Elastic Fibre Proteins in Elastogenesis and Wound Healing
Abstract
:1. Introduction
2. Elastic Fibre Proteins and Their Roles in Elastogenesis
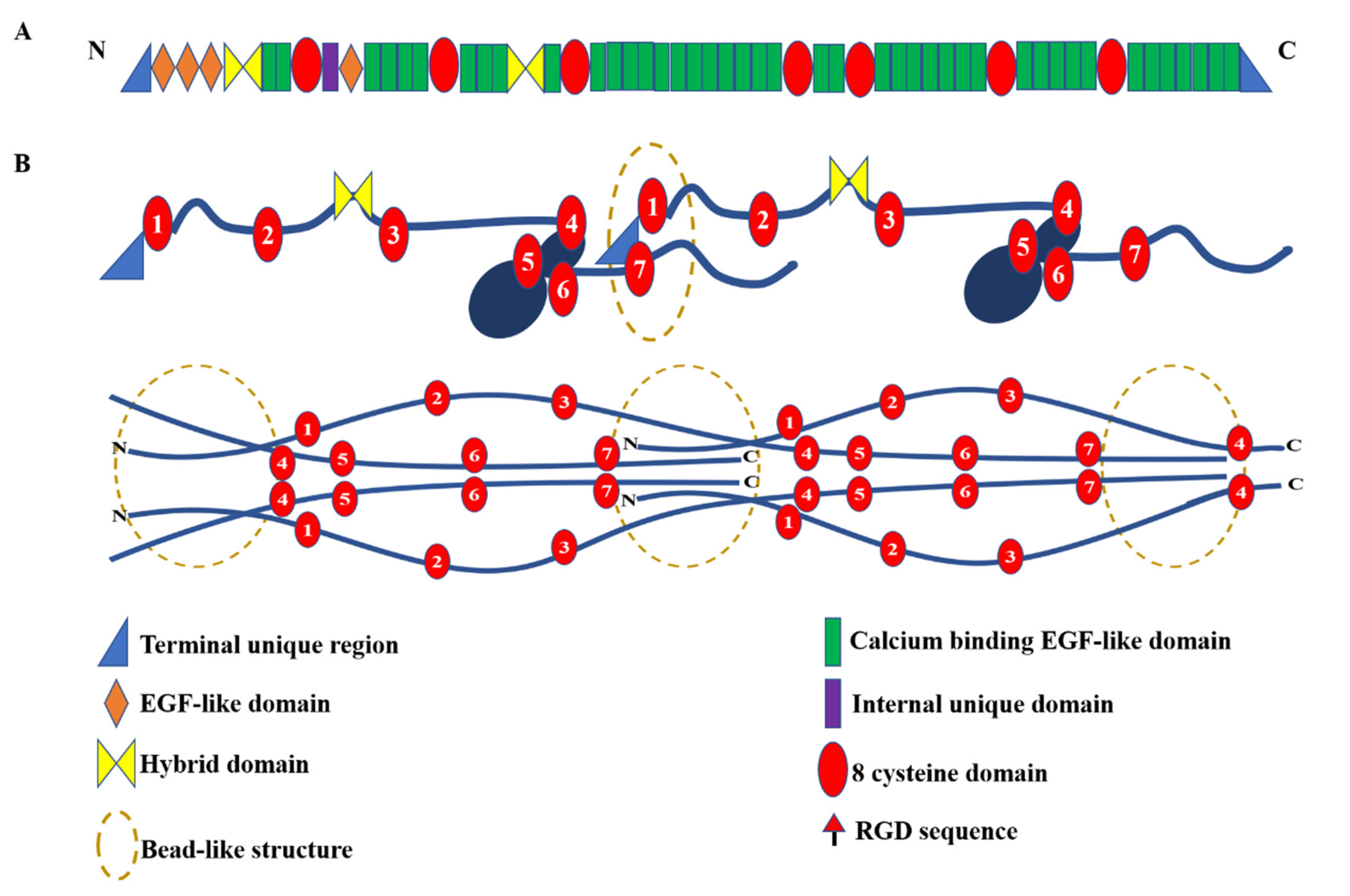
2.1. Fibrillin-1
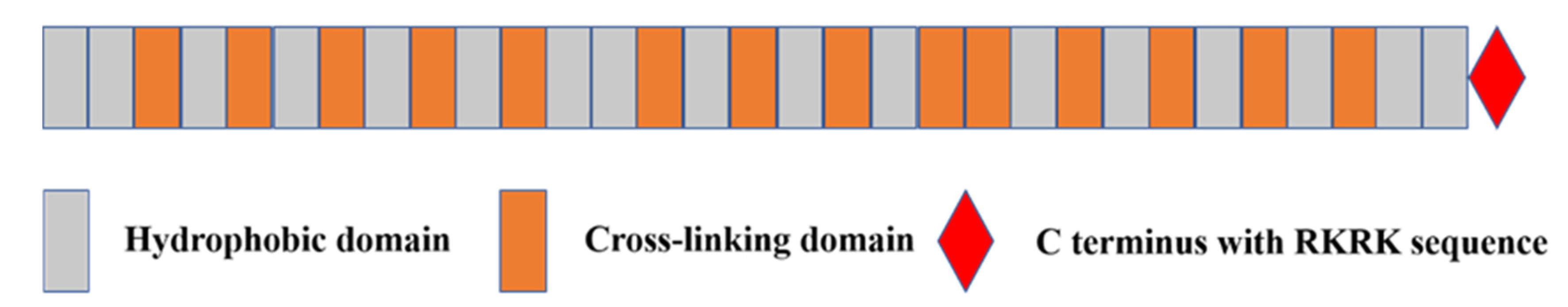
2.2. Tropoelastin
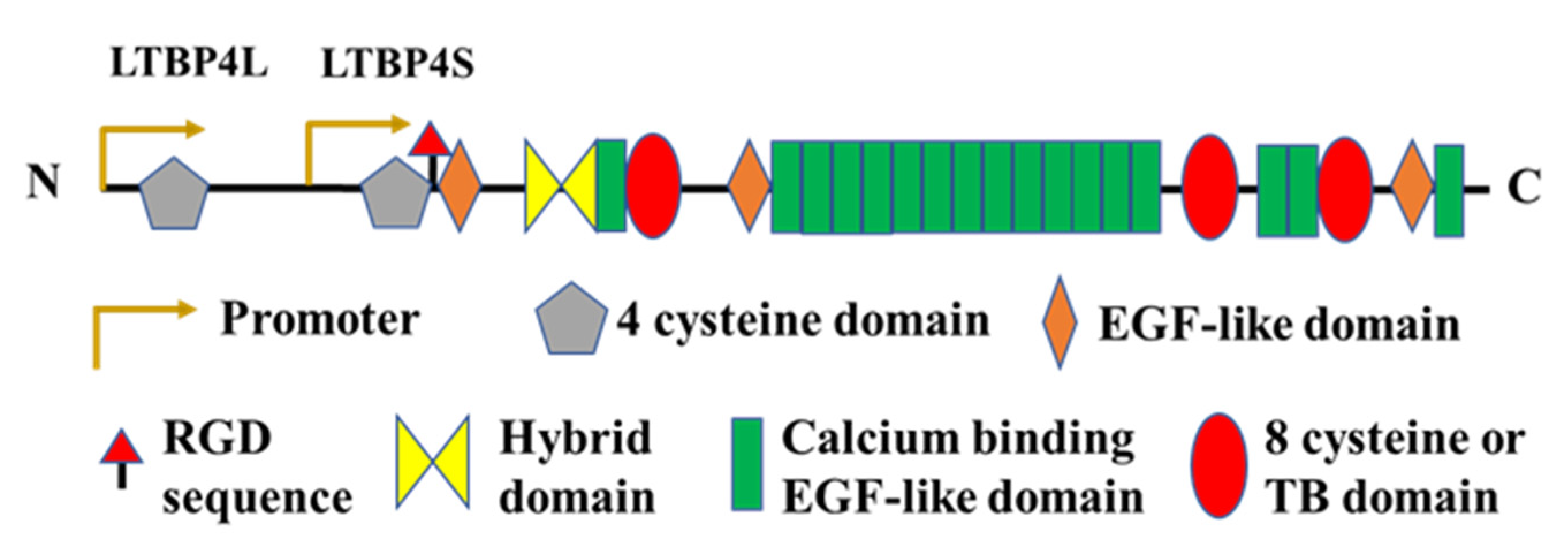
2.3. Latent TGFβ Binding Protein (LTBP)-4
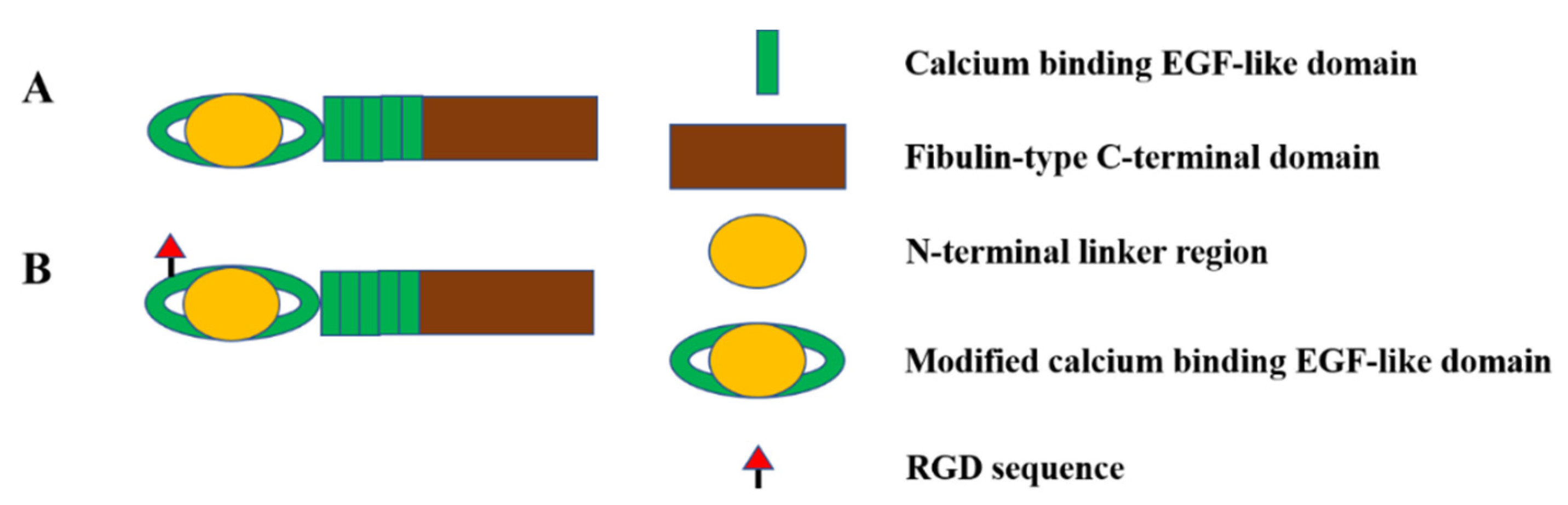
2.4. Fibulin-4 and Fibulin-5
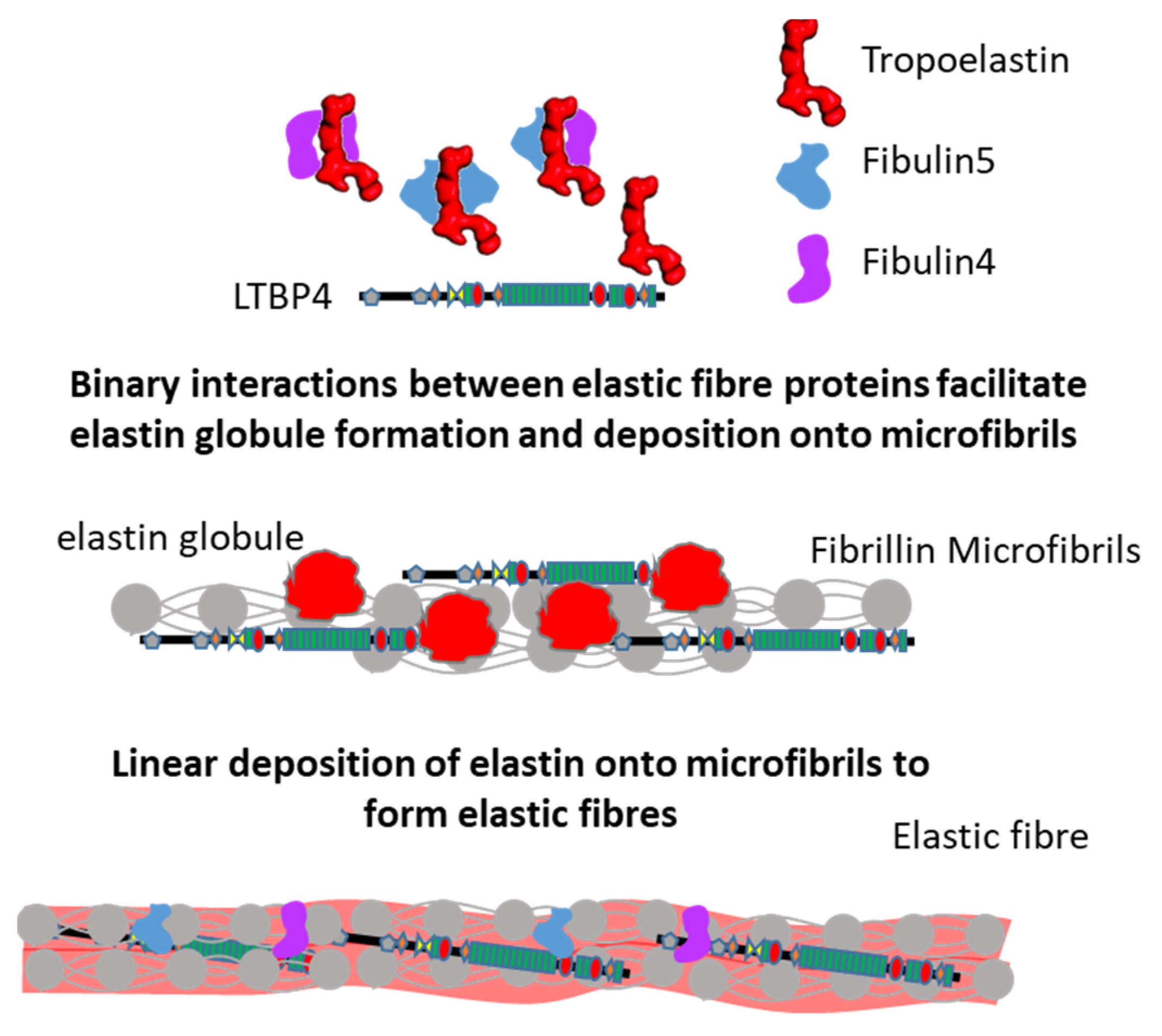
2.5. Interactions Supporting Elastic Fibre Assembly
| Interaction | Function |
|---|---|
| Fibrillin-1–fibulin-4 [58,70,71,78] | Tropoelastin cross-linking and deposition onto microfibrils |
| Fibrillin-1–fibulin-5 [58,70,71,73] | |
| Tropoelastin–fibulin-4 [58,75] | |
| Tropoelastin–fibulin-5 [58,71,72,73,74] | |
| Tropoelastin–fibrillin-1 [80] | Tropoelastin deposition and elastic fibre formation |
| Fibrillin-1–LTBP4 [79] | Deposition and sequestering of latent TGFβ in the extracellular matrix |
| Fibulin-4–fibulin-5 [58] | Unknown: Might contribute after initial elastin cross-linking |
| Tropoelastin–LTBP4 [77] | Unknown: Might contribute to elastic fibre formation |
| LTBP4–fibulin-5 [50] | Deposition of fibulin-5 and tropoelastin on microfibrils |
| LTBP4–fibulin-4 [48,63] | Conformational switch of LTBP4 structure, deposition of tropoelastin onto the elongated LTBP4, and deposition of fibulin-4 on microfibrils |
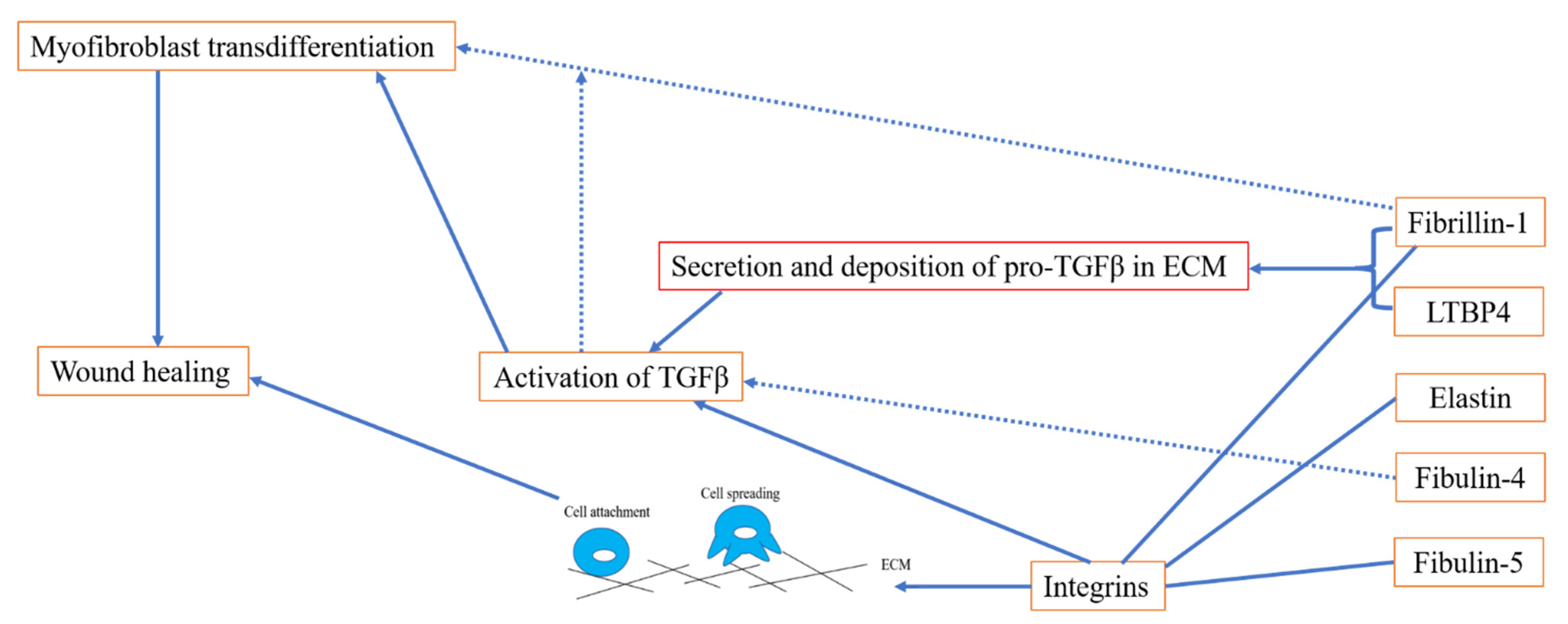
3. The Role of Elastic Fibre Proteins in Wound Repair
3.1. Elastic Fibre Proteins and TGFβ Signalling
3.2. Role of Elastic Fibre Proteins Supporting Integrin-Mediated Cell Adhesion
4. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Ramirez, F.; Caescu, C.; Wondimu, E.; Galatioto, J. Marfan syndrome; A connective tissue disease at the crossroads of mechanotransduction, TGFbeta signaling and cell stemness. Matrix. Biol. 2018, 71–72, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Halabi, C.M.; Kozel, B.A. Vascular elastic fiber heterogeneity in health and disease. Curr. Opin. Hematol. 2020, 27, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.; Singh, M.; Eckersley, A.; Cain, S.A.; Sherratt, M.J.; Baldock, C. Fibrillin microfibrils and elastic fibre proteins: Functional interactions and extracellular regulation of growth factors. Semin. Cell Dev. Biol. 2019, 89, 109–117. [Google Scholar] [CrossRef]
- Ghazawi, F.M.; Zargham, R.; Gilardino, M.S.; Sasseville, D.; Jafarian, F. Insights into the pathophysiology of hypertrophic scars and keloids: How do they differ? Adv. Skin Wound Care 2018, 31, 582–595. [Google Scholar] [CrossRef]
- Jumper, N.; Paus, R.; Bayat, A. Functional histopathology of keloid disease. Histol. Histopathol. 2015, 30, 11033–11057. [Google Scholar] [CrossRef]
- Godwin, A.; Singh, M.; Lockhart-Cairns, M.P.; Alanazi, Y.F.; Cain, S.A.; Baldock, C. The role of fibrillin and microfibril binding proteins in elastin and elastic fibre assembly. Matrix. Biol. 2019, 84, 17–30. [Google Scholar] [CrossRef]
- Charbonneau, N.L.; Dzamba, B.J.; Ono, R.N.; Keene, D.R.; Corson, G.M.; Reinhardt, D.P.; Sakai, L.Y. Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J. Biol. Chem. 2003, 278, 2740–2749. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, B.C.; Sakai, L.Y.; Little, C.D. Fibrillin delineates the primary axis of the early avian embryo. Dev. Dyn. 1993, 196, 70–78. [Google Scholar] [CrossRef]
- Milewicz, D.M.; Urban, Z.; Boyd, C. Genetic disorders of the elastic fiber system. Matrix. Biol. 2000, 19, 471–480. [Google Scholar] [CrossRef]
- Faivre, L.; Gorlin, R.J.; Wirtz, M.K.; Godfrey, M.; Dagoneau, N.; Samples, J.R.; Le Merrer, M.; Collod-Beroud, G.; Boileau, C.; Munnich, A.; et al. In frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J. Med. Genet. 2003, 40, 34–36. [Google Scholar] [CrossRef] [Green Version]
