Analysis of microRNAs in Small Urinary Extracellular Vesicles and Their Potential Roles in Pathogenesis of Renal ANCA-Associated Vasculitis
Abstract
1. Introduction
2. Results
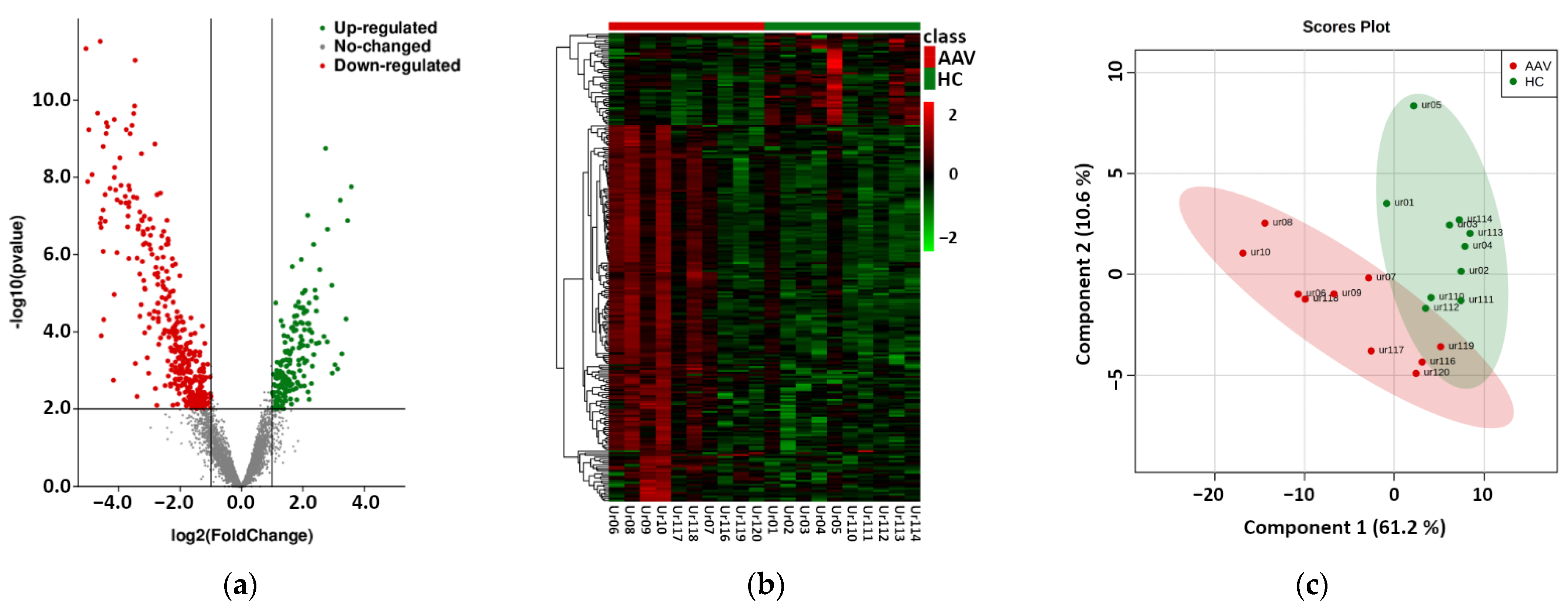
2.1. Next Generation Sequencing Based Phase
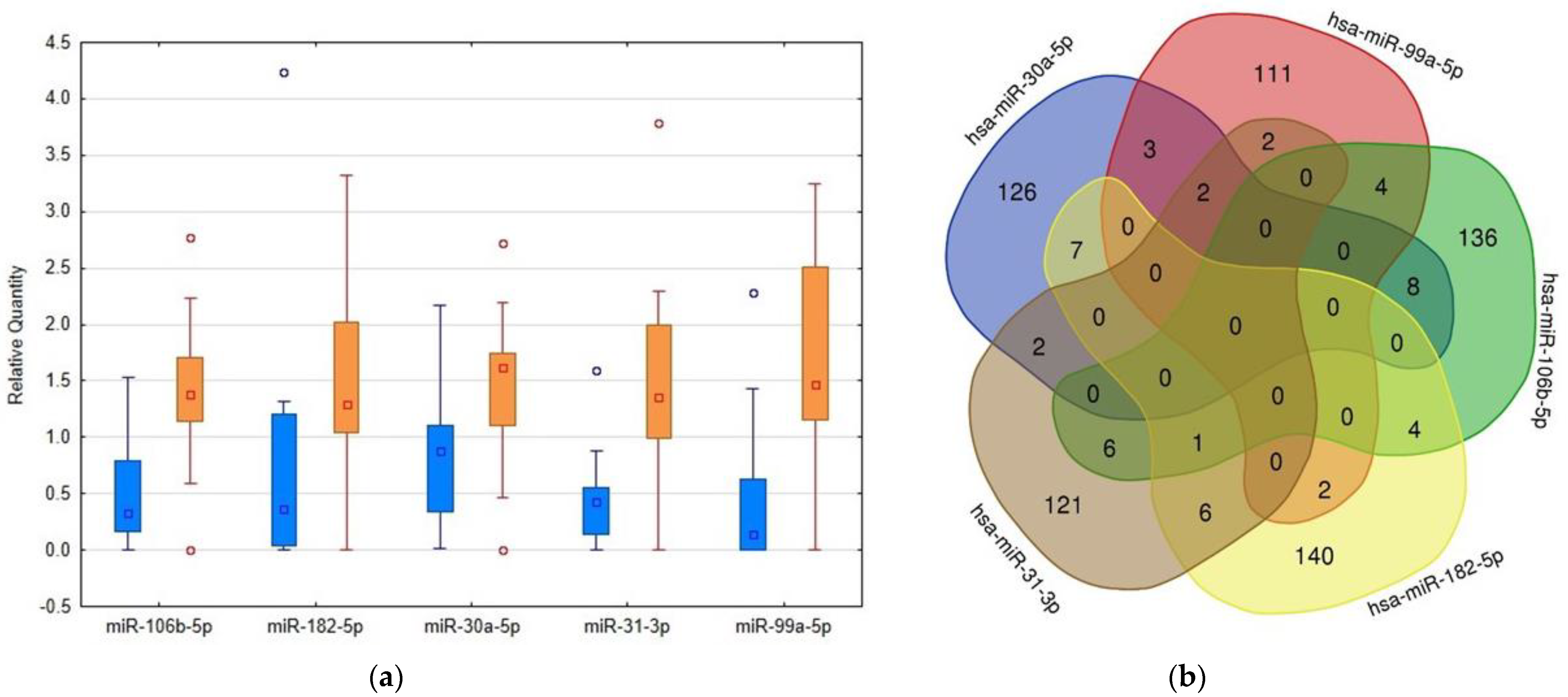
2.2. RT-PCR Based Confirmation Phase
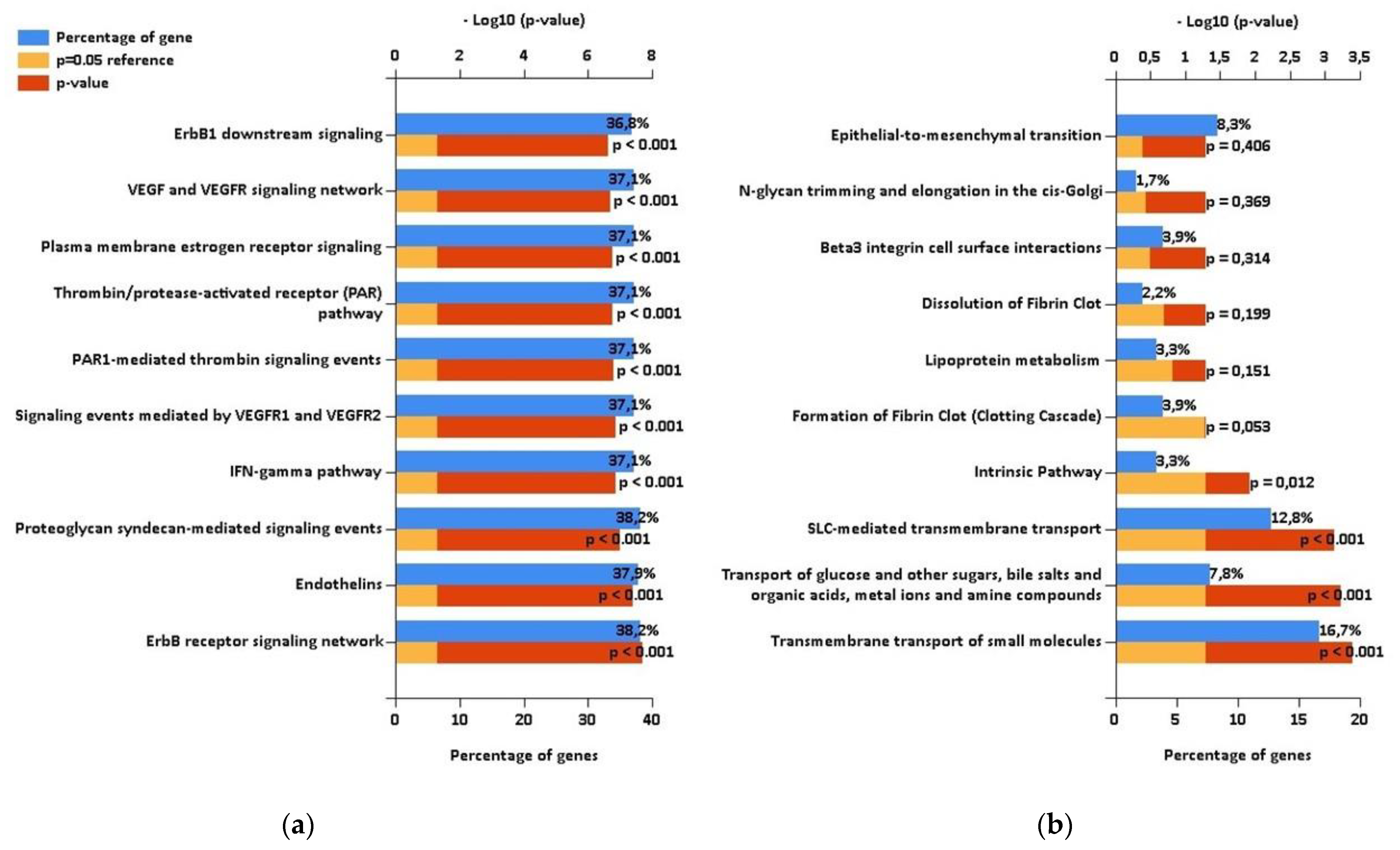
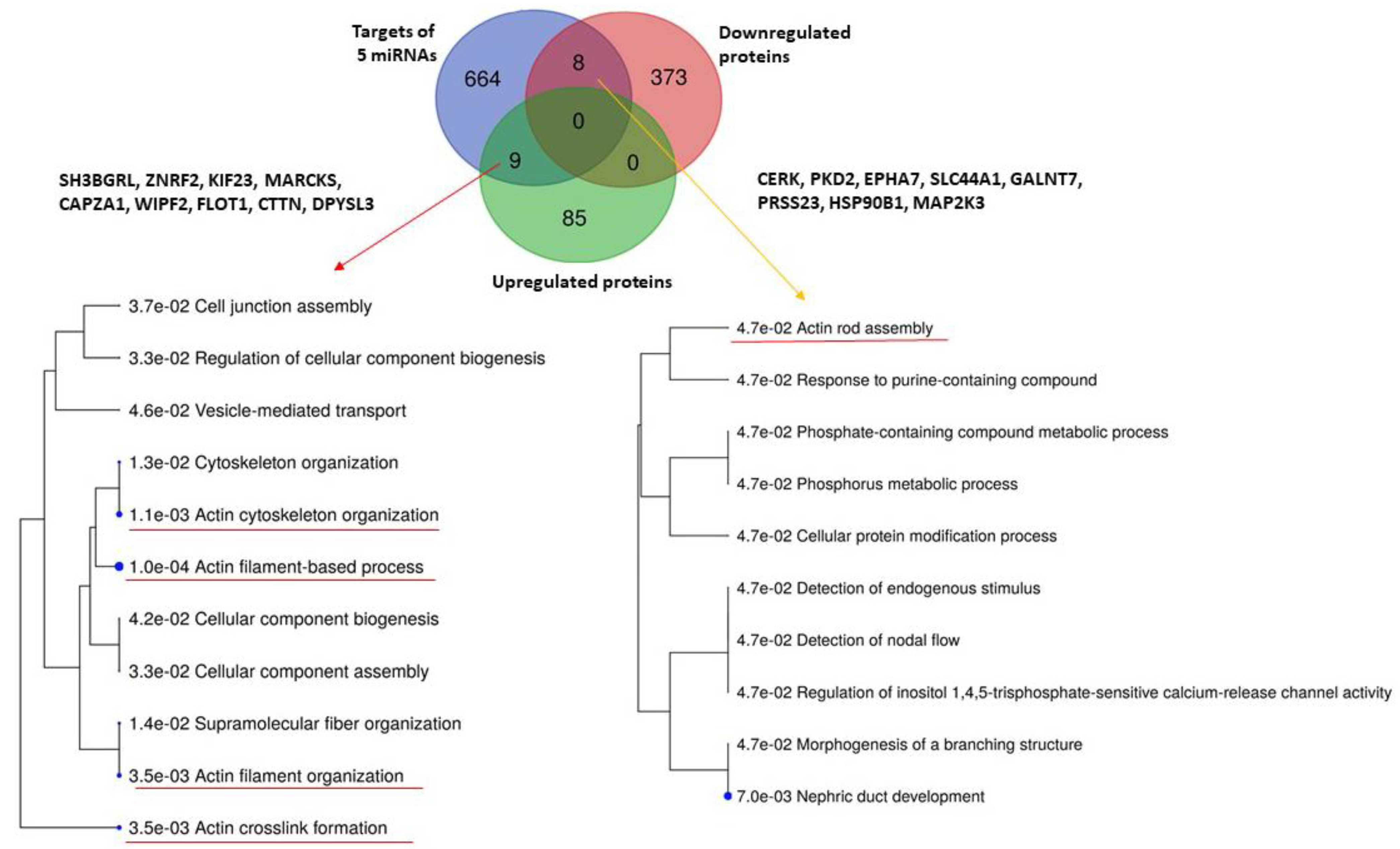
2.3. Comparison with Proteomic Results and Bioinformatic Analysis
3. Discussion
4. Materials and Methods
4.1. Patients and Urine Sample Collection
4.2. Urine Sample Preparation
4.3. Urinary Extracellular Vesicle Enrichment and miRNA Isolation
4.4. Next Generation Sequencing Based Phase
4.5. Single Target RT-PCR Based Confirmation Phase
4.6. Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Hussain, T.; Hussein, M.H.; Conca, W.; Al Mana, H.; Akhtar, M. Pathophysiology of ANCA-associated vasculitis. Adv. Anat. Pathol. 2017, 4, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Korabecna, M.; Tesar, V. NETosis provides the link between activation of neutrophils on hemodialysis membrane and comorbidities in dialyzed patients. Inflamm. Res. 2017, 66, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Bertram, A.; Lovric, S.; Engel, A.; Beese, M.; Wyss, K.; Hertel, B.; Park, J.K.; Becker, J.U.; Kegel, J.; Haller, H.; et al. Circulating ADAM17 level reflects disease activity in proteinase-3 ANCA-associated vasculitis. J. Am. Soc. Nephrol 2015, 26, 2860–2870. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Ferreira, R.; Guedes, S.; Amado, F.; Thongboonkerd, V. What can urinary exosomes tell us? Cell Mol. Life Sci. 2021, 78, 3265–3283. [Google Scholar] [CrossRef]
