Functional Dissection of P1 Bacteriophage Holin-like Proteins Reveals the Biological Sense of P1 Lytic System Complexity
Abstract
1. Introduction
2. Results
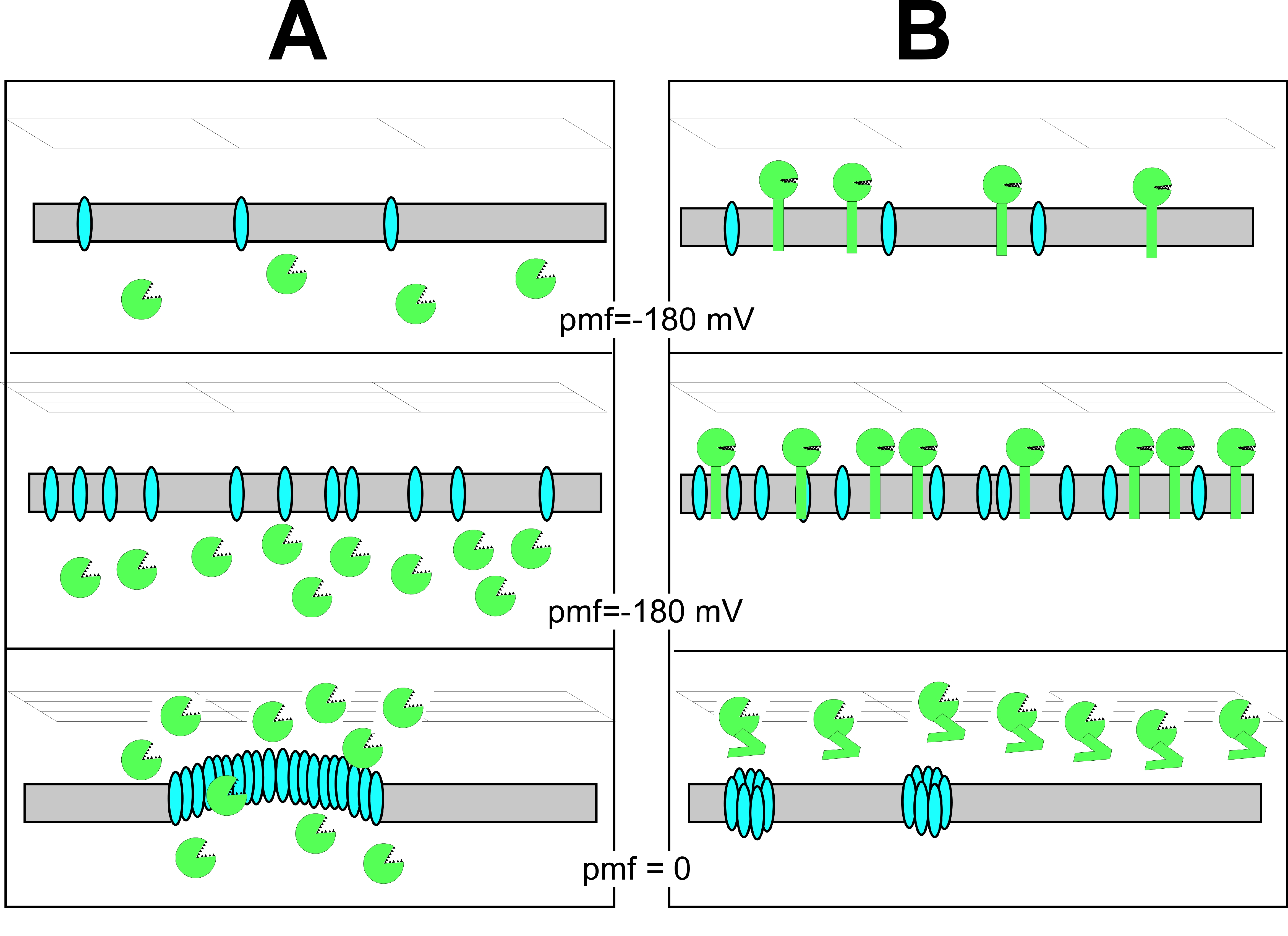
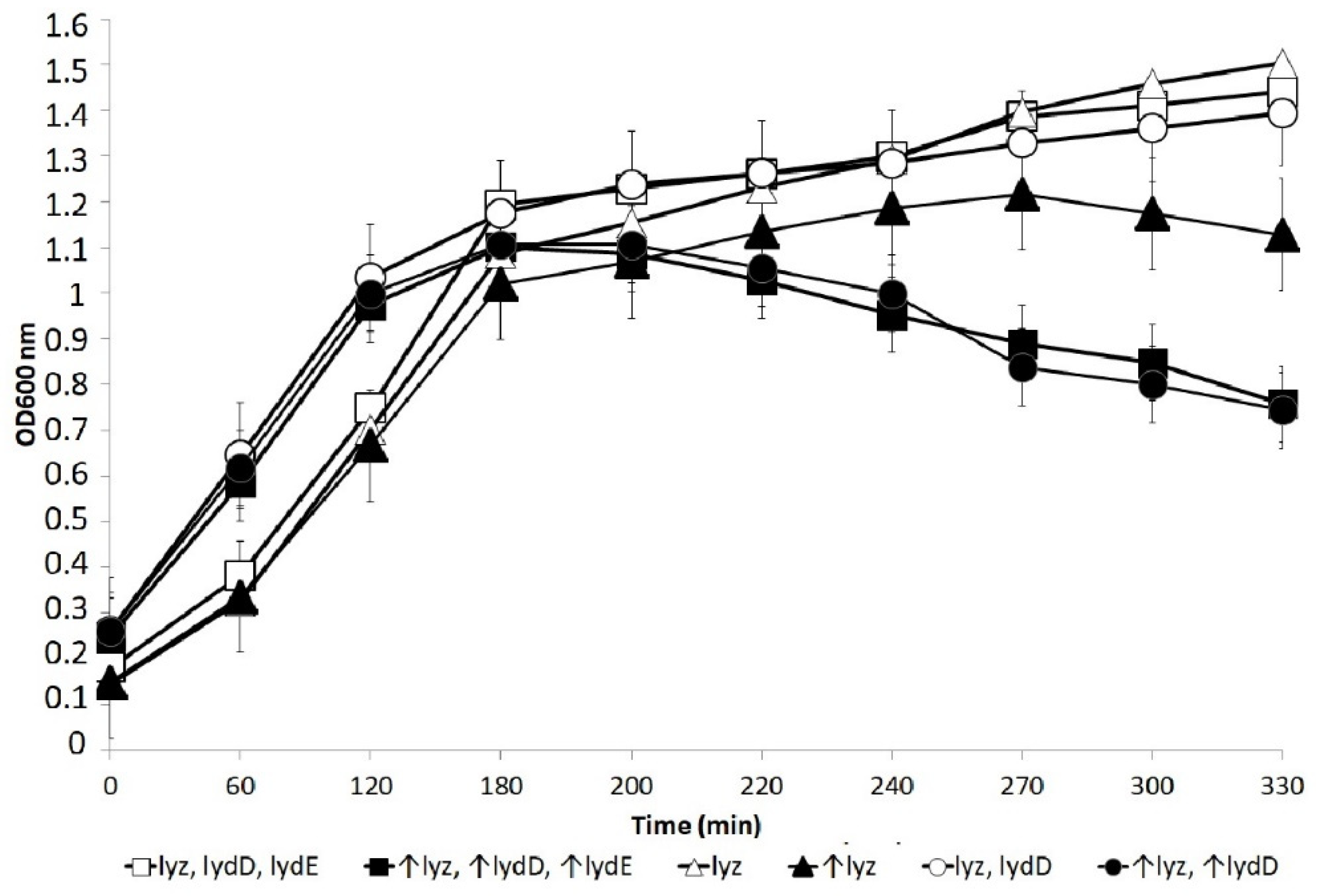
2.1. Influence of LydD on the Phenotype of Cells Producing the P1 Lyz Endolysin
2.2. Lethal Effect of Cloned lydC or lydA on E. coli Cells
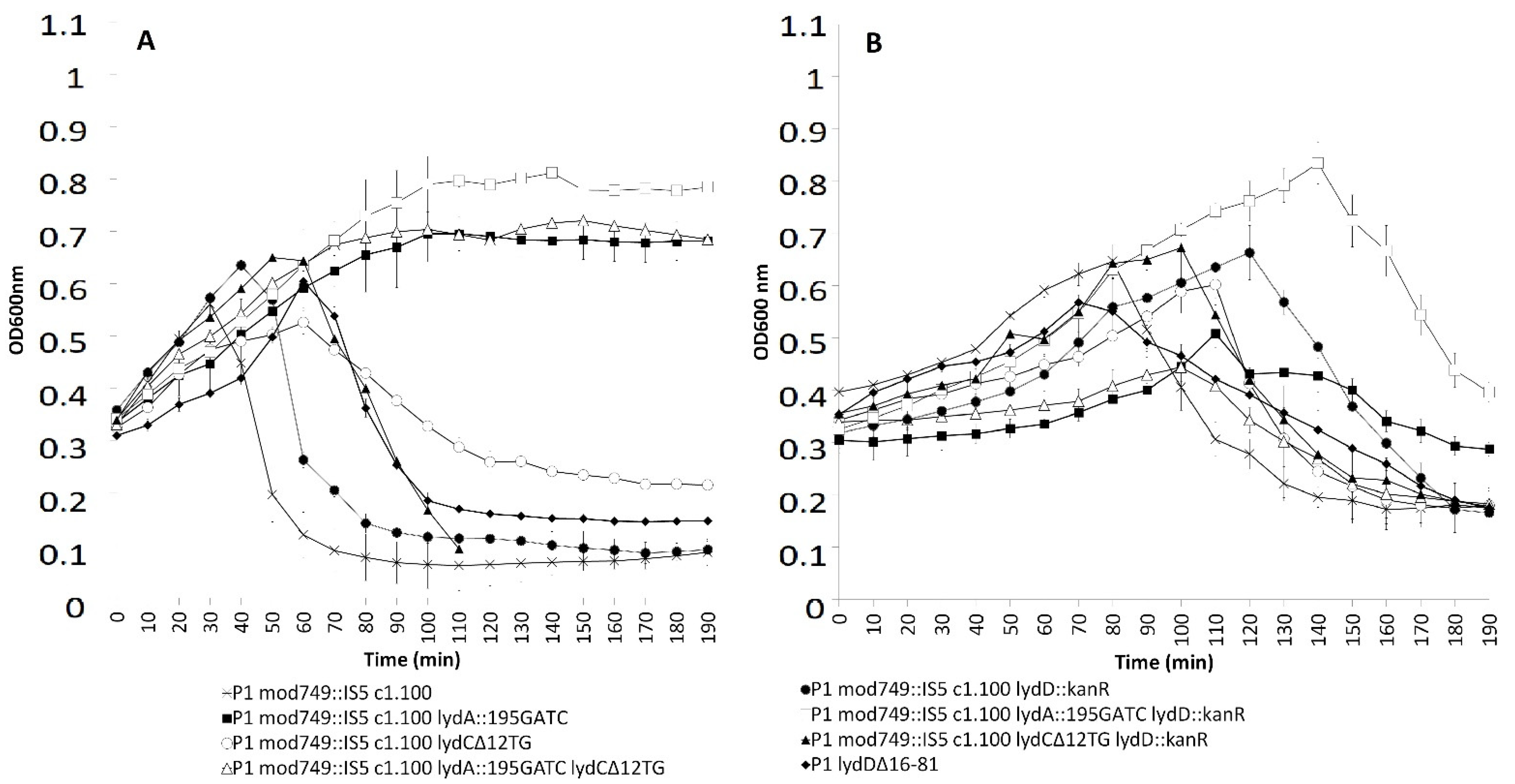
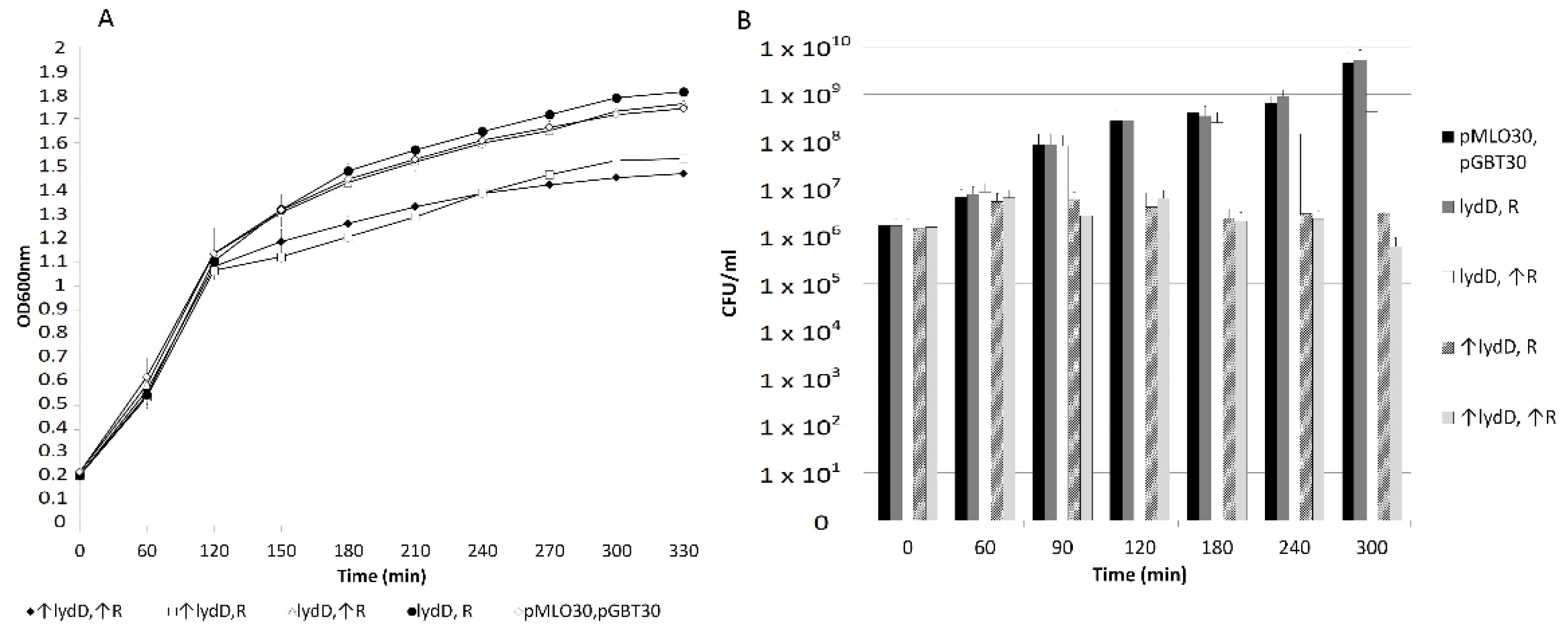
2.3. Effects of LydA, LydC, or LydD Depletion on P1-Mediated Cell Lysis
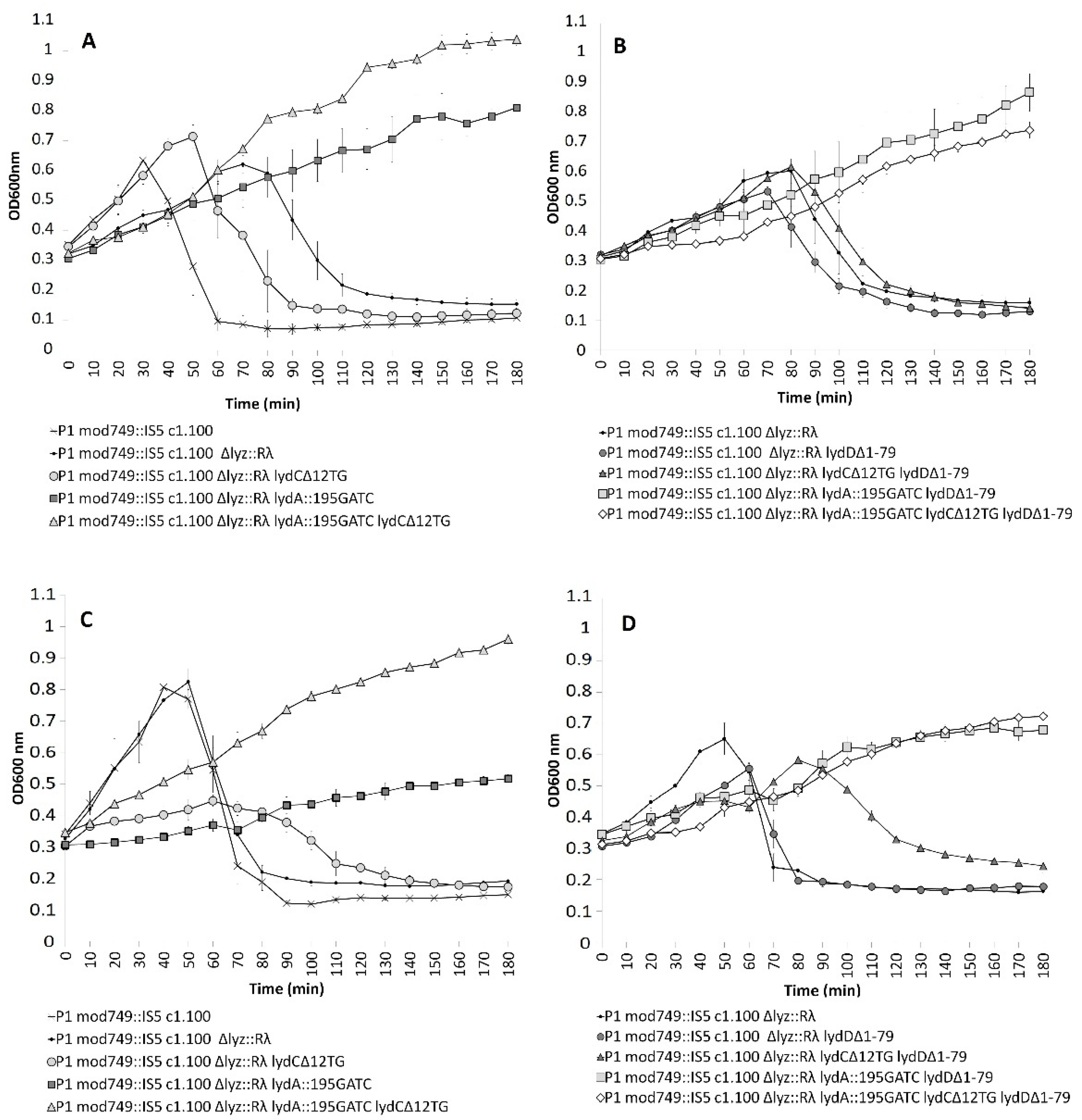
2.4. Influence of the P1 lyz Gene Replacement with the λ R Gene on P1-Mediated Cell Lysis
2.5. Influence of LydD on E. coli Cells Producing R Endolysin
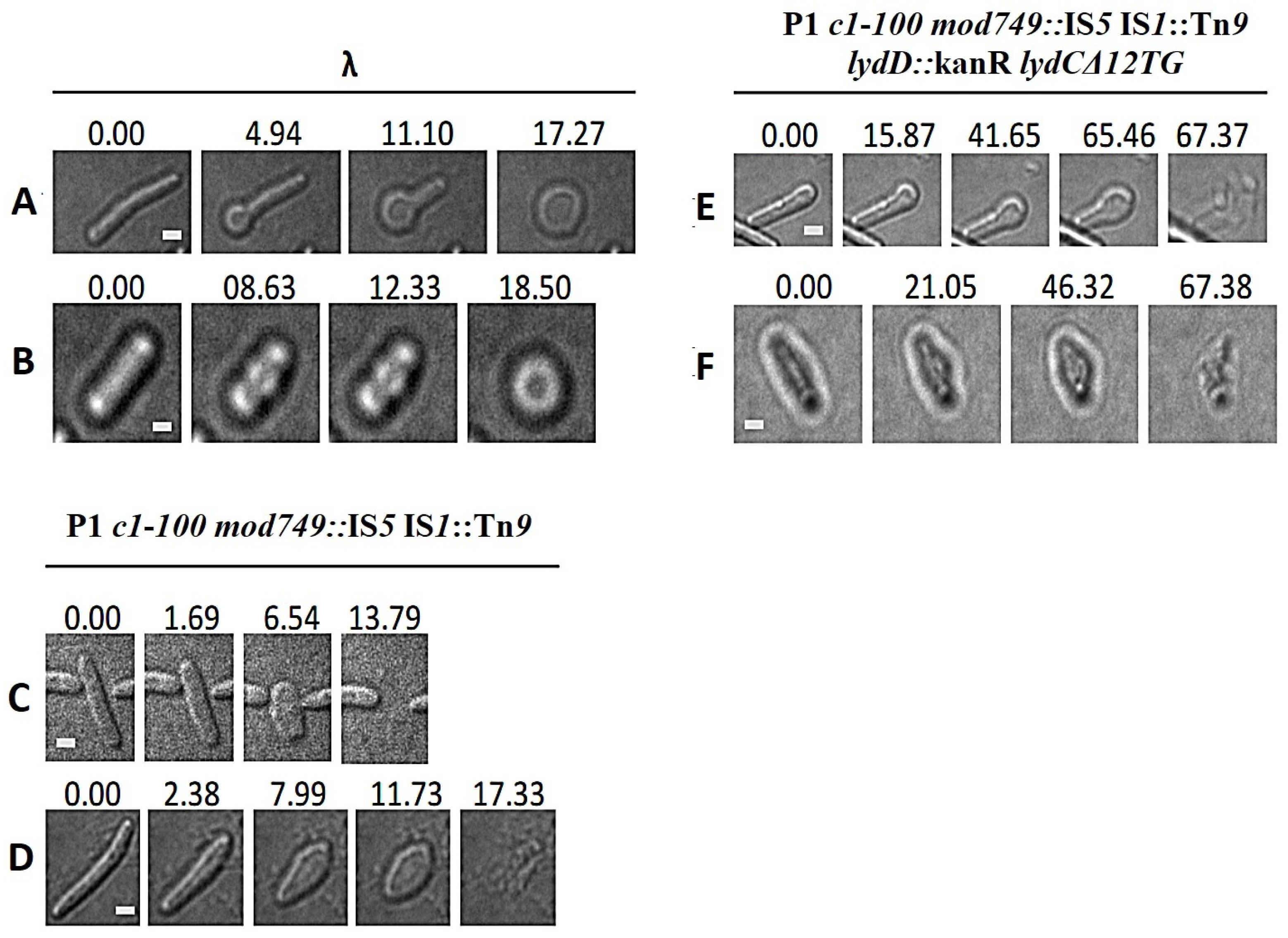
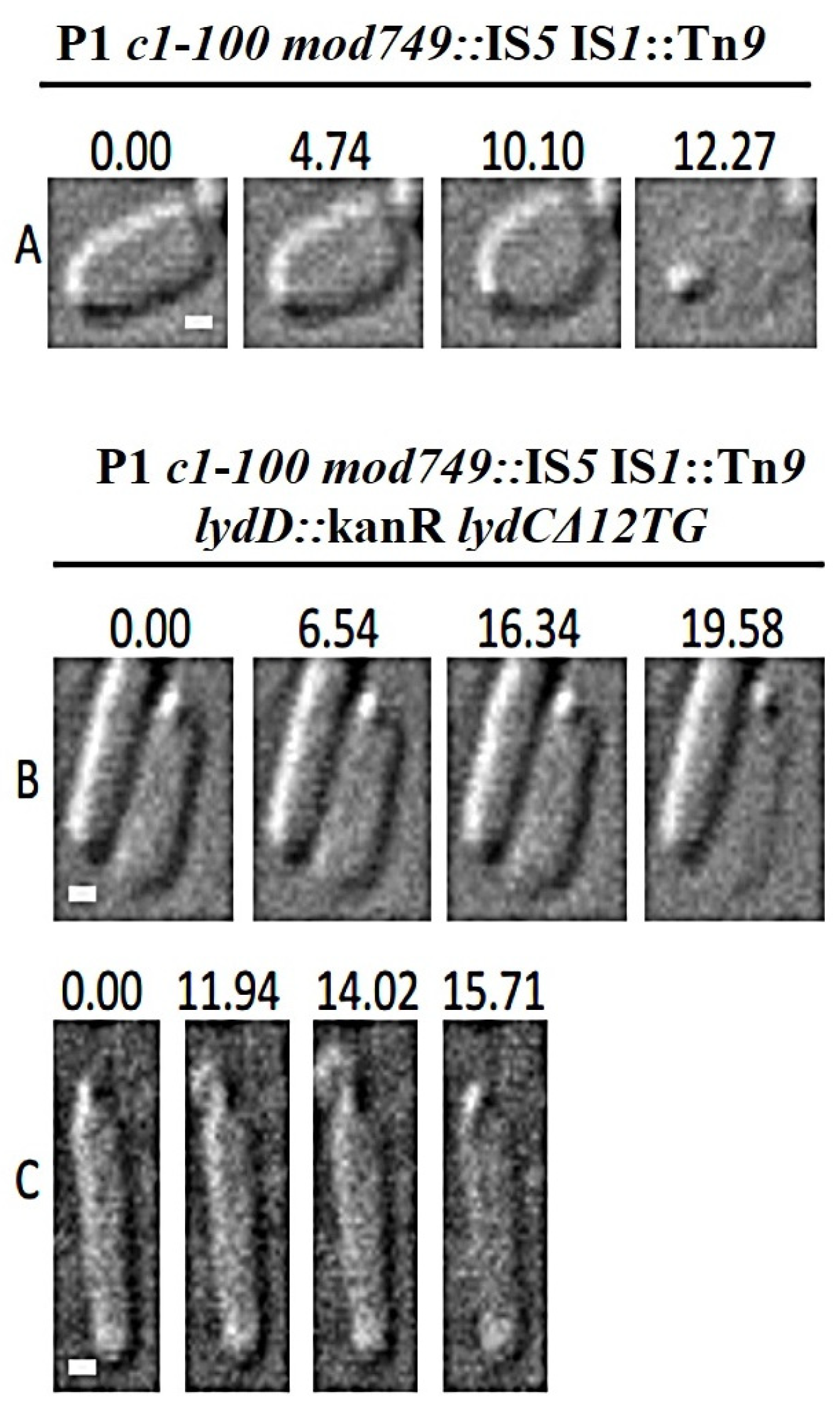
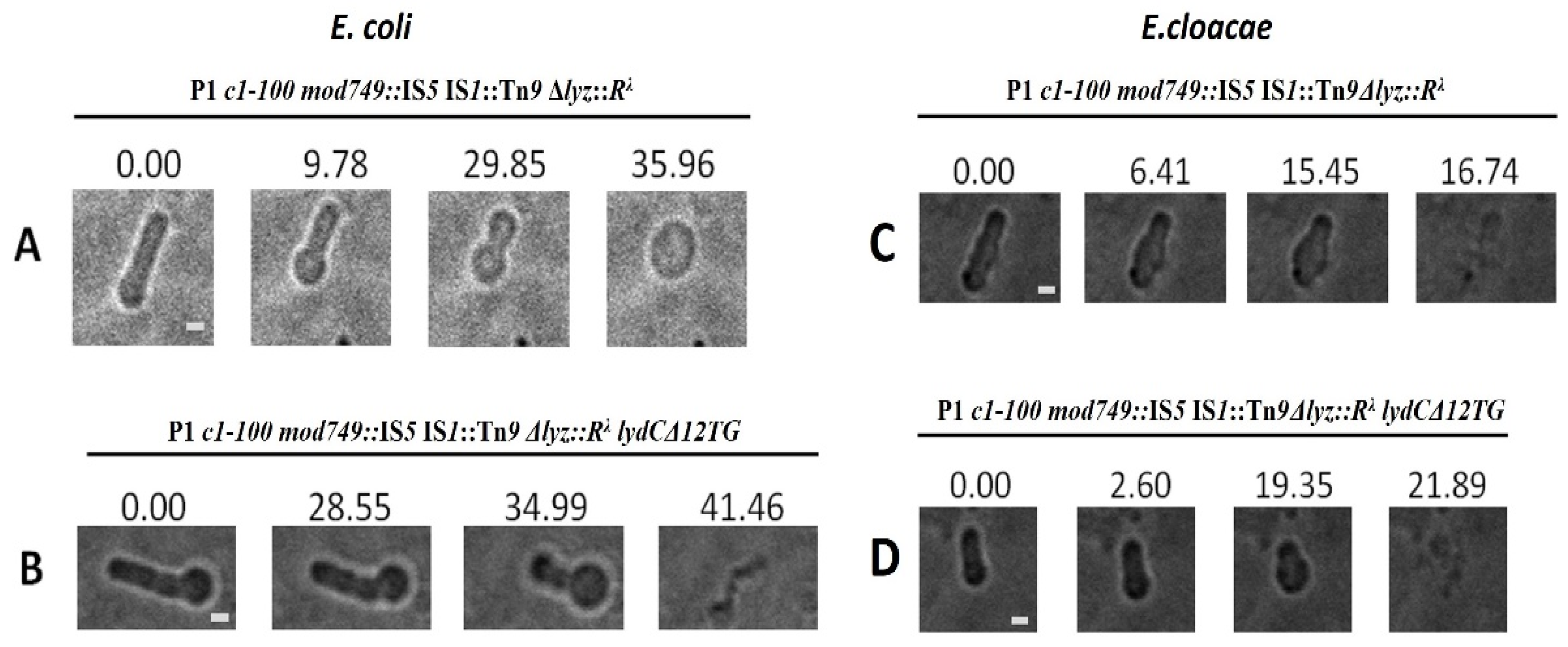
2.6. Morphological Transitions during E. coli and E. cloacae Cell Lysis Mediated by P1 or Its Mutants
2.7. Influence of P1 Lyz Gene Replacement with λ R on Cell Morphological Transitions during Lysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Bacteriophages, and Plasmids
4.2. Bacterial Growth Conditions and Bacteriophage Propagation
4.3. DNA Manipulation
4.4. Lysogenization
4.5. Construction of P1 Phage Mutants
4.6. Assays of Lysis Kinetics
4.7. Time-Lapse Microscopy of Living Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The Protein Clocks of Bacteriophage Infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.-N.; Deaton, J.; Young, R. Sizing the Holin Lesion with an Endolysin-β-Galactosidase Fusion. J. Bacteriol. 2003, 185, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Melo, L.D.R.; Santos, S.B.; Nobrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular Aspects and Comparative Genomics of Bacteriophage Endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Dewey, J.S.; Savva, C.G.; White, R.L.; Vitha, S.; Holzenburg, A.; Young, R. Micron-Scale Holes Terminate the Phage Infection Cycle. Proc. Natl. Acad. Sci. USA 2010, 107, 2219–2223. [Google Scholar] [CrossRef]
- Xu, M.; Struck, D.K.; Deaton, J.; Wang, I.-N.; Young, R. A Signal-Arrest-Release Sequence Mediates Export and Control of the Phage P1 Endolysin. Proc. Natl. Acad. Sci. USA 2004, 101, 6415–6420. [Google Scholar] [CrossRef]
- Kuty, G.F.; Xu, M.; Struck, D.K.; Summer, E.J.; Young, R. Regulation of a Phage Endolysin by Disulfide Caging. J. Bacteriol. 2010, 192, 5682–5687. [Google Scholar] [CrossRef]