- Loeys, B.L.; Gerber, E.E.; Riegert-Johnson, D.; Iqbal, S.; Whiteman, P.; McConnell, V.; Chillakuri, C.R.; Macaya, D.; Coucke, P.J.; De Paepe, A.; et al. Mutations in fibrillin-1 cause congenital scleroderma: Stiff skin syndrome. Sci. Transl. Med. 2010, 2, 23ra20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Goff, C.; Mahaut, C.; Wang, L.W.; Allali, S.; Abhyankar, A.; Jensen, S.; Zylberberg, L.; Collod-Beroud, G.; Bonnet, D.; Alanay, Y.; et al. Mutations in the TGFbeta binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am. J. Hum. Genet. 2011, 89, 7–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passarge, E.; Robinson, P.N.; Graul-Neumann, L.M. Marfanoid-progeroid-lipodystrophy syndrome: A newly recognized fibrillinopathy. Eur. J. Hum. Genet. 2016, 24, 1244–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.; D’Alessio, M.; Ramirez, F.; Lynch, J.R.; Sykes, B.; Pangilinan, T.; Bonadio, J. Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum. Mol. Genet. 1993, 2, 961–968. [Google Scholar] [CrossRef]
- Corson, G.M.; Chalberg, S.C.; Dietz, H.C.; Charbonneau, N.L.; Sakai, L.Y. Fibrillin binds calcium and is coded by cDNAs that reveal a multidomain structure and alternatively spliced exons at the 5′ end. Genomics 1993, 17, 476–484. [Google Scholar] [CrossRef]
- Muthu, M.L.; Reinhardt, D.P. Fibrillin-1 and fibrillin-1-derived asprosin in adipose tissue function and metabolic disorders. J. Cell Commun. Signal. 2020, 14, 159–173. [Google Scholar] [CrossRef]
- Lin, G.; Tiedemann, K.; Vollbrandt, T.; Peters, H.; Batge, B.; Brinckmann, J.; Reinhardt, D.P. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J. Biol. Chem. 2002, 277, 50795–50804. [Google Scholar] [CrossRef] [Green Version]
- Marson, A.; Rock, M.J.; Cain, S.A.; Freeman, L.J.; Morgan, A.; Mellody, K.; Shuttleworth, C.A.; Baldock, C.; Kielty, C.M. Homotypic fibrillin-1 interactions in microfibril assembly. J. Biol. Chem. 2005, 280, 5013–5021. [Google Scholar] [CrossRef] [Green Version]
- Hubmacher, D.; El-Hallous, E.I.; Nelea, V.; Kaartinen, M.T.; Lee, E.R.; Reinhardt, D.P. Biogenesis of extracellular microfibrils: Multimerization of the fibrillin-1 C terminus into bead-like structures enables self-assembly. Proc. Natl. Acad. Sci. USA 2008, 105, 6548–6553. [Google Scholar] [CrossRef] [Green Version]
- Tiedemann, K.; Bätge, B.; Müller, P.K.; Reinhardt, D.P. Interactions of fibrillin-1 with heparin/heparan sulfate, implications for microfibrillar assembly. J. Biol. Chem. 2001, 276, 36035–36042. [Google Scholar] [CrossRef] [Green Version]
- Cain, S.A.; Baldock, C.; Gallagher, J.; Morgan, A.; Bax, D.V.; Weiss, A.S.; Shuttleworth, C.A.; Kielty, C.M. Fibrillin-1 interactions with heparin. Implications for microfibril and elastic fiber assembly. J. Biol. Chem. 2005, 280, 30526–30537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadin, D.A.; Robertson, I.B.; McNaught-Davis, J.; Evans, P.; Stoddart, D.; Handford, P.A.; Jensen, S.A.; Redfield, C. Structure of the fibrillin-1 N-terminal domains suggests that heparan sulfate regulates the early stages of microfibril assembly. Structure 2013, 21, 1743–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penner, A.S.; Rock, M.J.; Kielty, C.M.; Shipley, J.M. Microfibril-associated glycoprotein-2 interacts with fibrillin-1 and fibrillin-2 suggesting a role for MAGP-2 in elastic fiber assembly. J. Biol. Chem. 2002, 277, 35044–35049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatier, L.; Chen, D.; Fagotto-Kaufmann, C.; Hubmacher, D.; McKee, M.D.; Annis, D.S.; Mosher, D.F.; Reinhardt, D.P. Fibrillin assembly requires fibronectin. Mol. Biol. Cell 2009, 20, 846–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.; Andrikopoulos, K.; Tian, J.; Lee, S.Y.; Keene, D.R.; Ono, R.; Reinhardt, D.P.; Sakai, L.Y.; Biery, N.J.; Bunton, T.; et al. Targetting of the gene encoding fibrillin-1 recapitulates the vascular aspect of Marfan syndrome. Nat. Genet. 1997, 17, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Lima, B.L.; Santos, E.J.; Fernandes, G.R.; Merkel, C.; Mello, M.R.; Gomes, J.P.; Soukoyan, M.; Kerkis, A.; Massironi, S.M.; Visintin, J.A.; et al. A new mouse model for marfan syndrome presents phenotypic variability associated with the genetic background and overall levels of Fbn1 expression. PLoS ONE 2010, 5, e14136. [Google Scholar] [CrossRef] [PubMed]
