Squalene through Its Post-Squalene Metabolites Is a Modulator of Hepatic Transcriptome in Rabbits
Abstract
:1. Introduction
2. Results
2.1. Body Weight and Hepatic Parameters
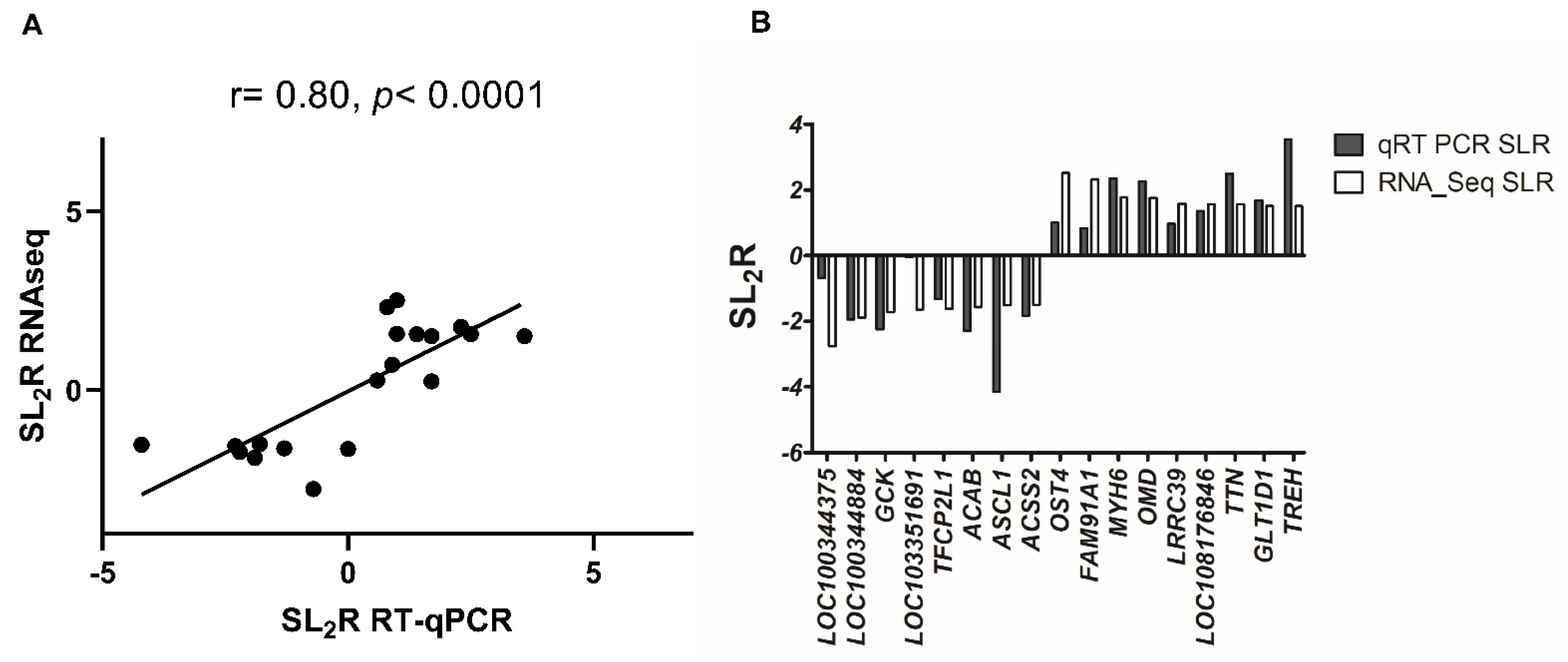
2.2. Hepatic Gene Expression
3. Discussion
4. Materials and Methods
4.1. Animal Models and Experimental Design
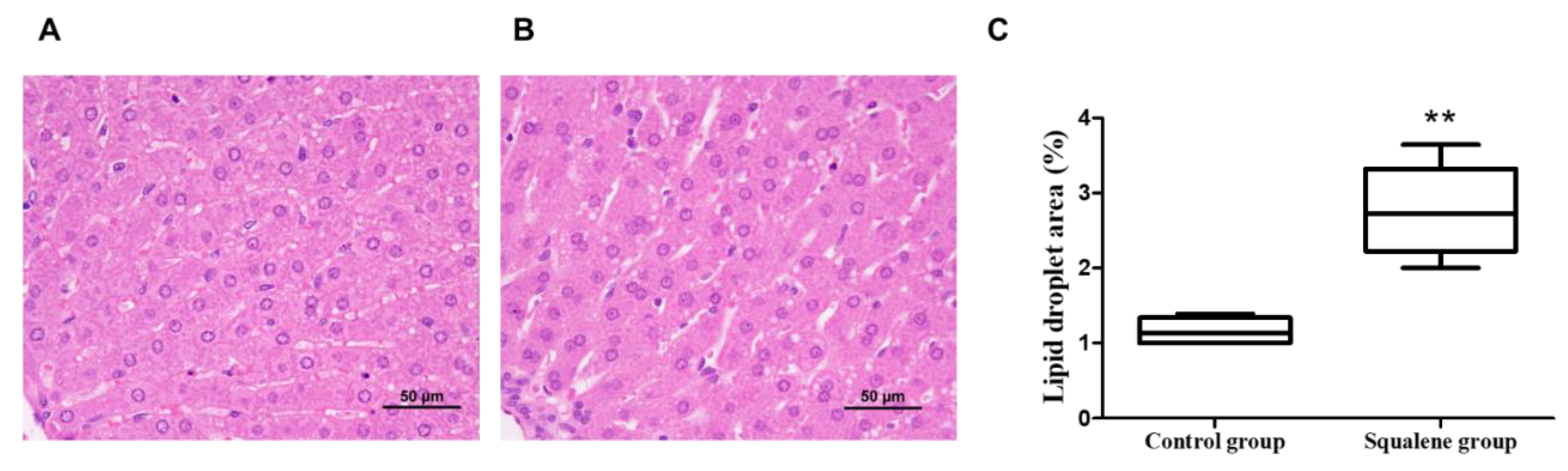
4.2. Liver Histology Analyses
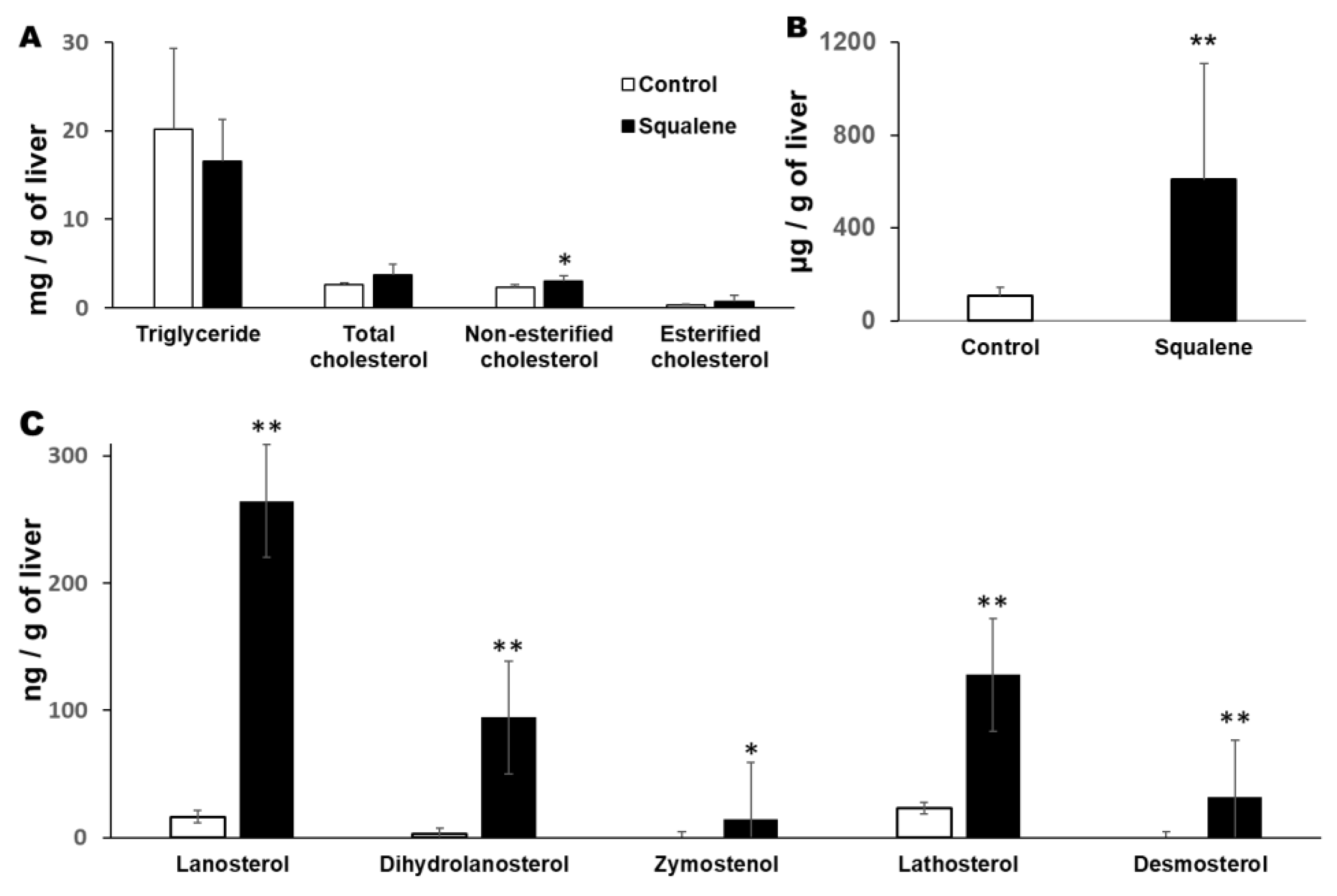
4.3. Quantification of Hepatic Lipids
4.4. Quantification of Hepatic Sterols
4.5. Quantification of Hepatic Squalene
4.6. RNA Extraction
4.7. RNAseq and Data Analyses
4.7.1. Library Construction and Sequencing Was Carried Out by BGI (Shenzhen, China) Service
4.7.2. Bioinformatics Workflow
4.8. Quantification of mRNA
4.9. AML12 Cell Culture
4.10. Quality Control and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keys, A.; Menotti, A.; Karvonen, M.J.; Aravanis, C.; Blackburn, H.; Buzina, R.; Djordjevic, B.S.; Dontas, A.S.; Fidanza, F.; Keys, M.H.; et al. The diet and 15-year death rate in the seven countries study. Am. J. Epidemiol. 1986, 124, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Estruch, R.; Corella, D.; Fito, M.; Ros, E.; Predimed, I. Benefits of the Mediterranean Diet: Insights from the PREDIMED Study. Prog. Cardiovasc. Dis. 2015, 58, 50–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaforio, J.J.; Visioli, F.; Alarcon-de-la-Lastra, C.; Castaner, O.; Delgado-Rodriguez, M.; Fito, M.; Hernandez, A.F.; Huertas, J.R.; Martinez-Gonzalez, M.A.; Menendez, J.A.; et al. Virgin Olive Oil and Health: Summary of the III International Conference on Virgin Olive Oil and Health Consensus Report, JAEN (Spain) 2018. Nutrients 2019, 11, 2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto-Alarcon, S.A.; Valenzuela, R.; Valenzuela, A.; Videla, L.A. Liver protective effects of extra virgin olive oil: Interaction between its chemical composition and the cell-signaling pathways involved in protection. Endocr. Metab. Immune Disord.-Drug Targets (Former. Curr. Drug Targets-Immune Endocr. Metab. Disord.) 2018, 18, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Vitaglione, P.; Savarese, M.; Paduano, A.; Scalfi, L.; Fogliano, V.; Sacchi, R. Healthy virgin olive oil: A matter of bitterness. Crit. Rev. Food Sci. Nutr. 2015, 55, 1808–1818. [Google Scholar] [CrossRef]