- São-José, C.; Parreira, R.; Vieira, G.; Santos, M.A. The N-Terminal Region of the Oenococcus Oeni Bacteriophage FOg44 Lysin Behaves as a Bona Fide Signal Peptide in Escherichia Coli and as a Cis -Inhibitory Element, Preventing Lytic Activity on Oenococcal Cells. J. Bacteriol. 2000, 182, 5823–5831. [Google Scholar] [CrossRef]
- Zampara, A.; Ahern, S.J.; Briers, Y.; Brøndsted, L.; Sørensen, M.C.H. Two Distinct Modes of Lysis Regulation in Campylobacter Fletchervirus and Firehammervirus Phages. Viruses 2020, 12, 1247. [Google Scholar] [CrossRef]
- Cahill, J.; Young, R. Phage Lysis: Multiple Genes for Multiple Barriers. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2019; Volume 103, pp. 33–70. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Gontijo, M.T.P.; Vidigal, P.M.P.; Lopez, M.E.S.; Brocchi, M. Bacteriophages That Infect Gram-Negative Bacteria as Source of Signal-Arrest-Release Motif Lysins. Res. Microbiol. 2021, 172, 103794. [Google Scholar] [CrossRef]
- Briers, Y.; Peeters, L.M.; Volckaert, G.; Lavigne, R. The Lysis Cassette of Bacteriophage ΦKMV Encodes a Signal-Arrest-Release Endolysin and a Pinholin. Bacteriophage 2011, 1, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk-Popławska, M.; Tracz-Gaszewska, Z.; Lasota, P.; Kwiatek, A.; Piekarowicz, A. Haemophilus Influenzae HP1 Bacteriophage Encodes a Lytic Cassette with a Pinholin and a Signal-Arrest-Release Endolysin. Int. J. Mol. Sci. 2020, 21, 4013. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Born, Y.; Lucid, A.; Loessner, M.J.; Fieseler, L.; Coffey, A. Erwinia Amylovora Phage VB_EamM_Y3 Represents Another Lineage of Hairy Myoviridae. Res. Microbiol. 2018, 169, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Lucid, A.; Neve, H.; Franz, C.; O’Mahony, J.; Turner, D.; Lavigne, R.; Coffey, A. Pectobacterium Atrosepticum Phage VB_PatP_CB5: A Member of the Proposed Genus ‘Phimunavirus’. Viruses 2018, 10, 394. [Google Scholar] [CrossRef]
- Cahil, E.J.; Enderle, C.J.; Ahern, S.J.; Gill, J.J.; Torres, C.P.; Appel, D.N.; Black, M.C.; Young, R.; Gonzalez, C.F. Genomic and Biological Analysis of Phage Xfas53 and Related Prophages of Xylella fastidiosa. J. Bacteriol. 2010, 192, 179–190. [Google Scholar] [CrossRef]
- Savva, C.G.; Dewey, J.S.; Moussa, S.H.; To, K.H.; Holzenburg, A.; Young, R. Stable Micron-Scale Holes Are a General Feature of Canonical Holins: Canonical Holins Form Micron-Scale Holes. Mol. Microbiol. 2014, 91, 57–65. [Google Scholar] [CrossRef]
- Pang, T.; Savva, C.G.; Fleming, K.G.; Struck, D.K.; Young, R. Structure of the Lethal Phage Pinhole. Proc. Natl. Acad. Sci. USA 2009, 106, 18966–18971. [Google Scholar] [CrossRef]