- Feneck, E.M.; Souza, R.B.; Lewis, P.N.; Hayes, S.; Pereira, L.V.; Meek, K.M. Developmental abnormalities in the cornea of a mouse model for Marfan syndrome. Exp. Eye Res. 2020, 194, 108001. [Google Scholar] [CrossRef]
- Judge, D.P.; Biery, N.J.; Keene, D.R.; Geubtner, J.; Myers, L.; Huso, D.L.; Sakai, L.Y.; Dietz, H.C. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J. Clin. Investig. 2004, 114, 172–181. [Google Scholar] [CrossRef] [Green Version]
- Sakai, L.Y.; Keene, D.R.; Renard, M.; De Backer, J. FBN1: The disease-causing gene for Marfan syndrome and other genetic disorders. Gene 2016, 591, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Sengle, G.; Tsutsui, K.; Keene, D.R.; Tufa, S.F.; Carlson, E.J.; Charbonneau, N.L.; Ono, R.N.; Sasaki, T.; Wirtz, M.K.; Samples, J.R.; et al. Microenvironmental regulation by fibrillin-1. PLoS Genet. 2012, 8, e1002425. [Google Scholar] [CrossRef] [Green Version]
- Baldock, C.; Siegler, V.; Bax, D.V.; Cain, S.A.; Mellody, K.T.; Marson, A.; Haston, J.L.; Berry, R.; Wang, M.C.; Grossmann, J.G.; et al. Nanostructure of fibrillin-1 reveals compact conformation of EGF arrays and mechanism for extensibility. Proc. Natl. Acad. Sci. USA 2006, 103, 11922–11927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldock, C.; Koster, A.J.; Ziese, U.; Rock, M.J.; Sherratt, M.J.; Kadler, K.E.; Shuttleworth, C.A.; Kielty, C.M. The supramolecular organization of fibrillin-rich microfibrils. J. Cell Biol. 2001, 152, 1045–1056. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.S.; Knott, V.; Jovanović, J.; Harlos, K.; Grimes, J.M.; Choulier, L.; Mardon, H.J.; Stuart, D.I.; Handford, P.A. Structure of the integrin binding fragment from fibrillin-1 gives new insights into microfibril organization. Structure 2004, 12, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.L.; Isogai, Z.; Keene, D.R.; Hazeki, N.; Ono, R.N.; Sengle, G.; Bächinger, H.P.; Sakai, L.Y. Effects of fibrillin-1 degradation on microfibril ultrastructure. J. Biol. Chem. 2007, 282, 4007–4020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozsvar, J.; Yang, C.; Cain, S.A.; Baldock, C.; Tarakanova, A.; Weiss, A.S. Tropoelastin and Elastin Assembly. Front. Bioeng. Biotechnol. 2021, 9, 643110. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.C.; Keeley, F.W.; Weiss, A.S. Coacervation of tropoelastin. Adv. Colloid Interface Sci. 2011, 167, 94–103. [Google Scholar] [CrossRef]
- Schmelzer, C.; Hedtke, T.; Heinz, A. Unique molecular networks: Formation and role of elastin cross-links. IUBMB Life 2020, 72, 842–854. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.J.; Yanagisawa, H. Recent updates on the molecular network of elastic fiber formation. Essays Biochem. 2019, 63, 365–376. [Google Scholar] [CrossRef]
- Kozel, B.A.; Mecham, R.P. Elastic fiber ultrastructure and assembly. Matrix. Biol. 2019, 84, 31–40. [Google Scholar] [CrossRef]
- Sephel, G.C.; Buckley, A.; Davidson, J.M. Developmental initiation of elastin gene expression by human fetal skin fibroblasts. J. Investig. Dermatol. 1987, 88, 732–735. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, S.D.; Endicott, S.K.; Province, M.A.; Pierce, J.A.; Campbell, E.J. Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of D-aspartate and nuclear weapons-related radiocarbon. J. Clin. Investig. 1991, 87, 1828–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldock, C.; Oberhauser, A.F.; Ma, L.; Lammie, D.; Siegler, V.; Mithieux, S.M.; Tu, Y.; Chow, J.Y.; Suleman, F.; Malfois, M.; et al. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc. Natl. Acad. Sci. USA 2011, 108, 4322–4327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarakanova, A.; Yeo, G.C.; Baldock, C.; Weiss, A.S.; Buehler, M.J. Molecular model of human tropoelastin and implications of associated mutations. Proc. Natl. Acad. Sci. USA 2018, 115, 7338–7343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, G.C.; Tarakanova, A.; Baldock, C.; Wise, S.G.; Buehler, M.J.; Weiss, A.S. Subtle balance of tropoelastin molecular shape and flexibility regulates dynamics and hierarchical assembly. Sci. Adv. 2016, 2, e1501145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, M.; Reichheld, S.E.; Muiznieks, L.D.; Sitarz, E.E.; Sharpe, S.; Keeley, F.W. Single nucleotide polymorphisms and domain/splice variants modulate assembly and elastomeric properties of human elastin. Implications for tissue specificity and durability of elastic tissue. Biopolymers 2017, 107, e23007. [Google Scholar] [CrossRef]