- Foscolou, A.; Critselis, E.; Panagiotakos, D. Olive oil consumption and human health: A narrative review. Maturitas 2018, 118, 60–66. [Google Scholar] [CrossRef]
- Lou-Bonafonte, J.M.; Arnal, C.; Navarro, M.A.; Osada, J. Efficacy of bioactive compounds from extra virgin olive oil to modulate atherosclerosis development. Mol. Nutr. Food Res. 2012, 56, 1043–1057. [Google Scholar] [CrossRef]
- Covas, M.-I.; Ruiz-Gutiérrez, V.; De La Torre, R.; Kafatos, A.; Lamuela-Raventós, R.M.; Osada, J.; Owen, R.W.; Visioli, F. Minor Components of Olive Oil: Evidence to Date of Health Benefits in Humans. Nutr. Rev. 2006, 64, 20–30. [Google Scholar] [CrossRef]
- Gabas-Rivera, C.; Jurado-Ruiz, E.; Sanchez-Ortiz, A.; Romanos, E.; Martinez-Beamonte, R.; Navarro, M.A.; Surra, J.C.; Arnal, C.; Rodriguez-Yoldi, M.J.; Andres-Lacueva, C.; et al. Dietary Squalene Induces Cytochromes Cyp2b10 and Cyp2c55 Independently of Sex, Dose, and Diet in Several Mouse Models. Mol. Nutr. Food Res. 2020, 64, e2000354. [Google Scholar] [CrossRef]
- Lanzon, A.; Albi, T.; Cert, A.; Gracian, J. The Hydrocarbon Fraction of Virgin Olive Oil and Changes Resulting from Refining. J. Am. Oil Chem. Soc. 1994, 71, 285–291. [Google Scholar] [CrossRef]
- Martinez-Beamonte, R.; Sanclemente, T.; Surra, J.C.; Osada, J. Could squalene be an added value to use olive by-products? J. Sci. Food Agric. 2020, 100, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Reddy, L.H.; Couvreur, P. Squalene: A natural triterpene for use in disease management and therapy. Adv. Drug Deliv. Rev. 2009, 61, 1412–1426. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Torres, A. Squalene: Current Knowledge and Potential Therapeutical Uses; Nova Science Publishers, Incorporated: Hauppauge, NY, USA, 2011. [Google Scholar]
- Tilvis, R.S.; Miettinen, T.A. Absorption and metabolic fate of dietary 3H-squalene in the rat. Lipids 1983, 18, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Gabas-Rivera, C.; Barranquero, C.; Martinez-Beamonte, R.; Navarro, M.A.; Surra, J.C.; Osada, J. Dietary squalene increases high density lipoprotein-cholesterol and paraoxonase 1 and decreases oxidative stress in mice. PLoS ONE 2014, 9, e104224. [Google Scholar] [CrossRef]
