The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration
Abstract
1. Introduction
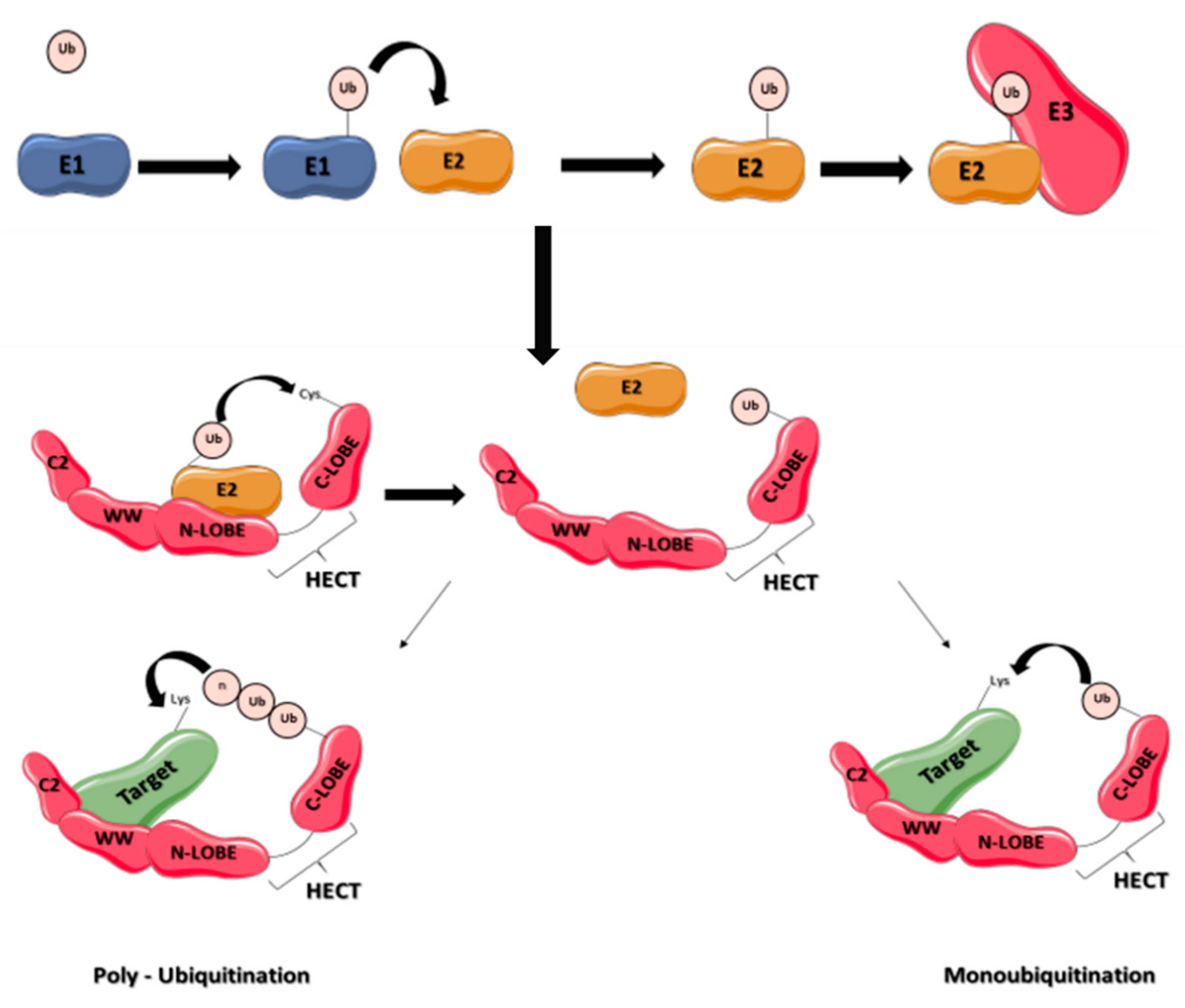
2. Overview of the Ubiquitin System
3. The NEDD4 E3 Ligases Subfamily
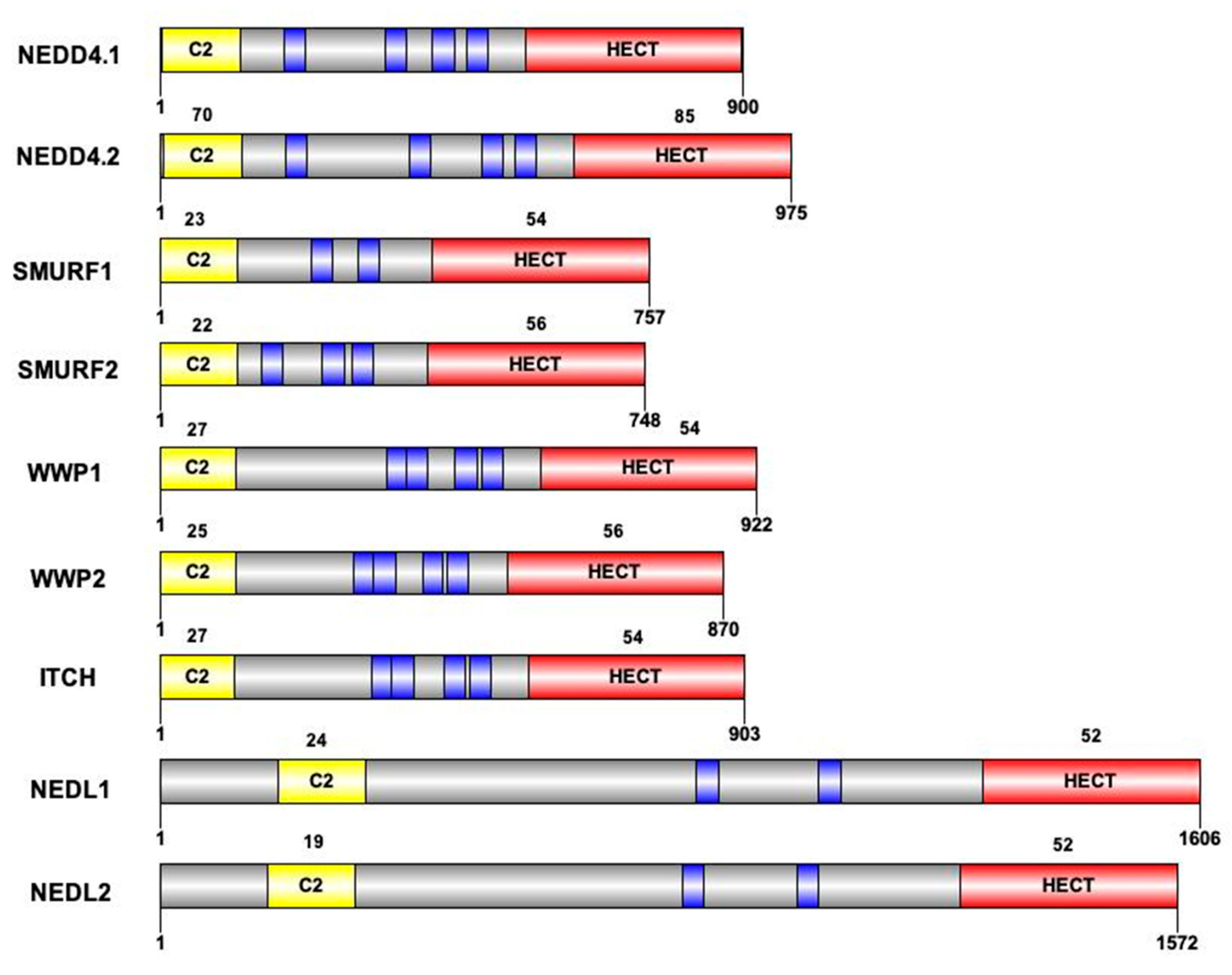
3.1. Structure and Diversity
3.2. Functions and Regulations
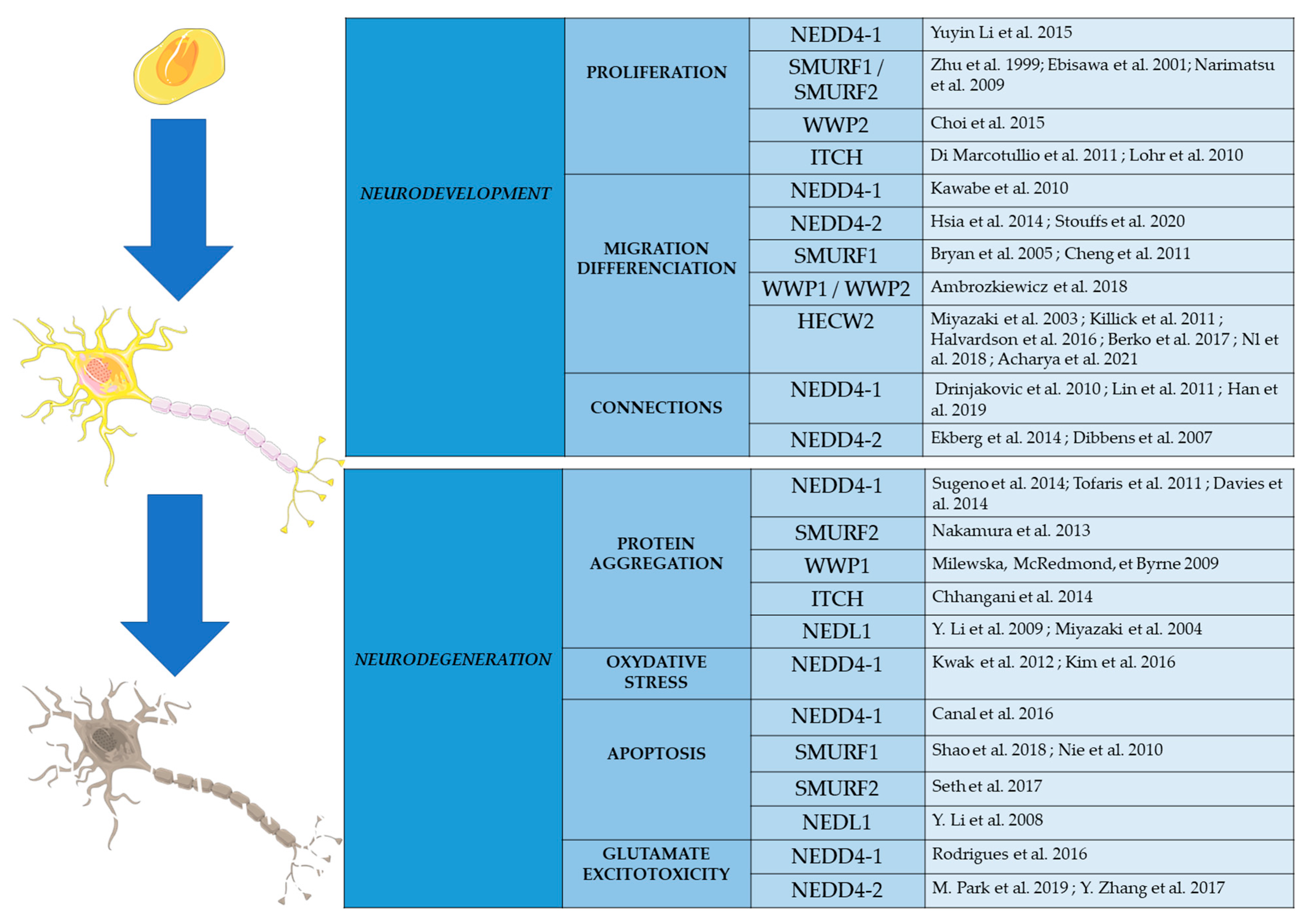
4. NEDD4 E3 Ligases in Neurodevelopment
5. NEDD4 E3 Ligases in Neurodegeneration
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbert, S.L.; Dobyns, W.B.; Lahn, B.T. Genetic links between brain development and brain evolution. Nat. Rev. Genet. 2005, 6, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Joshi, V.; Amanullah, A.; Mishra, R.; Arora, N.; Prasad, A.; Mishra, A. E3 Ubiquitin Ligases Neurobiological Mechanisms: Development to Degeneration. Front. Mol. Neurosci. 2017, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Ramocki, M.B.; Zoghbi, H.Y. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature 2008, 455, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dong, K.; Wang, Y.; Peng, G.; Song, X.; Yu, Y.; Shen, P.; Cui, X. Integrating HECW1 expression into the clinical indicators exhibits high accuracy in assessing the prognosis of patients with clear cell renal cell carcinoma. BMC Cancer 2021, 21, 890. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Wong, B.R.; Parlati, F.; Qu, K.; Demo, S.; Pray, T.; Huang, J.; Payan, D.G.; Bennett, M.K. Drug discovery in the ubiquitin regulatory pathway. Drug Discov. Today 2003, 8, 746–754. [Google Scholar] [CrossRef]
- Jana, N.R. Protein homeostasis and aging: Role of ubiquitin protein ligases. Neurochem. Int. 2012, 60, 443–447. [Google Scholar] [CrossRef]
- Shang, F.; Taylor, A. Ubiquitin–proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011, 51, 5–16. [Google Scholar] [CrossRef]
- Su, V.; Lau, A.F. Ubiquitination, intracellular trafficking, and degradation of connexins. Arch. Biochem. Biophys. 2012, 524, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.D. Ubiquitin-Dependent Signaling: The Role of Ubiquitination in the Response of Cells to Their Environment. J. Nutr. 1999, 129, 1933–1936. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Popelka, H.; Lei, Y.; Yang, Y.; Klionsky, D.J. The Roles of Ubiquitin in Mediating Autophagy. Cells 2020, 9, 2025. [Google Scholar] [CrossRef] [PubMed]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef]