- Qi, Y.; Shu, C.; Liu, S.; Chen, H.; Zhang, W. Association between single nucleotide polymorphisms of tropoelastin gene and aortic dissection. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2021, 46, 458–466. [Google Scholar] [CrossRef]
- Saracini, C.; Bolli, P.; Sticchi, E.; Pratesi, G.; Pulli, R.; Sofi, F.; Pratesi, C.; Gensini, G.F.; Abbate, R.; Giusti, B. Polymorphisms of genes involved in extracellular matrix remodeling and abdominal aortic aneurysm. J. Vasc. Surg. 2012, 55, 171–179.e2. [Google Scholar] [CrossRef] [Green Version]
- Bultmann-Mellin, I.; Conradi, A.; Maul, A.C.; Dinger, K.; Wempe, F.; Wohl, A.P.; Imhof, T.; Wunderlich, F.T.; Bunck, A.C.; Nakamura, T.; et al. Modeling autosomal recessive cutis laxa type 1C in mice reveals distinct functions for Ltbp-4 isoforms. Dis. Model Mech. 2015, 8, 403–415. [Google Scholar] [CrossRef] [Green Version]
- Bultmann-Mellin, I.; Essers, J.; van Heijingen, P.M.; von Melchner, H.; Sengle, G.; Sterner-Kock, A. Function of Ltbp-4L and fibulin-4 in survival and elastogenesis in mice. Dis. Model Mech. 2016, 9, 1367–1374. [Google Scholar] [CrossRef] [Green Version]
- Noda, K.; Dabovic, B.; Takagi, K.; Inoue, T.; Horiguchi, M.; Hirai, M.; Fujikawa, Y.; Akama, T.O.; Kusumoto, K.; Zilberberg, L.; et al. Latent TGF-beta binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. Proc. Natl. Acad. Sci. USA 2013, 110, 2852–2857. [Google Scholar] [CrossRef] [Green Version]
- Saharinen, J.; Taipale, J.; Monni, O.; Keski-Oja, J. Identification and characterization of a new latent transforming growth factor-beta-binding protein, LTBP-4. J. Biol. Chem. 1998, 273, 18459–18469. [Google Scholar] [CrossRef] [Green Version]
- Kantola, A.K.; Ryynänen, M.J.; Lhota, F.; Keski-Oja, J.; Koli, K. Independent regulation of short and long forms of latent TGF-beta binding protein (LTBP)-4 in cultured fibroblasts and human tissues. J. Cell Physiol. 2010, 223, 727–736. [Google Scholar] [CrossRef]
- Flanigan, K.M.; Ceco, E.; Lamar, K.M.; Kaminoh, Y.; Dunn, D.M.; Mendell, J.R.; King, W.M.; Pestronk, A.; Florence, J.M.; Mathews, K.D.; et al. LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann. Neurol. 2013, 73, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Bello, L.; Kesari, A.; Gordish-Dressman, H.; Cnaan, A.; Morgenroth, L.P.; Punetha, J.; Duong, T.; Henricson, E.K.; Pegoraro, E.; McDonald, C.M.; et al. Genetic modifiers of ambulation in the Cooperative International Neuromuscular Research Group Duchenne Natural History Study. Ann. Neurol. 2015, 77, 684–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban, Z.; Hucthagowder, V.; Schürmann, N.; Todorovic, V.; Zilberberg, L.; Choi, J.; Sens, C.; Brown, C.W.; Clark, R.D.; Holland, K.E.; et al. Mutations in LTBP4 cause a syndrome of impaired pulmonary, gastrointestinal, genitourinary, musculoskeletal, and dermal development. Am. J. Hum. Genet. 2009, 85, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Callewaert, B.; Su, C.T.; Van Damme, T.; Vlummens, P.; Malfait, F.; Vanakker, O.; Schulz, B.; Mac Neal, M.; Davis, E.C.; Lee, J.G.; et al. Comprehensive clinical and molecular analysis of 12 families with type 1 recessive cutis laxa. Hum. Mutat. 2013, 34, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, D.; Kancharla, S.; Kolli, P.; Sharma, A.K.; Singh, S.; Kumar, S.; Mohanty, A.K.; Jena, M.K. Role of fibulins in embryonic stage development and their involvement in various diseases. Biomolecules 2021, 11, 685. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, R.; McGovern, A.; Ridley, C.; Cain, S.A.; Baldwin, A.; Wang, M.C.; Guo, C.; Mironov, A.; Drymoussi, Z., Jr.; Trump, D.; et al. Differential regulation of elastic fiber formation by fibulin-4 and -5. J. Biol. Chem. 2009, 284, 24553–24567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hucthagowder, V.; Sausgruber, N.; Kim, K.H.; Angle, B.; Marmorstein, L.Y.; Urban, Z. Fibulin-4: A novel gene for an autosomal recessive cutis laxa syndrome. Am. J. Hum. Genet. 2006, 78, 1075–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasouki, M.; Markova, D.; Garola, R.; Sasaki, T.; Charbonneau, N.L.; Sakai, L.Y.; Chu, M.L. Compound heterozygous mutations in fibulin-4 causing neonatal lethal pulmonary artery occlusion, aortic aneurysm, arachnodactyly, and mild cutis laxa. Am. J. Med. Genet. A 2007, 143A, 2635–2641. [Google Scholar] [CrossRef] [PubMed]
- Kappanayil, M.; Nampoothiri, S.; Kannan, R.; Renard, M.; Coucke, P.; Malfait, F.; Menon, S.; Ravindran, H.K.; Kurup, R.; Faiyaz-Ul-Haque, M.; et al. Characterization of a distinct lethal arteriopathy syndrome in twenty-two infants associated with an identical, novel mutation in FBLN4 gene, confirms fibulin-4 as a critical determinant of human vascular elastogenesis. Orphanet. J. Rare Dis. 2012, 7, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirulis, J.T.; Bellingham, C.M.; Davis, E.C.; Hubmacher, D.; Reinhardt, D.P.; Mecham, R.P.; Keeley, F.W. Fibrillins, fibulins, and matrix-associated glycoprotein modulate the kinetics and morphology of in vitro self-assembly of a recombinant elastin-like polypeptide. Biochemistry 2008, 47, 12601–12613. [Google Scholar] [CrossRef] [Green Version]
- Kumra, H.; Nelea, V.; Hakami, H.; Pagliuzza, A.; Djokic, J.; Xu, J.; Yanagisawa, H.; Reinhardt, D.P. Fibulin-4 exerts a dual role in LTBP-4L-mediated matrix assembly and function. Proc. Natl. Acad. Sci. USA 2019, 116, 20428–20437. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Ruiz-Lozano, P.; Lindner, V.; Yabe, D.; Taniwaki, M.; Furukawa, Y.; Kobuke, K.; Tashiro, K.; Lu, Z.; Andon, N.L.; et al. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J. Biol. Chem. 1999, 274, 22476–22483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markova, D.; Zou, Y.; Ringpfeil, F.; Sasaki, T.; Kostka, G.; Timpl, R.; Uitto, J.; Chu, M.L. Genetic heterogeneity of cutis laxa: A heterozygous tandem duplication within the fibulin-5 (FBLN5) gene. Am. J. Hum. Genet. 2003, 72, 998–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Megarbane, H.; Florence, J.; Sass, J.O.; Schwonbeck, S.; Foglio, M.; de Cid, R.; Cure, S.; Saker, S.; Mégarbané, A.; Fischer, J. An autosomal-recessive form of cutis laxa is due to homozygous elastin mutations, and the phenotype may be modified by a heterozygous fibulin 5 polymorphism. J. Investig. Dermatol. 2009, 129, 1650–1655. [Google Scholar] [CrossRef] [Green Version]
- Stone, E.M.; Braun, T.A.; Russell, S.R.; Kuehn, M.H.; Lotery, A.J.; Moore, P.A.; Eastman, C.G.; Casavant, T.L.; Sheffield, V.C. Missense variations in the fibulin 5 gene and age-related macular degeneration. N. Engl. J. Med. 2004, 351, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Lotery, A.J.; Baas, D.; Ridley, C.; Jones, R.P.; Klaver, C.C.; Stone, E.; Nakamura, T.; Luff, A.; Griffiths, H.; Wang, T.; et al. Reduced secretion of fibulin 5 in age-related macular degeneration and cutis laxa. Hum. Mutat. 2006, 27, 568–574. [Google Scholar] [CrossRef] [Green Version]
- Schneider, R.; Jensen, S.A.; Whiteman, P.; McCullagh, J.S.; Redfield, C.; Handford, P.A. Biophysical characterisation of fibulin-5 proteins associated with disease. J. Mol. Biol. 2010, 401, 605–617. [Google Scholar] [CrossRef]
- El-Hallous, E.; Sasaki, T.; Hubmacher, D.; Getie, M.; Tiedemann, K.; Brinckmann, J.; Bätge, B.; Davis, E.C.; Reinhardt, D.P. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J. Biol. Chem. 2007, 282, 8935–8946. [Google Scholar] [CrossRef] [Green Version]