- Yang, B.C.; Phillips, M.I.; Mohuczy, D.; Meng, H.; Shen, L.; Mehta, P.; Mehta, J.L. Increased Angiotensin II Type 1 Receptor Expression in Hypercholesterolemic Atherosclerosis in Rabbits. Arterioscler. Thromb. Vasc. Biol. 1998, 18, 1433–1439. [Google Scholar] [CrossRef] [Green Version]
- Brousseau, M.E.; Hoeg, J.M. Transgenic rabbits as models for atherosclerosis research. J. Lipid Res. 1999, 40, 365–375. [Google Scholar] [CrossRef]
- Fan, J.; Kitajima, S.; Watanabe, T.; Xu, J.; Zhang, J.; Liu, E.; Chen, Y.E. Rabbit models for the study of human atherosclerosis: From pathophysiological mechanisms to translational medicine. Pharmacol. Ther. 2015, 146, 104–119. [Google Scholar] [CrossRef] [Green Version]
- Kritchevsky, D.; Moyer, A.W.; Tesar, W.C.; Logan, J.B.; Brown, R.A.; Richmond, G. Squalene feeding in experimental atherosclerosis. Circ. Res. 1954, 2, 340–343. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Beamonte, R.; Alda, O.; Sanclemente, T.; Felices, M.J.; Escusol, S.; Arnal, C.; Herrera-Marcos, L.V.; Gascon, S.; Surra, J.C.; Osada, J.; et al. Hepatic subcellular distribution of squalene changes according to the experimental setting. J. Physiol. Biochem. 2018, 74, 531–538. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Beamonte, R.; Sanchez-Marco, J.; Felices, M.J.; Barranquero, C.; Gascon, S.; Arnal, C.; Burillo, J.C.; Lasheras, R.; Busto, R.; Lasuncion, M.A.; et al. Dietary squalene modifies plasma lipoproteins and hepatic cholesterol metabolism in rabbits. Food Funct. 2021, 12, 8141–8153. [Google Scholar] [CrossRef]
- Gabas-Rivera, C.; Martinez-Beamonte, R.; Rios, J.L.; Navarro, M.A.; Surra, J.C.; Arnal, C.; Rodriguez-Yoldi, M.J.; Osada, J. Dietary oleanolic acid mediates circadian clock gene expression in liver independently of diet and animal model but requires apolipoprotein A1. J. Nutr. Biochem. 2013, 24, 2100–2109. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, F.; Zeigerer, A.; Kory, N.; Krahmer, N. Hepatic lipid droplet homeostasis and fatty liver disease. Semin. Cell Dev. Biol. 2020, 108, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Fromme, J.C.; Munson, M. Capturing endosomal vesicles at the Golgi. Nat. Cell Biol. 2017, 19, 1384–1386. [Google Scholar] [CrossRef] [PubMed]