- Abbasian, N.; Herbert, K.E.; Pawluczyk, I.; Burton, J.O.; Bevington, A. Vesicles bearing gifts: The functional importance of micro-RNA transfer in extracellular vesicles in chronic kidney disease. Am. J. Physiol. Renal. Physiol. 2018, 315, F1430–F1443. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, H.; Long, Y.; Li, P.; Gu, Y. The main sources of circulating cell-free DNA: Apoptosis, necrosis, and active secretion. Crit. Rev. Oncol. Hematol. 2021, 157, 103166. [Google Scholar] [CrossRef]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis, and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Karpman, D.; Ståhl, A.L.; Arvidsson, I. Extracellular vesicles in renal disease. Nat. Rev. Nephrol. 2017, 13, 545–562. [Google Scholar] [CrossRef]
- Solé, C.; Moliné, T.; Vidal, M.; Ordi-Ros, J.; Cortés-Hernández, J. An exosomal urinary miRNA signature for early diagnosis of renal fibrosis in lupus nephritis. Cells 2019, 8, 773. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vives, E.; Solé, C.; Moliné, T.; Vidal, M.; Agraz, I.; Ordi-Ros, J.; Cortés-Hernández, J. The urinary exosomal miRNA expression profile is predictive of clinical response in lupus nephritis. Int. J. Mol. Sci. 2020, 21, 1372. [Google Scholar] [CrossRef] [PubMed]
- Prikryl, P.; Satrapova, V.; Frydlova, J.; Hruskova, Z.; Zima, T.; Tesar, V.; Vokurka, M. Mass spectrometry-based proteomic exploration of the small urinary extracellular vesicles in ANCA-associated vasculitis in comparison with total urine. J. Proteomics. 2021, 233, 104067. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.; Grossman, J.; Hahn, B.H.; La Cava, A. Cutaneous vasculitis in breast cancer treated with chemotherapy. Clin. Immunol. 2008, 129, 3–9. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034.1. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Lv, L.L.; Feng, Y.; Tang, T.T.; Liu, B.C. New insight into the role of extracellular vesicles in kidney disease. J. Cell Mol. Med. 2019, 23, 731–739. [Google Scholar] [CrossRef]