- Milewska, M.; McRedmond, J.; Byrne, P.C. Identification of novel spartin-interactors shows spartin is a multifunctional protein. J. Neurochem. 2009, 111, 1022–1030. [Google Scholar] [CrossRef]
- Wang, Y.; Argiles-Castillo, D.; Kane, E.I.; Zhou, A.; Spratt, D.E. HECT E3 ubiquitin ligases–emerging insights into their biological roles and disease relevance. J. Cell Sci. 2020, 133, jcs228072. [Google Scholar] [CrossRef]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Huang, X.; Chen, J.; Cao, W.; Yang, L.; Chen, Q.; He, J.; Yi, Q.; Huang, H.; Zhang, E.; Cai, Z. The many substrates and functions of NEDD4-1. Cell Death Dis. 2019, 10, 904. [Google Scholar] [CrossRef]
- Henshall, T.L.; Manning, J.; Alfassy, O.S.; Goel, P.; Boase, N.A.; Kawabe, H.; Kumar, S. Deletion of Nedd4-2 results in progressive kidney disease in mice. Cell Death Differ. 2017, 24, 2150–2160. [Google Scholar] [CrossRef]
- Jiang, C.; Kawabe, H.; Rotin, D. The Ubiquitin Ligase Nedd4L Regulates the Na/K/2Cl Co-transporter NKCC1/SLC12A2 in the Colon. J. Biol. Chem. 2017, 292, 3137–3145. [Google Scholar] [CrossRef]
- Leitz, D.H.W.; Duerr, J.; Mulugeta, S.; Agircan, A.S.; Zimmermann, S.; Kawabe, H.; Dalpke, A.H.; Beers, M.F.; Mall, M.A. Congenital Deletion of Nedd4-2 in Lung Epithelial Cells Causes Progressive Alveolitis and Pulmonary Fibrosis in Neonatal Mice. Int. J. Mol. Sci. 2021, 22, 6146. [Google Scholar] [CrossRef] [PubMed]
- Kamynina, E.; Debonneville, C.; Bens, M.; Vandewalle, A.; Staub, O. A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+channel. FASEB J. 2001, 15, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Rauh, R.; Dinudom, A.; Fotia, A.B.; Paulides, M.; Kumar, S.; Korbmacher, C.; Cook, D.I. Stimulation of the epithelial sodium channel (ENaC) by the serum- and glucocorticoid-inducible kinase (Sgk) involves the PY motifs of the channel but is independent of sodium feedback inhibition. Pflüg. Arch. Eur. J. Physiol. 2006, 452, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Alarcón, C.; Sapkota, G.; Rahman, S.; Chen, P.-Y.; Goerner, N.; Macias, M.J.; Erdjument-Bromage, H.; Tempst, P.; Massagué, J. Ubiquitin Ligase Nedd4L Targets Activated Smad2/3 to Limit TGF-β Signaling. Mol. Cell 2009, 36, 457–468. [Google Scholar] [CrossRef]