- Barr, F.A.; Short, B. Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 2003, 15, 405–413. [Google Scholar] [CrossRef]
- Kawano, Y.; Cohen, D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 2013, 48, 434–441. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Mao, Y.; Li, H.; Wang, X.; Xu, W.; Song, R.; Jia, J.; Lei, Z.; Irwin, D.M.; Niu, G.; et al. Characterization of the gene expression profile of heterozygous liver-specific glucokinase knockout mice at a young age. Biomed. Pharmacother. 2012, 66, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Kargbo, R.B. Inhibition of ACSS2 for Treatment of Cancer and Neuropsychiatric Diseases. ACS Med. Chem. Lett. 2019, 10, 1100–1101. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Zhang, M.; Plec, A.A.; Estill, S.J.; Cai, L.; Repa, J.J.; McKnight, S.L.; Tu, B.P. ACSS2 promotes systemic fat storage and utilization through selective regulation of genes involved in lipid metabolism. Proc. Natl. Acad. Sci. USA 2018, 115, E9499–E9506. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Luo, J.; Ma, G.; Zhang, X.; Yao, D.; Li, M.; Loor, J.J. Acyl-CoA synthetase short-chain family member 2 (ACSS2) is regulated by SREBP-1 and plays a role in fatty acid synthesis in caprine mammary epithelial cells. J. Cell Physiol. 2018, 233, 1005–1016. [Google Scholar] [CrossRef]
- Trepo, E.; Romeo, S.; Zucman-Rossi, J.; Nahon, P. PNPLA3 gene in liver diseases. J. Hepatol. 2016, 65, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, T.; Sumida, Y.; Umemura, A.; Matsuo, K.; Takahashi, M.; Takamura, T.; Yasui, K.; Saibara, T.; Hashimoto, E.; Kawanaka, M. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS ONE 2012, 7, e38322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packard, C.J.; Boren, J.; Taskinen, M.R. Causes and Consequences of Hypertriglyceridemia. Front. Endocrinol. 2020, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Oesterreicher, T.J.; Markesich, D.C.; Henning, S.J. Cloning, characterization and mapping of the mouse trehalase (Treh) gene. Gene 2001, 270, 211–220. [Google Scholar] [CrossRef]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef] [Green Version]