- Lv, L.L.; Feng, Y.; Wen, Y.; Wu, W.J.; Ni, H.F.; Li, Z.L.; Zhou, L.T.; Wang, B.; Zhang, J.D.; Crowley, S.D.; et al. Exosomal CCL2 from tubular epithelial cells is critical for albumin-induced tubulointerstitial inflammation. J. Am. Soc. Nephrol. 2018, 29, 919–935. [Google Scholar] [CrossRef]
- Gildea, J.J.; Seaton, J.E.; Victor, K.G.; Reyes, C.M.; Bigler Wang, D.; Pettigrew, A.C.; Courtner, C.E.; Shah, N.; Tran, H.T.; Van Sciver, R.E.; et al. Exosomal transfer from human renal proximal tubule cells to distal tubule and collecting duct cells. Clin. Biochem. 2014, 47, 89–94. [Google Scholar] [CrossRef]
- Mossberg, M.; Ståhl, A.L.; Kahn, R.; Kristoffersson, A.C.; Tati, R.; Heijl, C.; Segelmark, M.; Leeb-Lundberg, L.M.F.; Karpman, D. C1-inhibitor decreases the release of vasculitis-like chemotactic endothelial microvesicles. J. Am. Soc. Nephrol. 2017, 28, 2472–2481. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating extracellular vesicles in human disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Erozenci, L.A.; Piersma, S.R.; Pham, T.V.; Bijnsdorp, I.V.; Jimenez, C.R. Longitudinal stability of urinary extracellular vesicle protein patterns within and between individuals. Sci. Rep. 2021, 11, 15629. [Google Scholar] [CrossRef]
- Gracia, T.; Wang, X.; Su, Y.; Norgett, E.E.; Williams, T.L.; Moreno, P.; Micklem, G.; Karet Frankl, F.E. Urinary exosomes contain microRNAs capable of paracrine modulation of tubular transporters in kidney. Sci. Rep. 2017, 7, 40601. [Google Scholar] [CrossRef]