- Lee, D.-E.; Yoo, J.E.; Kim, J.; Kim, S.; Kim, S.; Lee, H.; Cheong, H. NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. Cell Death Dis. 2020, 11, 38. [Google Scholar] [CrossRef]
- Tanksley, J.P.; Chen, X.; Coffey, R.J. NEDD4L Is Downregulated in Colorectal Cancer and Inhibits Canonical WNT Signaling. PLoS ONE 2013, 8, e81514. [Google Scholar] [CrossRef]
- Debonneville, C.; Flores, S.Y.; Kamynina, E.; Plant, P.J.; Tauxe, C.; Thomas, M.A.; Münster, C.; Chraïbi, A.; Pratt, J.; Horisberger, J.; et al. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na+ channel cell surface expression. EMBO J. 2001, 20, 7052–7059. [Google Scholar] [CrossRef]
- Lee, I.-H.; Dinudom, A.; Sanchez-Perez, A.; Kumar, S.; Cook, D.I. Akt Mediates the Effect of Insulin on Epithelial Sodium Channels by Inhibiting Nedd4-2. J. Biol. Chem. 2007, 282, 29866–29873. [Google Scholar] [CrossRef]
- Snyder, P.M.; Olson, D.R.; Thomas, B.C. Serum and Glucocorticoid-regulated Kinase Modulates Nedd4-2-mediated Inhibition of the Epithelial Na+ Channel. J. Biol. Chem. 2002, 277, 5–8. [Google Scholar] [CrossRef]
- Lang, F.; Pelzl, L.; Hauser, S.; Hermann, A.; Stournaras, C.; Schöls, L. To die or not to die SGK1-sensitive ORAI/STIM in cell survival. Cell Calcium 2018, 74, 29–34. [Google Scholar] [CrossRef]
- Fang, D.; Elly, C.; Gao, B.; Fang, N.; Altman, Y.; Joazeiro, C.; Hunter, T.; Copeland, N.; Jenkins, N.; Liu, Y.-C. Dysregulation of T lymphocyte function in itchy mice: A role for Itch in TH2 differentiation. Nat. Immunol. 2002, 3, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; De Simone, M.; Pollice, A.; Santoro, R.; La Mantia, G.; Guerrini, L.; Calabrò, V. Itch/AIP4 Associates with and Promotes p63 Protein Degradation. Cell Cycle 2006, 5, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; De Laurenzi, V.; Munarriz, E.; Green, D.; Liu, Y.-C.; Vousden, K.H.; Cesareni, G.; Melino, G. The ubiquitin–protein ligase Itch regulates p73 stability. EMBO J. 2005, 24, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Kathania, M.; Khare, P.; Zeng, M.; Cantarel, B.; Zhang, H.; Ueno, H.; Venuprasad, M.K.P.K.M.Z.K. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat. Immunol. 2016, 17, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H.; Jung, E.-H.; Kim, G.-Y.; Kim, B.-C.; Lim, J.H.; Woo, C.-H. Itch E3 ubiquitin ligase positively regulates TGF-β signaling to EMT via Smad7 ubiquitination. Mol. Cells 2015, 38, 20–25. [Google Scholar] [CrossRef]