- Park, T.; Struck, D.K.; Dankenbring, C.A.; Young, R. The Pinholin of Lambdoid Phage 21: Control of Lysis by Membrane Depolarization. J. Bacteriol. 2007, 189, 9135–9139. [Google Scholar] [CrossRef]
- Xu, M.; Arulandu, A.; Struck, D.K.; Swanson, S.; Sacchettini, J.C.; Young, R. Disulfide Isomerization After Membrane Release of Its SAR Domain Activates P1 Lysozyme. Science 2005, 307, 113–117. [Google Scholar] [CrossRef]
- Sun, Q.; Kuty, G.F.; Arockiasamy, A.; Xu, M.; Young, R.; Sacchettini, J.C. Regulation of a Muralytic Enzyme by Dynamic Membrane Topology. Nat. Struct. Mol. Biol. 2009, 16, 1192–1194. [Google Scholar] [CrossRef]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Gründling, A.; Manson, M.D.; Young, R. Holins Kill without Warning. Proc. Natl. Acad. Sci. USA 2001, 98, 9348–9352. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; São-José, C. More than a Hole: The Holin Lethal Function May Be Required to Fully Sensitize Bacteria to the Lytic Action of Canonical Endolysins: Holin Function and Endolysin Activity. Mol. Microbiol. 2016, 102, 92–106. [Google Scholar] [CrossRef]
- White, R.; Chiba, S.; Pang, T.; Dewey, J.S.; Savva, C.G.; Holzenburg, A.; Pogliano, K.; Young, R. Holin Triggering in Real Time. Proc. Natl. Acad. Sci. USA 2011, 108, 798–803. [Google Scholar] [CrossRef]
- Young, R. Phage Lysis: Do We Have the Hole Story Yet? Curr. Opin. Microbiol. 2013, 16, 790–797. [Google Scholar] [CrossRef]
- Summer, E.J.; Berry, J.; Tran, T.A.T.; Niu, L.; Struck, D.K.; Young, R. Rz/Rz1 Lysis Gene Equivalents in Phages of Gram-Negative Hosts. J. Mol. Biol. 2007, 373, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Summer, E.J.; Struck, D.K.; Young, R. The Final Step in the Phage Infection Cycle: The Rz and Rz1 Lysis Proteins Link the Inner and Outer Membranes. Mol. Microbiol. 2008, 70, 341–351. [Google Scholar] [CrossRef]
- Berry, J.; Rajaure, M.; Pang, T.; Young, R. The Spanin Complex Is Essential for Lambda Lysis. J. Bacteriol. 2012, 194, 5667–5674. [Google Scholar] [CrossRef]
- Rajaure, M.; Berry, J.; Kongari, R.; Cahill, J.; Young, R. Membrane fusion during phage lysis. Proc. Natl. Acad. Sci. USA 2015, 112, 5497–5502. [Google Scholar] [CrossRef]
- Guo, T.; Xin, Y.; Zhang, C.; Kong, J. A Cytoplasmic Antiholin Is Embedded in Frame with the Holin in a Lactobacillus Fermentum Bacteriophage. Appl. Environ. Microbiol. 2018, 84, e02518-17. [Google Scholar] [CrossRef]