- Ben-Dov, I.Z.; Whalen, V.M.; Goilav, B.; Max, K.E.; Tuschl, T. Cell and microvesicle urine microRNA deep sequencing profiles from healthy individuals: Observations with potential impact on biomarker studies. PLoS ONE 2016, 11, e0147249. [Google Scholar] [CrossRef]
- Sur, D.; Cainap, C.; Burz, C.; Havasi, A.; Chis, I.C.; Vlad, C.; Milosevic, V.; Balacescu, O.; Irimie, A. The role of miRNA -31-3p and miR-31-5p in the anti-EGFR treatment efficacy of wild-type K-RAS metastatic colorectal cancer. Is it really the next best thing in miRNAs? J. BUON 2019, 24, 1739–1746. [Google Scholar] [PubMed]
- Uil, M.; Hau, C.M.; Ahdi, M.; Mills, J.D.; Kers, J.; Saleem, M.A.; Florquin, S.; Gerdes, V.E.A.; Nieuwland, R.; Roelofs, J.J.T.H. Cellular origin and microRNA profiles of circulating extracellular vesicles in different stages of diabetic nephropathy. Clin. Kidney J. 2019, 14, 358–365. [Google Scholar] [CrossRef]
- Tsuji, K.; Kitamura, S.; Wada, J. MicroRNAs as biomarkers for nephrotic syndrome. Int. J. Mol. Sci. 2020, 22, 88. [Google Scholar] [CrossRef]
- Szeto, C.C. Urine miRNA in nephrotic syndrome. Clin. Chim. Acta 2014, 436, 308–313. [Google Scholar] [CrossRef]
- Chen, T.; Wang, C.; Yu, H.; Ding, M.; Zhang, C.; Lu, X.; Zhang, C.Y.; Zhang, C. Increased urinary exosomal microRNAs in children with idiopathic nephrotic syndrome. EBioMedicine 2019, 39, 552–561. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, C.; Chen, H.; Li, L.; Tu, Y.; Liu, C.; Shi, S.; Zen, K.; Liu, Z. Evaluation of microRNAs miR-196a, miR-30a-5P, and miR-490 as biomarkers of disease activity among patients with FSGS. Clin. J. Am. Soc. Nephrol. 