- Chastagner, P.; Israël, A.; Brou, C. AIP4/Itch Regulates Notch Receptor Degradation in the Absence of Ligand. PLoS ONE 2008, 3, e2735. [Google Scholar] [CrossRef]
- Goto, J.; Otaki, Y.; Watanabe, T.; Kobayashi, Y.; Aono, T.; Watanabe, K.; Wanezaki, M.; Kutsuzawa, D.; Kato, S.; Tamura, H.; et al. HECT (Homologous to the E6-AP Carboxyl Terminus)-Type Ubiquitin E3 Ligase ITCH Attenuates Cardiac Hypertrophy by Suppressing the Wnt/β-Catenin Signaling Pathway. Hypertension 2020, 76, 1868–1878. [Google Scholar] [CrossRef]
- Infante, P.; Severini, L.L.; Bernardi, F.; Bufalieri, F.; Di Marcotullio, L. Targeting Hedgehog Signalling through the Ubiquitylation Process: The Multiple Roles of the HECT-E3 Ligase Itch. Cells 2019, 8, 98. [Google Scholar] [CrossRef]
- Salah, Z.; Melino, G.; Aqeilan, R.I. Negative Regulation of the Hippo Pathway by E3 Ubiquitin Ligase ITCH Is Sufficient to Promote Tumorigenicity. Cancer Res. 2011, 71, 2010–2020. [Google Scholar] [CrossRef]
- Angers, A.; Ramjaun, A.R.; McPherson, P.S. The HECT Domain Ligase Itch Ubiquitinates Endophilin and Localizes to the trans-Golgi Network and Endosomal System. J. Biol. Chem. 2004, 279, 11471–11479. [Google Scholar] [CrossRef]
- Fang, D.; Kerppola, T.K. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 14782–14787. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Chen, C. WWP1: A versatile ubiquitin E3 ligase in signaling and diseases. Cell. Mol. Life Sci. 2012, 69, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Laine, A.; Ronai, Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 2007, 26, 1477–1483. [Google Scholar] [CrossRef]
- McDonald, F.J.; Western, A.H.; McNeil, J.D.; Thomas, B.C.; Olson, D.R.; Snyder, P.M. Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na+ channel. Am. J. Physiol.-Renal Physiol. 2002, 283, F431–F436. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Z.; Wang, B.; Zhang, J.; Zhao, Y.; Jin, Y. Wwp2-Mediated Ubiquitination of the RNA Polymerase II Large Subunit in Mouse Embryonic Pluripotent Stem Cells. Mol. Cell. Biol. 2007, 27, 5296–5305. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, X.; Luo, Z. WWP2: A Multifunctional Ubiquitin Ligase Gene. Pathol. Oncol. Res. 2014, 20, 799–803. [Google Scholar] [CrossRef]