- Dumax-Vorzet, A.; Roboti, P.; High, S. OST4 is a subunit of the mammalian oligosaccharyltransferase required for efficient N-glycosylation. J. Cell Sci. 2013, 126, 2595–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, J.D.; Novak, P.; Lake, A.D.; Hardwick, R.N.; Cherrington, N.J. Impaired N-linked glycosylation of uptake and efflux transporters in human non-alcoholic fatty liver disease. Liver Int. 2017, 37, 1074–1081. [Google Scholar] [CrossRef]
- Mohanty, S.; Chaudhary, B.P.; Zoetewey, D. Structural Insight into the Mechanism of N-Linked Glycosylation by Oligosaccharyltransferase. Biomolecules 2020, 10, 624. [Google Scholar] [CrossRef] [Green Version]
- van den Boogert, M.A.W.; Larsen, L.E.; Ali, L.; Kuil, S.D.; Chong, P.L.W.; Loregger, A.; Kroon, J.; Schnitzler, J.G.; Schimmel, A.W.M.; Peter, J.; et al. N-Glycosylation Defects in Humans Lower Low-Density Lipoprotein Cholesterol Through Increased Low-Density Lipoprotein Receptor Expression. Circulation 2019, 140, 280–292. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Lacey, M.; Ehrlich, M. Epigenetics of skeletal muscle-associated genes in the ASB, LRRC, TMEM, and OSBPL gene families. Epigenomes 2020, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Cappellari, G.G.; Zanetti, M.; Semolic, A.; Vinci, P.; Ruozi, G.; Falcione, A.; Filigheddu, N.; Guarnieri, G.; Graziani, A.; Giacca, M. Unacylated ghrelin reduces skeletal muscle reactive oxygen species generation and inflammation and prevents high-fat diet–induced hyperglycemia and whole-body insulin resistance in rodents. Diabetes 2016, 65, 874–886. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Bermudez, J.; Baudrier, L.; Bayraktar, E.C.; Shen, Y.; La, K.; Guarecuco, R.; Yucel, B.; Fiore, D.; Tavora, B.; Freinkman, E.; et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 2019, 567, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J. Squalene: Potential chemopreventive agent. Expert Opin. Investig. Drugs 2000, 9, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Ran, G.; Wang, X.; Jiang, N.; Liang, J.; Lin, X.; Ling, C.; Zhao, B. Gene manipulation in liver ductal organoids by optimized recombinant adeno-associated virus vectors. J. Biol. Chem. 2019, 294, 14096–14104. [Google Scholar] [CrossRef] [PubMed]