- Ziermann, R.; Bartlett, B.; Calendar, R.; Christie, G.E. Functions Involved in Bacteriophage P2-Induced Host Cell Lysis and Identification of a New Tail Gene. J. Bacteriol. 1994, 176, 4974–4984. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, C.Y.; Nam, K.; Young, R. S Gene Expression and the Timing of Lysis by Bacteriophage Lambda. J. Bacteriol. 1995, 177, 3283–3294. [Google Scholar] [CrossRef] [PubMed]
- Pontarollo, R.A.; Rioux, C.R.; Potter, A.A. Cloning and Characterization of Bacteriophage-like DNA from Haemophilus Somnus Homologous to Phages P2 and HP1. J. Bacteriol. 1997, 179, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- Takáč, M.; Witte, A.; Bläsi, U. Functional Analysis of the Lysis Genes of Staphylococcus Aureus Phage P68 in Escherichia coli. Microbiology 2005, 151, 2331–2342. [Google Scholar] [CrossRef] [PubMed]
- Srividhya, K.V.; Krishnaswamy, S. Sub Classification and Targeted Characterization of Prophage-Encoded Two-Component Cell Lysis Cassette. J. Biosci. 2007, 32, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Kongari, R.; Rajaure, M.; Cahill, J.; Rasche, E.; Mijalis, E.; Berry, J.; Young, R. Phage spanins: Diversity, topological dynamics and gene co vergence. BMC Bioinform. 2018, 19, 326. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage Lysis: Mechanism and Regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Yarmolinsky, M.B.; Sternberg, N. Bacteriophage P1. In The Bacteriophages; Calendar, R., Ed.; (The Viruses); Springer: Boston, MA, USA, 1988; pp. 291–438. [Google Scholar] [CrossRef]
- Łobocka, M.B.; Rose, D.J.; Plunkett, G.; Rusin, M.; Samojedny, A.; Lehnherr, H.; Yarmolinsky, M.B.; Blattner, F.R. Genome of Bacteriophage P1. J. Bacteriol. 2004, 186, 7032–7068. [Google Scholar] [CrossRef]
- Giermasińska, K.; Łobocka, M. Interakcje bakteriofaga P1 z komórkami wybranych patogenów roślin z rodzaju Erwinia i rodzajów pokrewnych. In W: Wybrane Zagadnienia z Zakresu Chemii, Biologii i Fizyki; Zdunek, B., Olszówka, M., Eds.; Wydawnictwo Naukowe Tygiel: Lublin, Poland, 2016; pp. 48–67. [Google Scholar]
- Schmidt, C.; Velleman, M.; Arber, W. Three Functions of Bacteriophage P1 Involved in Cell Lysis. J. Bacteriol. 1996, 178, 1099–1104. [Google Scholar] [CrossRef]
- Walker, J.T.; Walker, D.H. Mutations in Coliphage P1 Affecting Host Cell Lysis. J. Virol. 1980, 35, 519–530. [Google Scholar] [CrossRef]
- Walker, J.T.; Walker, D.H. Coliphage P1 Morphogenesis: Analysis of Mutants by Electron Microscopy. J. Virol. 1983, 45, 1118–1139. [Google Scholar] [CrossRef] [PubMed]