2014, 9, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.Y.; Cai, G.Y.; Li, J.J.; Bu, R.; Chen, X.M. Urinary erythrocyte-derived miRNAs: Emerging role in IgA nephropathy. Kidney Blood Press. Res. 2017, 42, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Magayr, T.A.; Song, X.; Streets, A.J.; Vergoz, L.; Chang, L.; Valluru, M.K.; Yap, H.L.; Lannoy, M.; Haghighi, A.; Simms, R.J.; et al. Global microRNA profiling in human urinary exosomes reveals novel disease biomarkers and cellular pathways for autosomal dominant polycystic kidney disease. Kidney Int. 2020, 98, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Ghai, V.; Wu, X.; Bheda-Malge, A.; Argyropoulos, C.P.; Bernardo, J.F.; Orchard, T.; Galas, D.; Wang, K. Genome-wide profiling of urinary extracellular vesicle microRNAs associated with diabetic nephropathy in type 1 diabetes. Kidney Int. Rep. 2017, 3, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Shihana, F.; Wong, W.K.M.; Joglekar, M.V.; Mohamed, F.; Gawarammana, I.B.; Isbister, G.K.; Hardikar, A.A.; Seth, D.; Buckley, N.A. Urinary microRNAs as non-invasive biomarkers for toxic acute kidney injury in humans. Sci. Rep. 2021, 11, 9165. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Ding, X.; Zheng, J.; Wang, B.; Li, Y.; Xiang, H.; Dou, M.; Qiao, Y.; Tian, P.; Xue, W. miR-182-5p and miR-378a-3p regulate ferroptosis in I/R-induced renal injury. Cell Death Dis. 2020, 11, 929. [Google Scholar] [CrossRef]
- Wilflingseder, J.; Regele, H.; Perco, P.; Kainz, A.; Soleiman, A.; Mühlbacher, F.; Mayer, B.; Oberbauer, R. miRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation 2013, 95, 835–841. [Google Scholar] [CrossRef]
- Wilflingseder, J.; Jelencsics, K.; Bergmeister, H.; Sunzenauer, J.; Regele, H.; Eskandary, F.; Reindl-Schwaighofer, R.; Kainz, A.; Oberbauer, R. miR-182-5p inhibition ameliorates ischemic acute kidney injury. Am. J. Pathol. 2017, 187, 70–79. [Google Scholar] [CrossRef]