- Kotarba, G.; Krzywinska, E.; Grabowska, A.I.; Taracha, A.; Wilanowski, T. TFCP2/TFCP2L1/UBP1 transcription factors in cancer. Cancer Lett. 2018, 420, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Shastri, B. Functional Characterisation of Glycosyltransferase 1 Domain Containing 1 Gene in Mammary Carcinoma Cells. ResearchSpace@ Auckland. Dissertation Thesis, University of Auckland, Auckland, New Zealand, 2013. [Google Scholar]
- Canfran-Duque, A.; Casado, M.E.; Pastor, O.; Sanchez-Wandelmer, J.; de la Pena, G.; Lerma, M.; Mariscal, P.; Bracher, F.; Lasuncion, M.A.; Busto, R. Atypical antipsychotics alter cholesterol and fatty acid metabolism in vitro. J. Lipid Res. 2013, 54, 310–324. [Google Scholar] [CrossRef] [Green Version]

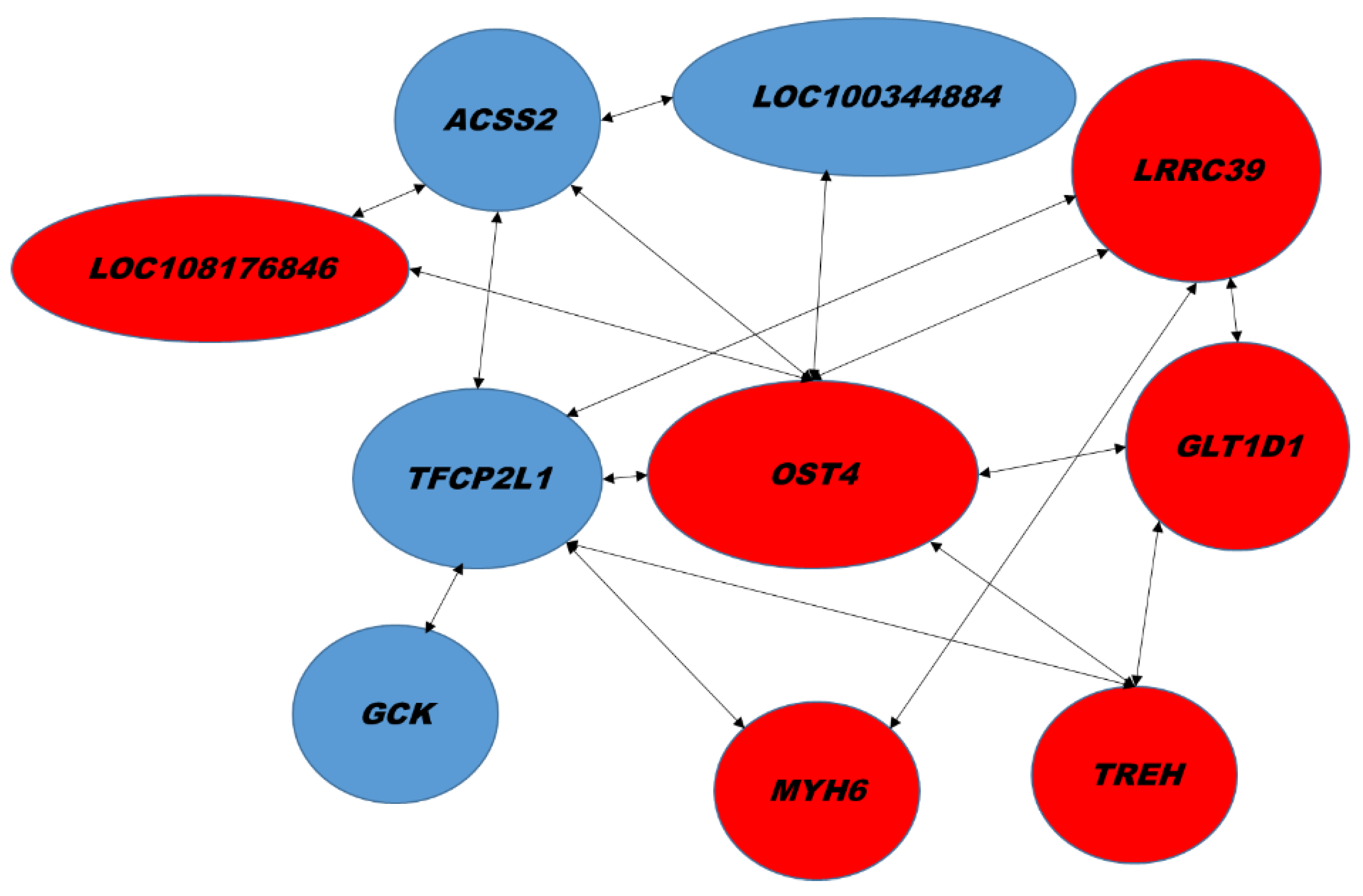


| Biological Process | GenBank | Name | Gene Symbol | Signal log2 Ratio | p-Value |
|---|---|---|---|---|---|
| Intracellular protein transport | XM_017339423.1 | B-cell receptor-associated protein 29, BCAP29 | LOC100344375 | −2.8 | 0.00000 |
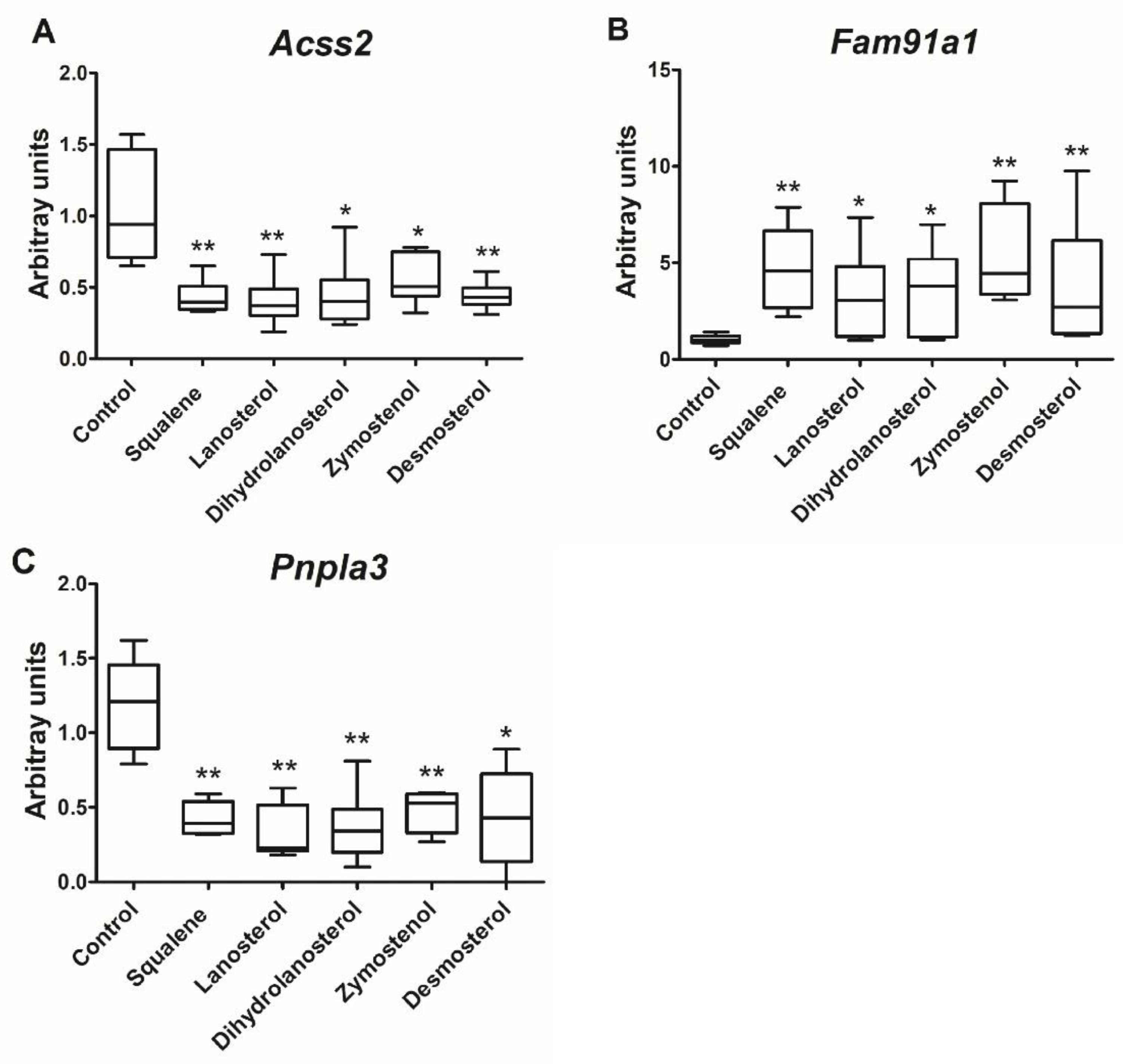
| Hydrolysis of triglycerides | XM_017339724.1 XM_008252198.2 XM_008252200.2 XM_017339725.1 XM_008252201.2 | Patatin-like phospholipase domain-containing protein 3, PNPLA3 | LOC100344884(PNPLA3) | −1.9 | 0.00018 |
| Glucose metabolism | XM_008261818.2 | Glucokinase | GCK | −1.7 | 0.00061 |
| LncRNA | XR_001795369.1 XR_001795370.1 XR_519422.2 | Uncharacterized LOC103351691 | LOC103351691 | −1.6 | 0.00026 |
| Regulation of transcription | XM_008251077.2 | Transcription factor CP2 like 1 | TFCP2L1 | −1.6 | 0.00001 |
| Fatty acid biosynthesis | XM_017339196.1 | Acetyl-CoA carboxylase beta | ACAB | −1.6 | 0.00004 |
| Transcription activity | XM_002711229.3 | Achaete-scute family bHLH transcription factor 1 | ASCL1 | −1.5 | 0.00246 |
| NA | XM_017348395.1 | E3 ubiquitin-protein ligase HERC2-like | LOC108178363 | −1.5 | 0.00225 |
