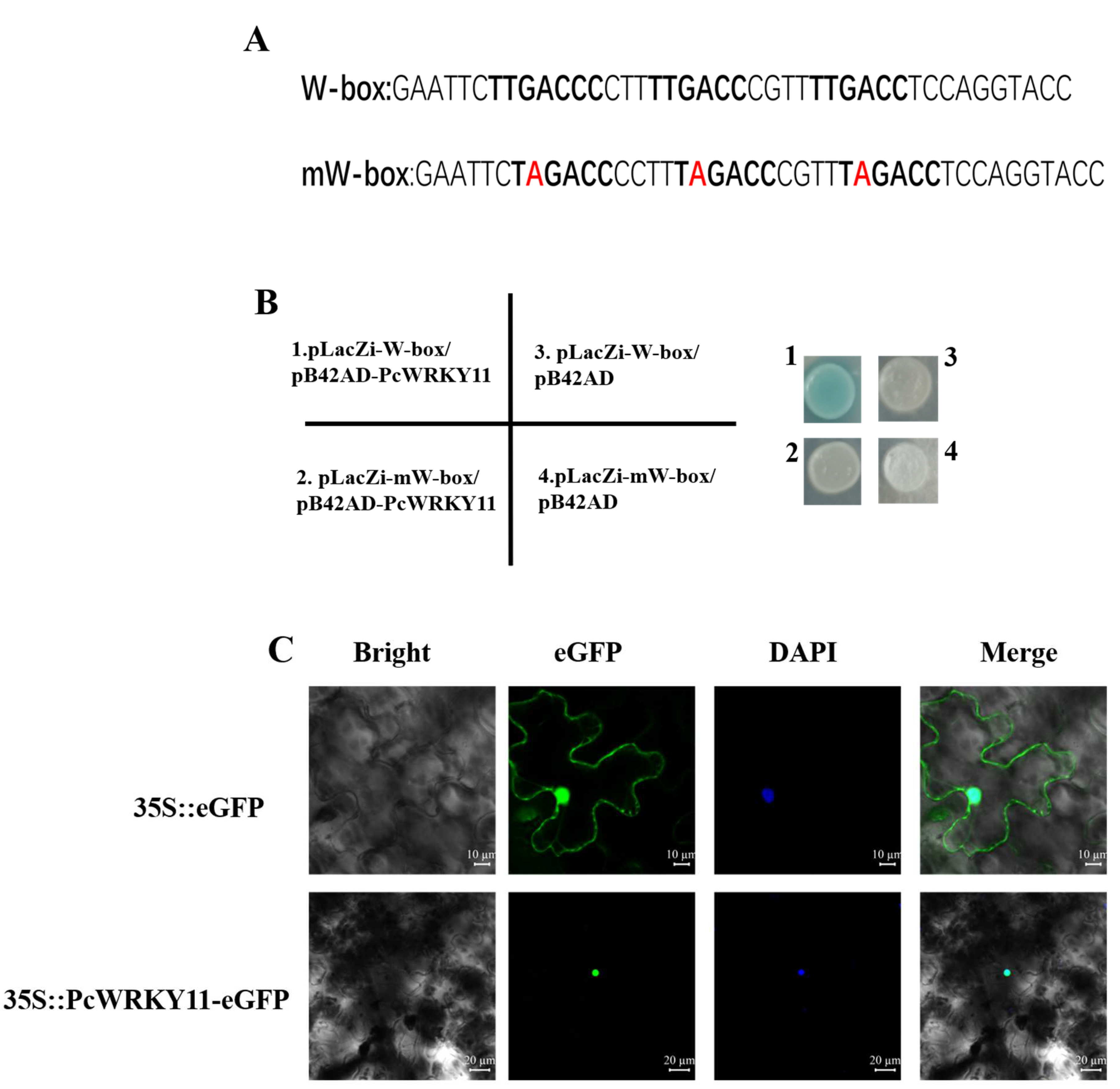
2.3. Characterization of PcWRKY11 as A Transcription Factor
2.4. PcWRKY11 Enhances Salt Tolerance of Transgenic Arabidopsis
2.5. Analysis of Oxidative Damage in PcWRKY11 Overexpressed Plants
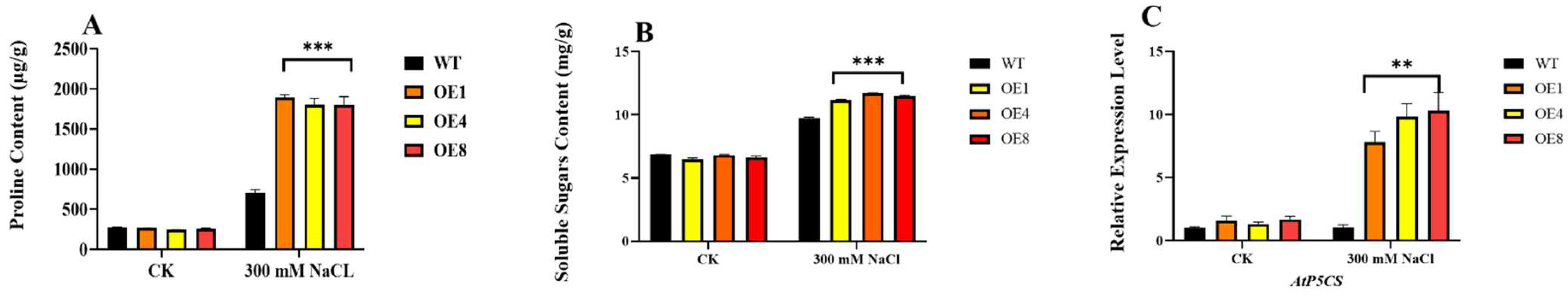
2.6. Analysis of Osmotic Stress in PcWRKY11 Overexpressed Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. Gene Cloning and Sequence Analysis
4.3. Vector Construction and Agrobacterium-Mediated A. thaliana Transformation
4.4. Subcellular Localization Analysis of PcWRKY11
4.5. Yeast One-Hybrid Assay
4.6. Quantitative Real-Time PCR Analysis
4.7. Determination of Related Physiological Indexes
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J. Oxidative stress. In Free Radicals in Biology and Medicine, 3rd ed.; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Sharma, P.; Dubey, R.S. Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 2007, 26, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants: Functional domains evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Luking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genom. 2009, 282, 503–516. [Google Scholar] [CrossRef]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Park, C.Y.; Lee, J.H.; Yoo, J.H.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; Lee, S.M.; Kim, H.S.; Kang, K.Y.; Chung, W.S.; et al. WRKY group II-d transcription factors interact with calmodulin. FEBS Lett. 2005, 28, 1545–1550. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54 confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species Production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, Y.; Guo, Y.; Huang, J.; Qiao, L. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 2021, 160, 175–183. [Google Scholar] [CrossRef]
- Zhu, H.; Zhou, Y.; Zhai, H.; He, S.; Liu, Q. A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis. Biomolecules 2020, 10, 506. [Google Scholar] [CrossRef]
- Zhou, S.; Zheng, W.J.; Liu, B.H.; Zheng, J.C.; Dong, F.S.; Liu, Z.F.; Wen, Z.Y.; Yang, F.; Wang, H.B.; Xu, Z.S.; et al. Characterizing the role of TaWRKY13 in salt tolerance. Int. J. Mol. Sci. 2019, 20, 5712. [Google Scholar] [CrossRef]
- Murrell, C.; Gerber, E.; Krebs, C.; Parepa, M.; Schaffner, U.; Bossdorf, O. Invasive knotweed affects native plants through allelopathy. Am. J. Bot. 2011, 98, 38–43. [Google Scholar] [CrossRef]
- Dommanget, F.; Cavaillé, P.; Evette, A.; Martin, F. Asian knotweeds-An example of a raising threat. In Introduced Tree Species in European Forests: Opportunities and Challenges; European Forest Institute: Joensuu, Finland, 2016; pp. 202–211. [Google Scholar]