- Kavsak, P.; Rasmussen, R.K.; Causing, C.G.; Bonni, S.; Zhu, H.; Thomsen, G.H.; Wrana, J.L. Smad7 Binds to Smurf2 to Form an E3 Ubiquitin Ligase that Targets the TGFβ Receptor for Degradation. Mol. Cell 2000, 6, 1365–1375. [Google Scholar] [CrossRef]
- Zhu, H.; Kavsak, P.; Abdollah, S.; Wrana, J.L.; Thomsen, G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999, 400, 687–693. [Google Scholar] [CrossRef]
- Koganti, P.; Levy-Cohen, G.; Blank, M. Smurfs in Protein Homeostasis, Signaling, and Cancer. Front. Oncol. 2018, 8, 295. [Google Scholar] [CrossRef]
- Lu, K.; Yin, X.; Weng, T.; Xi, S.; Li, L.; Xing, G.; Cheng, X.; Yang, X.; Zhang, L.; He, F. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat. Cell Biol. 2008, 10, 994–1002. [Google Scholar] [CrossRef]
- Nie, J.; Xie, P.; Liu, L.; Xing, G.; Chang, Z.; Yin, Y.; Tian, C.; He, F.; Zhang, L. Smad Ubiquitylation Regulatory Factor 1/2 (Smurf1/2) Promotes p53 Degradation by Stabilizing the E3 Ligase MDM2. J. Biol. Chem. 2010, 285, 22818–22830. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Liu, H.; Xu, C. Estrogen degrades Scribble in endometrial epithelial cells through E3 ubiquitin ligase HECW1 in the development of diffuse adenomyosis. Biol. Reprod. 2020, 102, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Fujita, T.; Ozaki, T.; Kato, C.; Kurose, Y.; Sakamoto, M.; Kato, S.; Goto, T.; Itoyama, Y.; Aoki, M.; et al. NEDL1, a Novel Ubiquitin-protein Isopeptide Ligase for Dishevelled-1, Targets Mutant Superoxide Dismutase-1. J. Biol. Chem. 2004, 279, 11327–11335. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ning, G.; Si, P.; Zhang, C.; Liu, W.; Ge, W.; Cui, K.; Zhang, R.; Ge, S. E3 ubiquitin ligase HECW1 promotes the metastasis of non-small cell lung cancer cells through mediating the ubiquitination of Smad4. Biochem. Cell Biol. 2021, 99, 675–681. [Google Scholar] [CrossRef]
- Lu, L.; Hu, S.; Wei, R.; Qiu, X.; Lu, K.; Fu, Y.; Li, H.; Xing, G.; Li, D.; Peng, R.; et al. The HECT Type Ubiquitin Ligase NEDL2 Is Degraded by Anaphase-promoting Complex/Cyclosome (APC/C)-Cdh1, and Its Tight Regulation Maintains the Metaphase to Anaphase Transition. J. Biol. Chem. 2013, 288, 35637–35650. [Google Scholar] [CrossRef]