| Acetate-CoA ligase activity | XM_002710791.3 XM_002710792.3 | Acyl-CoA synthetase short chain family member 2 | ACSS2 | −1.5 | 0.00273 |
| Biological Process | GenBank | Name | Gene Symbol | Signal log2 Ratio | p-Value |
|---|---|---|---|---|---|
| Protein glycosylation | XM_017346007.1 | Dolichyl-diphosphooligosaccharide―protein glycosyltransferase subunit 4, OST4 | LOC108177690 (OST4) | 2.5 | 2.60 × 10−7 |
| Intracellular protein transport, vesicle tethering to Golgi | XM_002710763.3 XM_008255883.2 XM_008255884.2 XM_017341515.1 | Family with sequence similarity 91 member A1 | FAM91A1 | 2.3 | 4.39 × 10−6 |
| NA | XR_515397.2 | Uncharacterized LOC103345531 | LOC103345531 | 2 | 0.00009 |
| Actin binding, ATP binding | XM_017348206.1 | Myosin-6 | MYH6 | 1.8 | 0.00041 |
| Regulate bone mineralization | NM_001101695.1 | Osteomodulin | OMD | 1.8 | 0.00013 |
| Negative Regulation of translational initiation | NM_001204114.1 | Eukaryotic translation initiation factor 4E binding protein 3 | EIF4EBP3 | 1.7 | 0.00046 |
| Integral component of membrane | XM_008249188.2 XM_017337996.1 XM_008249187.2 XM_008249192.2 XM_008249193.2 XM_008249194.2 | Leucine rich repeat and Ig domain containing 1 | LINGO1 | 1.7 | 0.00022 |
| NA | XM_002715469.3 | Leucine rich repeat containing 39 | LRRC39 | 1.6 | 0.00086 |
| Nucleoside triphosphate catabolic process, immune response | XM_008263526.2 XM_017345377.1 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | ENPP3 | 1.6 | 0.00022 |
| lncRNA | XR_001793580.1 XR_001793581.1 | Uncharacterized LOC108176846 | LOC108176846 | 1.6 | 0.00009 |
| Muscle contraction | XM_017343215.1 | Titin | TTN | 1.6 | 0.00133 |
| Glycosylation | XM_008251202.1 | Glycosyltransferase 1 domain containing 1 | GLT1D1 | 1.5 | 0.00002 |
| Trehalose metabolism | NM_001082290.1 | Trehalase | TREH | 1.5 | 0.00057 |
| Gene Symbol | Control (n = 5) | Squalene (n = 5) | Fold Change | Signal log2 Ratio |
|---|---|---|---|---|
| ASCL1 | 2.4 ± 3.4 | 0.1 ± 0.2 * | 0.1 | −4.2 |
| ACAB | 2.3 ± 3.3 | 0.5 ± 0.3 | 0.2 | −2.3 |
| GCK | 1.5 ± 1.4 | 0.3 ± 0.2 * | 0.2 | −2.2 |
| LOC100344884 (PNPLA3) | 1.2 ± 0.6 | 0.3 ± 0.3 * | 0.3 | −1.9 |
| ACSS2 | 1.0 ± 0.2 | 0.3 ± 0.2 ** | 0.3 | −1.8 |