- Guidolin, A.; Zingg, J.M.; Lehnherr, H.; Arber, W. Bacteriophage P1 tail-fibre and dar operons are expressed from homologous phage-specific late promoter sequences. J. Mol. Biol. 1989, 208, 615–622. [Google Scholar] [CrossRef]
- Iida, S.; Hiestand-Nauer, R.; Sandmeier, H.; Lehnherr, H.; Arber, W. Accessory Genes in the darA Operon of Bacteriophage P1 Affect Antirestriction Function, Generalized Transduction, Head Morphogenesis, and Host Cell Lysis. Virology 1998, 251, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Iida, S.; Streiff, M.B.; Bickle, T.A.; Arber, W. Two DNA Antirestriction Systems of Bacteriophage P1, DarA, and DarB: Characterization of DarA− Phages. Virology 1987, 157, 156–166. [Google Scholar] [CrossRef]
- Piya, D.; Vara, L.; Russell, W.K.; Young, R.; Gill, J.J. The Multicomponent Antirestriction System of Phage P1 Is Linked to Capsid Morphogenesis: P1 Antirestriction Is Linked to Morphogenesis. Mol. Microbiol. 2017, 105, 399–412. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Probing the Function of the Two Holin-like Proteins of Bacteriophage SPP1. Virology 2017, 500, 184–189. [Google Scholar] [CrossRef]
- Halgasova, N.; Ugorcakova, J.; Gerova, M.; Timko, J.; Bukovska, G. Isolation and Characterization of Bacteriophage ΦBP from Paenibacillus Polymyxa CCM 7400. FEMS Microbiol. Lett. 2010, 305, 128–135. [Google Scholar] [CrossRef]
- Lu, Z.; Altermann, E.; Breidt, F.; Kozyavkin, S. Sequence Analysis of Leuconostoc Mesenteroides Bacteriophage Φ1-A4 Isolated from an Industrial Vegetable Fermentation. Appl. Environ. Microbiol. 2010, 76, 1955–1966. [Google Scholar] [CrossRef]
- Nakonieczna, A.; Rutyna, P.; Fedorowicz, M.; Kwiatek, M.; Mizak, L.; Łobocka, M. Three Novel Bacteriophages, J5a, F16Ba, and z1a, Specific for Bacillus anthracis, Define a New Clade of Historical Wbeta Phage Relatives. Viruses 2022, 14, 213. [Google Scholar] [CrossRef]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; São-José, C.; Pimentel, M. Diversity in Bacterial Lysis Systems: Bacteriophages Show the Way. FEMS Microbiol. Rev. 2013, 37, 554–571. [Google Scholar] [CrossRef]
- Delisle, A.L.; Barcak, G.J.; Guo, M. Isolation and Expression of the Lysis Genes of Actinomyces naeslundii Phage Av-1. Appl. Environ. Microbiol. 2006, 72, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Woźnica, W.M.; Bigos, J.; Łobocka, M.B. Lysis of bacterial cells in the process of bacteriophage release-canonical and newly discovered mechanisms. Postepy Hig. Med. Dosw. 2015, 69, 114–126. [Google Scholar]