- Qiu, X.; Wei, R.; Li, Y.; Zhu, Q.; Xiong, C.; Chen, Y.; Zhang, Y.; Lu, K.; He, F.; Zhang, L. NEDL2 regulates enteric nervous system and kidney development in its Nedd8 ligase activity-dependent manner. Oncotarget 2016, 7, 31440–31453. [Google Scholar] [CrossRef][Green Version]
- Ebisawa, T.; Fukuchi, M.; Murakami, G.; Chiba, T.; Tanaka, K.; Imamura, T.; Miyazono, K. Smurf1 Interacts with Transforming Growth Factor-β Type I Receptor through Smad7 and Induces Receptor Degradation. J. Biol. Chem. 2001, 276, 12477–12480. [Google Scholar] [CrossRef]
- Kuratomi, G.; Komuro, A.; Goto, K.; Shinozaki, M.; Miyazawa, K.; Miyazono, K.; Imamura, T. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor. Biochem. J. 2005, 386, 461–470. [Google Scholar] [CrossRef]
- Liu, X.; Elia, A.E.H.; Law, S.F.; Golemis, E.A.; Farley, J.; Wang, T. A novel ability of Smad3 to regulate proteasomal degradation of a Cas family member HEF1. EMBO J. 2000, 19, 6759–6769. [Google Scholar] [CrossRef]
- Murakami, G.; Watabe, T.; Takaoka, K.; Miyazono, K.; Imamura, T. Cooperative Inhibition of Bone Morphogenetic Protein Signaling by Smurf1 and Inhibitory Smads. Mol. Biol. Cell 2003, 14, 2809–2817. [Google Scholar] [CrossRef]
- Seo, S.R.; Lallemand, F.; Ferrand, N.; Pessah, M.; L’Hoste, S.; Camonis, J.; Atfi, A. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 2004, 23, 3780–3792. [Google Scholar] [CrossRef] [PubMed]
- Howitt, J.; Lackovic, J.; Low, L.-H.; Naguib, A.; MacIntyre, A.; Goh, C.-P.; Callaway, J.K.; Hammond, V.; Thomas, T.; Dixon, M.; et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. J. Cell Biol. 2012, 196, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Oliver, P.M.; Cao, X.; Worthen, G.S.; Shi, P.; Briones, N.; MacLeod, M.; White, J.; Kirby, P.; Kappler, J.; Marrack, P.; et al. Ndfip1 Protein Promotes the Function of Itch Ubiquitin Ligase to Prevent T Cell Activation and T Helper 2 Cell-Mediated Inflammation. Immunity 2006, 25, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Bruce, M.C.; Kanelis, V.; Fouladkou, F.; Debonneville, A.; Staub, O.; Rotin, D. Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 2008, 415, 155–163. [Google Scholar] [CrossRef]
- Sluimer, J.; Distel, B. Regulating the human HECT E3 ligases. Cell. Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef]