| TFCP2L1 | 1.0 ± 0.2 | 0.4 ± 0.2 ** | 0.4 | −1.3 |
| LOC100344375 | 1.5 ± 1.1 | 0.9 ± 0.5 | 0.6 | −0.7 |
| LOC103351691 | 2.0 ± 2.1 | 1.9 ± 1.7 | 1.0 | 0.0 |
| FAM91A1 | 1.1 ± 0.6 | 2.0 ± 0.9 * | 1.8 | 0.8 |
| LOC108177690 (OST4) | 1.1 ± 0.5 | 2.2 ± 0.8 * | 2.0 | 1.0 |
| LRRC39 | 1.1 ± 0.4 | 2.0 ± 0.9 * | 1.9 | 1.0 |
| LOC108176846 | 1.4 ± 1.2 | 3.6 ± 1.9 * | 2.6 | 1.4 |
| GLT1D1 | 1.4 ± 1.2 | 4.4 ± 2.3 * | 3.2 | 1.7 |
| MYH6 | 2.2 ± 1.9 | 11.2 ± 9.7 * | 5.1 | 2.3 |
| OMD | 4.1 ± 5.6 | 19.7 ± 28.2 | 4.8 | 2.3 |
| TTN | 1.0 ± 2.1 | 5.8 ± 5.3 | 5.6 | 2.6 |
| TREH | 1.4 ± 1.1 | 16.3 ± 15.2 * | 11.7 | 3.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abuobeid, R.; Sánchez-Marco, J.; Felices, M.J.; Arnal, C.; Burillo, J.C.; Lasheras, R.; Busto, R.; Lasunción, M.A.; Rodríguez-Yoldi, M.J.; Martínez-Beamonte, R.; et al. Squalene through Its Post-Squalene Metabolites Is a Modulator of Hepatic Transcriptome in Rabbits. Int. J. Mol. Sci. 2022, 23, 4172. https://doi.org/10.3390/ijms23084172
Abuobeid R, Sánchez-Marco J, Felices MJ, Arnal C, Burillo JC, Lasheras R, Busto R, Lasunción MA, Rodríguez-Yoldi MJ, Martínez-Beamonte R, et al. Squalene through Its Post-Squalene Metabolites Is a Modulator of Hepatic Transcriptome in Rabbits. International Journal of Molecular Sciences. 2022; 23(8):4172. https://doi.org/10.3390/ijms23084172
Chicago/Turabian StyleAbuobeid, Roubi, Javier Sánchez-Marco, María J. Felices, Carmen Arnal, Juan Carlos Burillo, Roberto Lasheras, Rebeca Busto, Miguel A. Lasunción, María Jesús Rodríguez-Yoldi, Roberto Martínez-Beamonte, and et al. 2022. "Squalene through Its Post-Squalene Metabolites Is a Modulator of Hepatic Transcriptome in Rabbits" International Journal of Molecular Sciences 23, no. 8: 4172. https://doi.org/10.3390/ijms23084172
APA StyleAbuobeid, R., Sánchez-Marco, J., Felices, M. J., Arnal, C., Burillo, J. C., Lasheras, R., Busto, R., Lasunción, M. A., Rodríguez-Yoldi, M. J., Martínez-Beamonte, R., & Osada, J. (2022). Squalene through Its Post-Squalene Metabolites Is a Modulator of Hepatic Transcriptome in Rabbits. International Journal of Molecular Sciences, 23(8), 4172. https://doi.org/10.3390/ijms23084172