- Park, T.; Struck, D.K.; Deaton, J.F.; Young, R. Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. USA 2006, 103, 19713–19718. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.; Park, T.; Young, R. Mapping the pinhole formation pathway of S21. Mol. Microbiol. 2010, 78, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef]
- Pang, T.; Park, T.; Young, R. Mutational analysis of the S21 pinholin. Mol. Microbiol. 2010, 76, 68–77. [Google Scholar] [CrossRef]
- Wang, I.N. Lysis timing and bacteriophage fitness. Genetics 2010, 172, 17–26. [Google Scholar] [CrossRef]
- Gallet, R.; Kannoly, S.; Wang, I.N. Effects of bacteriophage traits on plaque formation. BMC Microbiol. 2011, 11, 181. [Google Scholar] [CrossRef]
- Cornaglia, G.; Mazzariol, A.; Fontana, R.; Satta, G. Diffusion of Carbapenems Through the Outer Membrane of Enterobacteriaceae and Correlation of Their Activities with Their Periplasmic Concentrations. Microb. Drug Resist. 1996, 2, 273–276. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial Membrane Lipids: Diversity in Structures and Pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Gutiérrez, S.; Ferrara, L.; Pathania, M.; Masi, M.; Wang, J.; Bodrenko, I.; Zahn, M.; Winterhalter, M.; Stavenger, R.A.; Pagès, J.-M.; et al. Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins. ACS Infect. Dis. 2018, 4, 1487–1498. [Google Scholar] [CrossRef] [PubMed]
- Pomares, M.F.; Delgado, M.A.; Corbalán, N.S.; Farías, R.N.; Vincent, P.A. Sensitization of Microcin J25-Resistant Strains by a Membrane-Permeabilizing Peptide. Appl. Environ. Microbiol. 2010, 76, 6837–6842. [Google Scholar] [CrossRef]
- Neu, H.C.; Chou, J. Release of Surface Enzymes in Enterobacteriaceae by Osmotic Shock. J. Bacteriol. 1967, 94, 1934–1945. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, K.W.; Aspedon, A. Detergent-Shock Response in Enteric Bacteria. Mol. Microbiol. 1992, 6, 957–961. [Google Scholar] [CrossRef]
- Meyer, J.; Stålhammar-Carlemalm, M.; Streiff, M.; Iida, S.; Arber, W. Sequence Relations among the IncY Plasmid P15B, P1, and P7 Prophages. Plasmid 1986, 16, 81–89. [Google Scholar] [CrossRef]
- Yang, L.; Li, W.; Jiang, G.-Z.; Zhang, W.-H.; Ding, H.-Z.; Liu, Y.-H.; Zeng, Z.-L.; Jiang, H.-X. Characterization of a P1-like Bacteriophage Carrying CTX-M-27 in Salmonella Spp. Resistant to Third Generation Cephalosporins Isolated from Pork in China. Sci. Rep. 2017, 7, 40710. [Google Scholar] [CrossRef]