- Wang, X.; Wang, G.; Zhang, X.; Dou, Y.; Dong, Y.; Liu, D.; Xiao, J.; Zhao, Z. Inhibition of microRNA-182-5p contributes to attenuation of lupus nephritis via Foxo1 signaling. Exp. Cell Res. 2018, 373, 91–98. [Google Scholar] [CrossRef]
- Woo, Y.M.; Kim, D.Y.; Koo, N.J.; Kim, Y.M.; Lee, S.; Ko, J.Y.; Shin, Y.; Kim, B.H.; Mun, H.; Choi, S.; et al. Profiling of miRNAs and target genes related to cystogenesis in ADPKD mouse models. Sci. Rep. 2017, 7, 14151. [Google Scholar] [CrossRef]
- Pazourkova, E.; Pospisilova, S.; Svobodova, I.; Horinek, A.; Brisuda, A.; Soukup, V.; Hrbacek, J.; Capoun, O.; Mares, J.; Hanus, T.; et al. Comparison of microRNA content in plasma and urine indicates the existence of a transrenal passage of selected microRNAs. Adv. Exp. Med. Biol. 2016, 924, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Perego, S.; Sansoni, V.; Banfi, G.; Lombardi, G. Effect of collection matrix, platelet depletion, and storage conditions on plasma extracellular vesicles and extracellular vesicle-associated miRNAs measurements. Clin. Chem. Lab. Med. 2020, 59, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The non-canonical aspects of microRNAs: Many roads to gene regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Peng, C.H.; Lin, C.L.; Yang, C.W.; Shueh, S.; Huang, C.C. Vascular endothelial growth factor may provide additional values to C-reactive protein and anti-myeloperoxidase titer as a parameter for evaluating disease activity in anti-myeloperoxidase associated vasculitis. Ren. Fail. 2003, 25, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Antovic, A.; Svensson, E.; Lövström, B.; Illescas, V.B.; Nordin, A.; Börjesson, O.; Arnaud, L.; Bruchfeld, A.; Gunnarsson, I. Venous thromboembolism in antineutrophil cytoplasmic antibody-associated vasculitis: An underlying prothrombotic condition? Rheumatol. Adv. Pract. 