- Ayres, D.R.; Garcia-Rossi, D.; Davis, H.G.; Strong, D.R. Extent and degree of hybridization between exotic (Spartina alterniflora) and native (S. foliosa) cordgrass (Poaceae) in California, USA determined by random amplified polymorphic DNA (RAPDs). Mol. Ecol. 1999, 8, 1179–1186. [Google Scholar] [CrossRef]
- Daehler, C.C.; Strong, D.R. Status, prediction and prevention of introduced cordgrass Spartina spp. invasions in Pacific estuaries, USA. Biol. Conserv. 1996, 78, 51–55. [Google Scholar] [CrossRef]
- Miklovic, S.; Galatowitsch, S.M. Effect of NaCl and Typha angustifolia L. on marsh community establishment: A green-house study. Wetlands 2005, 25, 420–429. [Google Scholar] [CrossRef][Green Version]
- Vasquez, E.A.; Glenn, E.P.; Brown, J.J. Salt tolerance underlies the cryptic invasion of North American salt marshes by an introduced haplotype of the common reed Phragmites australis (Poaceae). Mar. Ecol. Prog. Ser. 2005, 298, 1–8. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.H.; Zhao, B.; Ma, Z.J.; Luo, Y.Q.; Chen, J.K.; Li, B. Effects of growing conditions on the growth of and interactions between salt marsh plants: Implications for invasibility of habitats. Biol. Invasions 2006, 8, 1547–1560. [Google Scholar] [CrossRef]
- Rouifed, S.; Byczek, C.; Laffray, D. Invasive knotweeds are highly tolerant to salt stress. Environ. Manag. 2012, 50, 1027–1034. [Google Scholar] [CrossRef]
- Bao, W.; Wang, X.; Chen, M.; Chai, T.; Wang, H. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant Cell Rep. 2018, 37, 1033–1048. [Google Scholar] [CrossRef]
- Hu, H.; Wang, X.; Wu, Z.; Chen, M.; Chai, T.; Wang, H. Overexpression of the Polygonum cuspidatum PcDREB2A gene encoding a DRE-Binding transcription factor enhances the drought tolerance of transgenic Arabidopsis thaliana. J. Plant Biol. 2021, 1–11. [Google Scholar] [CrossRef]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef]
- Clements, D.R.; Ditommaso, A. Predicting weed invasion in Canada under climate change: Evaluating evolutionary potential. Can. J. Plant Sci. 2012, 92, 1013–1020. [Google Scholar] [CrossRef]
- Michalet, S.; Rouifed, S.; Pellassa-Simon, T.; Fusade-Boyer, M.; Meiffren, G.; Nazaret, S.; Piola, F. Tolerance of Japanese knotweed s.l. to soil artificial polymetallic pollution: Early metabolic responses and performance during vegetative multiplication. Environ. Sci. Pollut. Res. 2017, 24, 20897–20907. [Google Scholar] [CrossRef]
- Yue, X.; Shi, P.; Tang, Y.; Zhang, H.; Ma, X.; Ju, Y.; Zhang, Z. Effects of methyl jasmonate on the monoterpenes of Muscat Hamburg grapes and wine. J. Sci. Food Agric. 2020, 101, 3665–3675. [Google Scholar] [CrossRef]