- Billard-Pomares, T.; Fouteau, S.; Jacquet, M.E.; Roche, D.; Barbe, V.; Castellanos, M.; Bouet, J.Y.; Cruveiller, S.; Médigue, C.; Blanco, J.; et al. Characterization of a P1-Like Bacteriophage Carrying an SHV-2 Extended-Spectrum β-Lactamase from an Escherichia Coli Strain. Antimicrob. Agents Chemother. 2014, 58, 6550–6557. [Google Scholar] [CrossRef]
- Hagbø, M.; Ravi, A.; Angell, I.L.; Sunde, M.; Ludvigsen, J.; Diep, D.B.; Foley, S.L.; Vento, M.; Collado, M.C.; Perez-Martinez, G.; et al. Experimental Support for Multidrug Resistance Transfer Potential in the Preterm Infant Gut Microbiota. Pediatr. Res. 2020, 88, 57–65. [Google Scholar] [CrossRef]
- Gottesman, M.E.; Yarmolinsky, M.B. Integration-Negative Mutants of Bacteriophage Lambda. J. Mol. Biol. 1968, 31, 487–505. [Google Scholar] [CrossRef]
- Łobocka, M.; Yarmolinsky, M. P1 Plasmid Partition: A Mutational Analysis of ParB. J. Mol. Biol. 1996, 259, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd. ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Adams, M.H. Bacteriophages; Interscience Publishers Inc.: New York, NY, USA, 1959. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 1. [Google Scholar]
- Helvoort, J.M.L.M.; Woldringh, C.L. Nucleoid Partitioning in Escherichia coli during Steady-State Growth and upon Recovery from Chloramphenicol Treatment. Mol. Microbiol. 1994, 13, 577–583. [Google Scholar] [CrossRef] [PubMed]

| Mutant of Bacteriophage P1 c1-100 mod749::IS5 IS1::Tn9 | P1 Gene(s) Inactivated [Gene Inserted] | Construction; Antibiotic Resistance Marker |
|---|---|---|
| lyz::kanR | lyz | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pWWO2 as the DNA donor; cmR, kanR |
| lydD::kanR | lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pAKI1 as the DNA donor; cmR, kanR |
| lydCΔ12TG | lydC | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pAKI2 as the DNA donor; cmR |
| lydA::195GATC | lydA | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pAKI3 as the DNA donor; cmR |
| lydD::kanRlydCΔ12TG | lydD, lydC | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydCΔ12_13TG as the recipient and plasmid pAKI1 as the DNA donor; cmR, kanR |
| lydD::kanRlydA::195GATC | lydA, lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydA::195_196GATC as the recipient and pAKI1 plasmid as the DNA donor; cmR, kanR |
| lydA::195GATC lydCΔ12TG | lydA, lydC | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydCΔ12_13TG as the recipient and plasmid pAKI3 as the DNA donor; cmR |
| Δlyz::Rλ | lyz [R] | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pAKI13 as the DNA donor; cmR |