- Freeman, L.J.; Lomas, A.; Hodson, N.; Sherratt, M.J.; Mellody, K.T.; Weiss, A.S.; Shuttleworth, A.; Kielty, C.M. Fibulin-5 interacts with fibrillin-1 molecules and microfibrils. Biochem. J. 2005, 388, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, H.; Davis, E.C.; Starcher, B.C.; Ouchi, T.; Yanagisawa, M.; Richardson, J.A.; Olson, E.N. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 2002, 415, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Loeys, B.L.; Coucke, P.J.; De Paepe, A.; Mecham, R.P.; Choi, J.; Davis, E.C.; Urban, Z. Fibulin-5 mutations: Mechanisms of impaired elastic fiber formation in recessive cutis laxa. Hum. Mol. Genet. 2006, 15, 3379–3386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Reymond, J.L.; Pinel, N.; Zabot, M.T.; Urban, Z. Inflammatory destruction of elastic fibers in acquired cutis laxa is associated with missense alleles in the elastin and fibulin-5 genes. J. Investig. Dermatol. 2006, 126, 283–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLaughlin, P.J.; Chen, Q.; Horiguchi, M.; Starcher, B.C.; Stanton, J.B.; Broekelmann, T.J.; Marmorstein, A.D.; McKay, B.; Mecham, R.; Nakamura, T.; et al. Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol. Cell Biol. 2006, 26, 1700–1709. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.P.; Wang, M.C.; Jowitt, T.A.; Ridley, C.; Mellody, K.T.; Howard, M.; Wang, T.; Bishop, P.N.; Lotery, A.J.; Kielty, C.M.; et al. Fibulin 5 forms a compact dimer in physiological solutions. J. Biol. Chem. 2009, 284, 25938–25943. [Google Scholar] [CrossRef] [Green Version]
- Alanazi, Y.F.; Lockhart-Cairns, M.P.; Cain, S.A.; Jowitt, T.A.; Weiss, A.S.; Baldock, C. Autosomal Recessive Cutis Laxa 1C Mutations Disrupt the Structure and Interactions of Latent TGFbeta Binding Protein-4. Front. Genet. 2021, 12, 706662. [Google Scholar] [CrossRef]
- Sasaki, T.; Hanisch, F.G.; Deutzmann, R.; Sakai, L.Y.; Sakuma, T.; Miyamoto, T.; Yamamoto, T.; Hannappel, E.; Chu, M.L.; Lanig, H.; et al. Functional consequence of fibulin-4 missense mutations associated with vascular and skeletal abnormalities and cutis laxa. Matrix. Biol. 2016, 56, 132–149. [Google Scholar] [CrossRef]
- Ono, R.N.; Sengle, G.; Charbonneau, N.L.; Carlberg, V.; Bächinger, H.P.; Sasaki, T.; Lee-Arteaga, S.; Zilberberg, L.; Rifkin, D.B.; Ramirez, F.; et al. Latent transforming growth factor beta-binding proteins and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J. Biol. Chem. 2009, 284, 16872–16881. [Google Scholar] [CrossRef] [Green Version]
- Rock, M.J.; Cain, S.A.; Freeman, L.J.; Morgan, A.; Mellody, K.; Marson, A.; Shuttleworth, C.A.; Weiss, A.S.; Kielty, C.M. Molecular basis of elastic fiber formation. Critical interactions and a tropoelastin-fibrillin-1 cross-link. J. Biol. Chem. 2004, 279, 23748–23758. [Google Scholar] [CrossRef] [Green Version]
- Handa, K.; Abe, S.; Suresh, V.V.; Fujieda, Y.; Ishikawa, M.; Orimoto, A.; Kobayashi, Y.; Yamada, S.; Yamaba, S.; Murakami, S.; et al. Fibrillin-1 insufficiency alters periodontal wound healing failure in a mouse model of Marfan syndrome. Arch. Oral. Biol. 2018, 90, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Yoshiba, N.; Yoshiba, K.; Ohkura, N.; Takei, E.; Edanami, N.; Oda, Y.; Hosoya, A.; Nakamura, H.; Okiji, T. Correlation between Fibrillin-1 Degradation and mRNA Downregulation and Myofibroblast Differentiation in Cultured Human Dental Pulp Tissue. J. Histochem. Cytochem. 2015, 63, 438–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.J.; Roy, N.K.; Mogford, J.E.; Schiemann, W.P.; Mustoe, T.A. Fibulin-5 promotes wound healing in vivo. J. Am. Coll. Surg. 2004, 199, 403–410. [Google Scholar] [CrossRef]
- Almine, J.F.; Wise, S.G.; Hiob, M.; Singh, N.K.; Tiwari, K.K.; Vali, S.; Abbasi, T.; Weiss, A.S. Elastin sequences trigger transient proinflammatory responses by human dermal fibroblasts. FASEB J. 2013, 27, 3455–3465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margadant, C.; Sonnenberg, A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010, 11, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Zilberberg, L.; Todorovic, V.; Dabovic, B.; Horiguchi, M.; Couroussé, T.; Sakai, L.Y.; Rifkin, D.B. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: Role of fibrillins and fibronectin. J. Cell. Physiol. 2012, 227, 3828–3836. [Google Scholar] [CrossRef] [Green Version]
- Miyazono, K.; Olofsson, A.; Colosetti, P.; Heldin, C.H. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991, 10, 1091–1101. [Google Scholar] [CrossRef]