- Ahmad, B.; Raina, A.; Naikoo, M.I.; Khan, S. Role of methyl jasmonates in salt stress tolerance in crop plants. Plant Signal. Mol. 2019, 24, 371–384. [Google Scholar]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997, 12, 1067–1078. [Google Scholar] [CrossRef]
- Wu, T.Y.; Goh, H.; Azodi, C.B.; Krishnamoorthi, S.; Liu, M.J.; Urano, D. Evolutionarily conserved hierarchical gene regulatory networks for plant salt stress response. Nat. Plants 2021, 7, 787–799. [Google Scholar] [CrossRef]
- Ali, M.A.; Azeem, F.; Nawaz, M.A.; Acet, T.; Abbas, A.; Imran, Q.M.; Shah, K.H.; Rehman, H.M.; Chung, G.; Yang, S.H.; et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018, 226, 12–21. [Google Scholar] [CrossRef]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.S.; Park, S.R.; Lee, S.; Hwang, D.J. Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Chen, J.; Tao, L.; An, Y.; Cai, H.; Guo, C. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS ONE 2018, 13, e0192382. [Google Scholar] [CrossRef]
- Wang, X.; Ajab, Z.; Liu, C.; Hu, S.; Liu, J.; Guan, Q. Overexpression of transcription factor SlWRKY28 improved the tolerance of Populus davidiana × P. bolleana to alkaline salt stress. BMC Genet. 2020, 21, 103. [Google Scholar] [CrossRef]
- Yan, J.; Li, J.; Zhang, H.; Liu, Y.; Zhang, A. ZmWRKY104 positively regulates salt tolerance by modulating ZmSOD4 expression in maize. Crop J. 2021, 10, 555–564. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Meena, M.; Divyanshu, K.; Kumar, S.; Swapnil, P.; Zehra, A.; Shukla, V.; Yadav, M.; Upadhyay, R.S. Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable environmental conditions. Heliyon 2019, 5, e02952. [Google Scholar] [CrossRef]
- Li, H.Y.; Li, D.H. Expression of AtP5CS1 gene enhanced drought tolerance of transgenic brassica oleracea plants. Plant Physiol. 2014, 50, 1009–1013. [Google Scholar]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Crooks, G.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2010, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Klei, K.; Fokkens, L.; Haring, M.A.; Schranz, M.E.; Testerink, C. Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J. Exp. Bot. 2016, 67, 2127–2138. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Bao, W.; Hu, H.; Chen, M.; Chai, T.; Wang, H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biol. 2019, 19, 498. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, M.X.; Chan, W.L.; Yang, F.; Tian, Y.; Song, T.; Xie, L.J.; Zhou, Y.; Xiao, S.; Zhang, J.; et al. SWATH-MS quantitative proteomic investigation of nitrogen starvation in Arabidopsis reveals new aspects of plant nitrogen stress responses. J. Proteom. 2018, 187, 161–170. [Google Scholar] [CrossRef]