- Ernst, C. Proliferation and Differentiation Deficits are a Major Convergence Point for Neurodevelopmental Disorders. Trends Neurosci. 2016, 39, 290–299. [Google Scholar] [CrossRef]
- Narimatsu, M.; Bose, R.; Pye, M.; Zhang, L.; Miller, B.; Ching, P.; Sakuma, R.; Luga, V.; Roncari, L.; Attisano, L.; et al. Regulation of Planar Cell Polarity by Smurf Ubiquitin Ligases. Cell 2009, 137, 295–307. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Zhou, J.; Luo, S.; Huang, R.; Zhao, C.; Diao, A. Nedd4 E3 ubiquitin ligase promotes cell proliferation and autophagy. Cell Prolif. 2015, 48, 338–347. [Google Scholar] [CrossRef]
- Choi, B.H.; Che, X.; Chen, C.; Lu, L.; Dai, W. WWP2 is required for normal cell cycle progression. Genes Cancer 2015, 6, 371–377. [Google Scholar] [CrossRef][Green Version]
- Persaud, A.; Alberts, P.; Hayes, M.; Guettler, S.; Clarke, I.; Sicheri, F.; Dirks, P.; Ciruna, B.; Rotin, D. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 2011, 30, 3259–3273. [Google Scholar] [CrossRef]
- Ohkubo, Y.; Uchida, A.O.; Shin, D.; Partanen, J.; Vaccarino, F.M. Fibroblast Growth Factor Receptor 1 Is Required for the Proliferation of Hippocampal Progenitor Cells and for Hippocampal Growth in Mouse. J. Neurosci. 2004, 24, 6057–6069. [Google Scholar] [CrossRef] [PubMed]
- Di Marcotullio, L.; Greco, A.; Mazzà, D.; Canettieri, G.; Pietrosanti, L.; Infante, P.; Coni, S.; Moretti, M.; De Smaele, E.; Ferretti, E.; et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene 2011, 30, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Lohr, N.J.; Molleston, J.P.; Strauss, K.A.; Torres-Martinez, W.; Sherman, E.A.; Squires, R.H.; Rider, N.L.; Chikwava, K.R.; Cummings, O.W.; Morton, D.H.; et al. Human ITCH E3 Ubiquitin Ligase Deficiency Causes Syndromic Multisystem Autoimmune Disease. Am. J. Hum. Genet. 2010, 86, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Ambrozkiewicz, M.C.; Schwark, M.; Kishimoto-Suga, M.; Borisova, E.; Hori, K.; Salazar-Lázaro, A.; Rusanova, A.; Altas, B.; Piepkorn, L.; Bessa, P.; et al. Polarity Acquisition in Cortical Neurons Is Driven by Synergistic Action of Sox9-Regulated Wwp1 and Wwp2 E3 Ubiquitin Ligases and Intronic miR-140. Neuron 2018, 100, 1097–1115.e15. [Google Scholar] [CrossRef]
- Bryan, B.; Cai, Y.; Wrighton, K.; Wu, G.; Feng, X.-H.; Liu, M. Ubiquitination of RhoA by Smurf1 promotes neurite outgrowth. FEBS Lett. 2005, 579, 1015–1019. [Google Scholar] [CrossRef]