- Annes, J.P.; Chen, Y.; Munger, J.S.; Rifkin, D.B. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J. Cell. Biol. 2004, 165, 723–734. [Google Scholar] [CrossRef]
- Lamar, K.M.; Miller, T.; Dellefave-Castillo, L.; McNally, E.M. Genotype-Specific Interaction of Latent TGFbeta Binding Protein 4 with TGFbeta. PLoS ONE 2016, 11, e0150358. [Google Scholar] [CrossRef] [Green Version]
- Su, C.T.; Huang, J.W.; Chiang, C.K.; Lawrence, E.C.; Levine, K.L.; Dabovic, B.; Jung, C.; Davis, E.C.; Madan-Khetarpal, S.; Urban, Z. Latent transforming growth factor binding protein 4 regulates transforming growth factor beta receptor stability. Hum. Mol. Genet. 2015, 24, 4024–4036. [Google Scholar] [CrossRef]
- Lu, J.; Liu, Q.; Wang, L.; Tu, W.; Chu, H.; Ding, W.; Jiang, S.; Ma, Y.; Shi, X.; Pu, W.; et al. Increased expression of latent TGF-beta-binding protein 4 affects the fibrotic process in scleroderma by TGF-beta/SMAD signaling. Lab. Investig. 2017, 97, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhu, J.; Wang, R.; Chen, X.; Mi, L.; Walz, T.; Springer, T.A. Latent TGF-beta structure and activation. Nature 2011, 474, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buscemi, L.; Ramonet, D.; Klingberg, F.; Formey, A.; Smith-Clerc, J.; Meister, J.J.; Hinz, B. The single-molecule mechanics of the latent TGF-beta1 complex. Curr. Biol. 2011, 21, 2046–2054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, M.G.; Cormier, A.; Ito, S.; Seed, R.I.; Bondesson, A.J.; Lou, J.; Marks, J.D.; Baron, J.L.; Cheng, Y.; Nishimura, S.L. Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta. Cell 2020, 180, 490–501.e16. [Google Scholar] [CrossRef] [PubMed]
- Isogai, Z.; Ono, R.N.; Ushiro, S.; Keene, D.R.; Chen, Y.; Mazzieri, R.; Charbonneau, N.L.; Reinhardt, D.P.; Rifkin, D.B.; Sakai, L.Y. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J. Biol. Chem. 2003, 278, 2750–2757. [Google Scholar] [CrossRef] [Green Version]
- Neptune, E.R.; Frischmeyer, P.A.; Arking, D.E.; Myers, L.; Bunton, T.E.; Gayraud, B.; Ramirez, F.; Sakai, L.Y.; Dietz, H.C. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003, 33, 407–411. [Google Scholar] [CrossRef]
- Nistala, H.; Lee-Arteaga, S.; Smaldone, S.; Siciliano, G.; Carta, L.; Ono, R.N.; Sengle, G.; Arteaga-Solis, E.; Levasseur, R.; Ducy, P.; et al. Fibrillin-1 and -2 differentially modulate endogenous TGF-beta and BMP bioavailability during bone formation. J. Cell. Biol. 2010, 190, 1107–1121. [Google Scholar] [CrossRef] [Green Version]
- Zeyer, K.A.; Zhang, R.M.; Kumra, H.; Hassan, A.; Reinhardt, D.P. The Fibrillin-1 RGD Integrin Binding Site Regulates Gene Expression and Cell Function through microRNAs. J. Mol. Biol. 2019, 431, 401–421. [Google Scholar] [CrossRef]
- Ramnath, N.W.; Hawinkels, L.J.; van Heijningen, P.M.; te Riet, L.; Paauwe, M.; Vermeij, M.; Danser, A.H.; Kanaar, R.; ten Dijke, P.; Essers, J. Fibulin-4 deficiency increases TGF-beta signalling in aortic smooth muscle cells due to elevated TGF-beta2 levels. Sci. Rep. 2015, 5, 16872. [Google Scholar] [CrossRef]
- Burger, J.; van Vliet, N.; van Heijningen, P.; Kumra, H.; Kremers, G.J.; Alves, M.; van Cappellen, G.; Yanagisawa, H.; Reinhardt, D.P.; Kanaar, R.; et al. Fibulin-4 deficiency differentially affects cytoskeleton structure and dynamics as well as TGFbeta signaling. Cell Signal. 2019, 58, 65–78. [Google Scholar] [CrossRef]
- Kuang, P.P.; Joyce-Brady, M.; Zhang, X.H.; Jean, J.C.; Goldstein, R.H. Fibulin-5 gene expression in human lung fibroblasts is regulated by TGF-beta and phosphatidylinositol 3-kinase activity. Am. J. Physiol. Cell Physiol. 2006, 291, C1412–C1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.H.; Albig, A.R.; Regner, M.; Schiemann, B.J.; Schiemann, W.P. Fibulin-5 initiates epithelial-mesenchymal transition (EMT) and enhances EMT induced by TGF-beta in mammary epithelial cells via a MMP-dependent mechanism. Carcinogenesis 2008, 29, 2243–2251. [Google Scholar] [CrossRef] [PubMed]