- Li, H.; Wang, L.; Luo, Y. Composition analysis by UPLC-PDA-ESI(-)-HRMS and antioxidant activity using Saccharomyces cerevisiae model of herbal teas and green teas from Hainan. Molecules 2018, 23, 2550. [Google Scholar] [CrossRef]
- Qiao, K.; Liang, S.; Wang, F.; Wang, H.; Hu, Z.; Chai, T. Effects of cadmium toxicity on diploid wheat (Triticum urartu) and the molecular mechanism of the cadmium response. J. Hazard. Mater. 2019, 374, 1–10. [Google Scholar] [CrossRef]
- Chen, F.Z.; Zhao, Y.; Chen, H.Z. MicroRNA-98 reduces amyloid β-protein production and improves oxidative stress and mitochondrial dysfunction through the Notch signaling pathway via HEY2 in Alzheimer’s disease mice. Int. J. Mol. Med. 2019, 43, 91–102. [Google Scholar] [CrossRef]
- Li, B.; Ding, Y.; Tang, X.; Wang, G.; Wu, S.; Li, X.; Huang, X.; Qu, T.; Chen, J.; Tang, X. Effect of L-arginine on maintaining storage quality of the white button mushroom (Agaricus bisporus). Food Bioprocess Technol. 2019, 12, 563–574. [Google Scholar] [CrossRef]
- Yin, Y.J.; Chen, C.J.; Guo, S.W.; Li, K.M.; Ma, Y.N.; Sun, W.M.; Xu, F.R.; Cheng, Y.X.; Dong, X. The fight against panax notoginseng root-rot disease using zingiberaceae essential oils as potential weapons. Front. Plant Sci. 2018, 9, 1346. [Google Scholar] [CrossRef]
- Wang, Q.; Xiao, G.; Chen, G.; Du, H.; Wang, L.; Guo, D.; Hu, T. Toxic effect of microcystin-LR on blood vessel development. Toxicol. Environ. Chem. 2019, 100, 737–752. [Google Scholar] [CrossRef]
- Liu, G.; Mei, H.; Chen, M.; Qin, S.; Li, K.; Zhang, W.; Chen, T. Protective effect of agmatine against hyperoxia-induced acute lung injury via regulating lncRNA gadd7. Biochem. Biophys. Res. Commun. 2019, 516, 68–74. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Wang, X.; Ma, H.; Fan, H.; Lin, F.; Chen, J.; Chai, T.; Wang, H. PcWRKY11, an II-d WRKY Transcription Factor from Polygonum cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 4357. https://doi.org/10.3390/ijms23084357
Wang G, Wang X, Ma H, Fan H, Lin F, Chen J, Chai T, Wang H. PcWRKY11, an II-d WRKY Transcription Factor from Polygonum cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(8):4357. https://doi.org/10.3390/ijms23084357
Chicago/Turabian StyleWang, Guowei, Xiaowei Wang, Hongping Ma, Haili Fan, Fan Lin, Jianhui Chen, Tuanyao Chai, and Hong Wang. 2022. "PcWRKY11, an II-d WRKY Transcription Factor from Polygonum cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 8: 4357. https://doi.org/10.3390/ijms23084357
APA StyleWang, G., Wang, X., Ma, H., Fan, H., Lin, F., Chen, J., Chai, T., & Wang, H. (2022). PcWRKY11, an II-d WRKY Transcription Factor from Polygonum cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences, 23(8), 4357. https://doi.org/10.3390/ijms23084357