2020, 4, rkaa056. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Feng, S.; Wu, Y.; Su, Y.; Jing, F.; Yi, Q. Serum levels of syndecan-1 in patients with Kawasaki disease. Pediatr. Infect. Dis. J. 2019, 38, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, Y.; Wu, Y.; Feng, M.; Zhao, X.; Gao, C.; Guo, H.; Luo, J. Double-negative T cells are absolutely elevated in patients with antineutrophil cytoplasmic autoantibody-associated vasculitis. Mol. Immunol. 2021, 132, 250–259. [Google Scholar] [CrossRef]
- Guan, Z.; VanBeusecum, J.P.; Inscho, E.W. Endothelin and the renal microcirculation. Semin. Nephrol. 2015, 35, 145–155. [Google Scholar] [CrossRef]
- Iwakiri, T.; Fujimoto, S.; Kitagawa, K.; Furuichi, K.; Yamahana, J.; Matsuura, Y.; Yamashita, A.; Uezono, S.; Shimao, Y.; Hisanaga, S.; et al. Validation of a newly proposed histopathological classification in Japanese patients with antineutrophil cytoplasmic antibody-associated glomerulonephritis. BMC Nephrol. 2013, 14, 125. [Google Scholar] [CrossRef][Green Version]
- Tse, W.Y.; Nash, G.B.; Hewins, P.; Savage, C.O.; Adu, D. ANCA-induced neutrophil F-actin polymerization: Implications for microvascular inflammation. Kidney Int. 2005, 67, 130–139. [Google Scholar] [CrossRef][Green Version]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef] [PubMed]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Bioinformatics & Evolutionary Genomics. Available online: https:bioinformatics.psb.ugent.be.webtools/Venn (accessed on 2 September 2021).