- Topalovski, M.; Hagopian, M.; Wang, M.; Brekken, R.A. Hypoxia and Transforming Growth Factor beta Cooperate to Induce Fibulin-5 Expression in Pancreatic Cancer. J. Biol. Chem. 2016, 291, 22244–22252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiemann, W.P.; Blobe, G.C.; Kalume, D.E.; Pandey, A.; Lodish, H.F. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J. Biol. Chem. 2002, 277, 27367–27377. [Google Scholar] [CrossRef] [Green Version]
- Pfaff, M.; Reinhardt, D.P.; Sakai, L.Y.; Timpl, R. Cell adhesion and integrin binding to recombinant human fibrillin-1. FEBS Lett. 1996, 384, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Bax, D.V.; Bernard, S.E.; Lomas, A.; Morgan, A.; Humphries, J.; Shuttleworth, C.A.; Humphries, M.J.; Kielty, C.M. Cell adhesion to fibrillin-1 molecules and microfibrils is mediated by alpha 5 beta 1 and alpha v beta 3 integrins. J. Biol. Chem. 2003, 278, 34605–34616. [Google Scholar] [CrossRef] [Green Version]
- Jovanovic, J.; Takagi, J.; Choulier, L.; Abrescia, N.G.; Stuart, D.I.; van der Merwe, P.A.; Mardon, H.J.; Handford, P.A. alphaVbeta6 is a novel receptor for human fibrillin-1. Comparative studies of molecular determinants underlying integrin-rgd affinity and specificity. J. Biol. Chem. 2007, 282, 6743–6751. [Google Scholar] [CrossRef] [Green Version]
- Del, C.J.; Reed, N.I.; Molnar, K.; Liu, S.; Dang, B.; Jensen, S.A.; DeGrado, W.; Handford, P.A.; Sheppard, D.; Sundaram, A.B. A disease-associated mutation in fibrillin-1 differentially regulates integrin-mediated cell adhesion. J. Biol. Chem. 2019, 294, 18232–18243. [Google Scholar] [CrossRef]
- Bax, D.V.; Rodgers, U.R.; Bilek, M.M.; Weiss, A. Cell adhesion to tropoelastin is mediated via the C-terminal GRKRK motif and integrin alphaVbeta3. J. Biol. Chem. 2009, 284, 28616–28623. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.; Bax, D.V.; Bilek, M.M.; Weiss, A.S. A novel cell adhesion region in tropoelastin mediates attachment to integrin alphaVbeta5. J. Biol. Chem. 2014, 289, 1467–1477. [Google Scholar] [CrossRef] [Green Version]
- Bochicchio, B.; Yeo, G.C.; Lee, P.; Emul, D.; Pepe, A.; Laezza, A.; Ciarfaglia, N.; Quaglino, D.; Weiss, A.S. Domains 12 to 16 of tropoelastin promote cell attachment and spreading through interactions with glycosaminoglycan and integrins alphaV and alpha5beta1. FEBS J. 2021, 288, 4024–4038. [Google Scholar] [CrossRef] [PubMed]
- Ozsvar, J.; Wang, R.; Tarakanova, A.; Buehler, M.J.; Weiss, A.S. Fuzzy binding model of molecular interactions between tropoelastin and integrin alphaVbeta3. Biophys. J. 2021, 120, 3138–3151. [Google Scholar] [CrossRef] [PubMed]
- Lomas, A.C.; Mellody, K.T.; Freeman, L.J.; Bax, D.V.; Shuttleworth, C.A.; Kielty, C.M. Fibulin-5 binds human smooth-muscle cells through alpha5beta1 and alpha4beta1 integrins, but does not support receptor activation. Biochem. J. 2007, 405, 417–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furie, N.; Shteynberg, D.; Elkhatib, R.; Perry, L.; Ullmann, Y.; Feferman, Y.; Preis, M.; Flugelman, M.Y.; Tzchori, I. Fibulin-5 regulates keloid-derived fibroblast-like cells through integrin beta-1. Int. J. Cosmet. Sci. 2016, 38, 35–40. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Alanazi, Y.F.; Jowitt, T.A.; Roseman, A.M.; Baldock, C. Elastic Fibre Proteins in Elastogenesis and Wound Healing. Int. J. Mol. Sci. 2022, 23, 4087. https://doi.org/10.3390/ijms23084087
Zhang X, Alanazi YF, Jowitt TA, Roseman AM, Baldock C. Elastic Fibre Proteins in Elastogenesis and Wound Healing. International Journal of Molecular Sciences. 2022; 23(8):4087. https://doi.org/10.3390/ijms23084087
Chicago/Turabian StyleZhang, Xinyang, Yasmene F. Alanazi, Thomas A. Jowitt, Alan M. Roseman, and Clair Baldock. 2022. "Elastic Fibre Proteins in Elastogenesis and Wound Healing" International Journal of Molecular Sciences 23, no. 8: 4087. https://doi.org/10.3390/ijms23084087
APA StyleZhang, X., Alanazi, Y. F., Jowitt, T. A., Roseman, A. M., & Baldock, C. (2022). Elastic Fibre Proteins in Elastogenesis and Wound Healing. International Journal of Molecular Sciences, 23(8), 4087. https://doi.org/10.3390/ijms23084087