| Δlyz::Rλ lydCΔ12TG | lyz [R], lydC | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydCΔ12_13TG as the recipient and plasmid pAKI13 as the DNA donor; cmR |
| Δlyz::Rλ lydA::195GATC lydCΔ12TG | lyz [R], lydA, lydC | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydA::195_196GATC lydCΔ12_13TG as the recipient and plasmid pAKI13 as the DNA donor; cmR |
| Δlyz::Rλ lydA::195GATC | lyz [R], lydA | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 Δlyz::Rλ as the recipient and plasmid pAKI3 as the DNA donor; cmR |
| Δlyz::Rλ lydDΔ1-79 * | lyz [R], lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pAKI23 as the DNA donor; cmR |
| Δlyz::Rλ lydA::195GATC lydDΔ1-79 * | lyz [R], lydA, lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydA::195_196GATC as the recipient and pAKI23 plasmid as the DNA donor; cmR, kanR |
| Δlyz::Rλ lydCΔ12TG lydDΔ1-79 * | lyz [R], lydC, lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydCΔ12_13TG as the recipient and plasmid pAKI23 as the DNA donor; cmR |
| Δlyz::Rλ lydA::195GATC lydCΔ12TG lydDΔ1-79 * | lyz [R], lydA, lydC, lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 lydA::195_196GATC lydCΔ12_13TG as the recipient and plasmid pAKI23 as the DNA donor; cmR |
| lydDΔ16-81 * | lydD | Recombinational replacement with the use of P1 c1-100 mod749::IS5 IS1::Tn9 as the recipient and plasmid pAKI26 as the DNA donor; cmR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bednarek, A.; Cena, A.; Izak, W.; Bigos, J.; Łobocka, M. Functional Dissection of P1 Bacteriophage Holin-like Proteins Reveals the Biological Sense of P1 Lytic System Complexity. Int. J. Mol. Sci. 2022, 23, 4231. https://doi.org/10.3390/ijms23084231
Bednarek A, Cena A, Izak W, Bigos J, Łobocka M. Functional Dissection of P1 Bacteriophage Holin-like Proteins Reveals the Biological Sense of P1 Lytic System Complexity. International Journal of Molecular Sciences. 2022; 23(8):4231. https://doi.org/10.3390/ijms23084231
Chicago/Turabian StyleBednarek, Agnieszka, Agata Cena, Wioleta Izak, Joanna Bigos, and Małgorzata Łobocka. 2022. "Functional Dissection of P1 Bacteriophage Holin-like Proteins Reveals the Biological Sense of P1 Lytic System Complexity" International Journal of Molecular Sciences 23, no. 8: 4231. https://doi.org/10.3390/ijms23084231
APA StyleBednarek, A., Cena, A., Izak, W., Bigos, J., & Łobocka, M. (2022). Functional Dissection of P1 Bacteriophage Holin-like Proteins Reveals the Biological Sense of P1 Lytic System Complexity. International Journal of Molecular Sciences, 23(8), 4231. https://doi.org/10.3390/ijms23084231