| miRNA | p-Value | FC (P/C) | SD (Controls) | SD (Patients) | Power |
|---|---|---|---|---|---|
| miR-26a-5p | 5.982 × 10−6 | 0.422 | 0.276 | 0.053 | 1.0000 |
| miR-192-5p | 5.417 × 10−4 | 0.312 | 0.546 | 0.099 | 1.0000 |
| miR-191-5p | 3.871 × 10−2 | 0.720 | 0.182 | 0.077 | 1.0000 |
| miR-31-3p | 2.439 × 10−4 | 2.933 | 0.166 | 0.232 | 1.0000 |
| miR-106b-5p | 4.253 × 10−4 | 2.841 | 0.124 | 0.214 | 1.0000 |
| miR-99a-5p | 4.257 × 10−3 | 4.329 | 0.161 | 0.282 | 1.0000 |
| miR-30a-5p | 9.190 × 10−3 | 2.551 | 0.154 | 0.172 | 1.0000 |
| miR-182-5p | 1.348× 10−2 | 3.175 | 0.190 | 0.171 | 1.0000 |
| miR-24-3p | 2.075 × 10−2 | 1.412 | 0.071 | 0.073 | 1.0000 |
| miR-200a-3p | 3.871 × 10−2 | 1.422 | 0.095 | 0.171 | 0.9999 |
| Parameters | NGS Initial Phase | RT-PCR Confirmation Phase | ||
|---|---|---|---|---|
| AVV Patients n = 10 | Healthy Controls n = 10 | AVV Patients n = 24 | Healthy Controls n = 16 | |
| Sex (Male/Female) | 6/4 | 6/4 | 12/12 | 8/8 |
| Age (median, range, years) | 65.5 (37–74) | 55 (42–74) | 63.5 (21–78) | 57 (42–74) |
| S-creatinine (median, range, μmol/L) | 241.5 (113.1–438.3) | 79.1 (64.4–86.1) | 302.0 (52.3–480.1) | 82.2 (64.4–91.3) |
| Proteinuria (median, range, g/24 h) | 1.62 (0.21–3.60) | 0.05 (0.03–0.13) | 1.44 (0.11–3.60) | 0.05 (0.03–0.13) |
| Hemoglobin (median, range, g/L) | 91.0 (79–142) | n/a | 98.5 (77–142) | n/a |
| C-reactive protein (median, range, mg/L) | 103.5 (1.1–153.0) | n/a | 12.0 (1.1–153.0) | n/a |
| Organ involvement (kidney/lung/ENT/eye/skin) | 10/3/0/1/0 | n/a | 24/9/4/1/2 | n/a |
| ANCA-subtypes (PR3/MPO/neg.) | 3/7 | n/a | 7/15/2 | n/a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frydlova, J.; Zednikova, I.; Satrapova, V.; Pazourkova, E.; Santorova, S.; Hruskova, Z.; Tesar, V.; Vokurka, M.; Prikryl, P.; Korabecna, M. Analysis of microRNAs in Small Urinary Extracellular Vesicles and Their Potential Roles in Pathogenesis of Renal ANCA-Associated Vasculitis. Int. J. Mol. Sci. 2022, 23, 4344. https://doi.org/10.3390/ijms23084344
Frydlova J, Zednikova I, Satrapova V, Pazourkova E, Santorova S, Hruskova Z, Tesar V, Vokurka M, Prikryl P, Korabecna M. Analysis of microRNAs in Small Urinary Extracellular Vesicles and Their Potential Roles in Pathogenesis of Renal ANCA-Associated Vasculitis. International Journal of Molecular Sciences. 2022; 23(8):4344. https://doi.org/10.3390/ijms23084344
Chicago/Turabian StyleFrydlova, Jana, Iveta Zednikova, Veronika Satrapova, Eva Pazourkova, Sarka Santorova, Zdenka Hruskova, Vladimir Tesar, Martin Vokurka, Petr Prikryl, and Marie Korabecna. 2022. "Analysis of microRNAs in Small Urinary Extracellular Vesicles and Their Potential Roles in Pathogenesis of Renal ANCA-Associated Vasculitis" International Journal of Molecular Sciences 23, no. 8: 4344. https://doi.org/10.3390/ijms23084344
APA StyleFrydlova, J., Zednikova, I., Satrapova, V., Pazourkova, E., Santorova, S., Hruskova, Z., Tesar, V., Vokurka, M., Prikryl, P., & Korabecna, M. (2022). Analysis of microRNAs in Small Urinary Extracellular Vesicles and Their Potential Roles in Pathogenesis of Renal ANCA-Associated Vasculitis. International Journal of Molecular Sciences, 23(8), 4344. https://doi.org/10.3390/ijms23084344