- Cheng, P.-L.; Lu, H.; Shelly, M.; Gao, H.; Poo, M.-M. Phosphorylation of E3 Ligase Smurf1 Switches Its Substrate Preference in Support of Axon Development. Neuron 2011, 69, 231–243. [Google Scholar] [CrossRef]
- Kawabe, H.; Neeb, A.; Dimova, K.; Young, S.; Takeda, M.; Katsurabayashi, S.; Mitkovski, M.; Malakhova, O.A.; Zhang, D.-E.; Umikawa, M.; et al. Regulation of Rap2A by the Ubiquitin Ligase Nedd4-1 Controls Neurite Development. Neuron 2010, 65, 358–372. [Google Scholar] [CrossRef]
- Hsia, H.-E.; Kumar, R.; Luca, R.; Takeda, M.; Courchet, J.; Nakashima, J.; Wu, S.; Goebbels, S.; An, W.; Eickholt, B.J.; et al. Ubiquitin E3 ligase Nedd4-1 acts as a downstream target of PI3K/PTEN-mTORC1 signaling to promote neurite growth. Proc. Natl. Acad. Sci. USA 2014, 111, 13205–13210. [Google Scholar] [CrossRef]
- Stouffs, K.; Verloo, P.; Brock, S.; Régal, L.; Beysen, D.; Ceulemans, B.; Jansen, A.; Meuwissen, M.E. Recurrent NEDD4L Variant in Periventricular Nodular Heterotopia, Polymicrogyria and Syndactyly. Front. Genet. 2020, 11, 26. [Google Scholar] [CrossRef]
- Miyazaki, K.; Ozaki, T.; Kato, C.; Hanamoto, T.; Fujita, T.; Irino, S.; Watanabe, K.-I.; Nakagawa, T.; Nakagawara, A. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem. Biophys. Res. Commun. 2003, 308, 106–113. [Google Scholar] [CrossRef]
- Killick, R.; Niklison-Chirou, M.; Tomasini, R.; Bano, D.; Rufini, A.; Grespi, F.; Velletri, T.; Tucci, P.; Sayan, B.S.; Conforti, F.; et al. p73: A Multifunctional Protein in Neurobiology. Mol. Neurobiol. 2011, 43, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Acharya, A.; Kavus, H.; Dunn, P.; Nasir, A.; Folk, L.; Withrow, K.; Wentzensen, I.M.; Ruzhnikov, M.R.Z.; Fallot, C.; Smol, T.; et al. Delineating the genotypic and phenotypic spectrum of HECW2-related neurodevelopmental disorders. J. Med. Genet. 2021. [Google Scholar] [CrossRef] [PubMed]
- Berko, E.R.; Cho, M.T.; Eng, C.; Shao, Y.; Sweetser, D.; Waxler, J.; Robin, N.H.; Brewer, F.; Donkervoort, S.; Mohassel, P.; et al. De novo missense variants inHECW2are associated with neurodevelopmental delay and hypotonia. J. Med. Genet. 2017, 54, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Halvardson, J.; Zhao, J.J.; Zaghlool, A.; Wentzel, C.; Georgii-Hemming, P.; Månsson, E.; Sävmarker, H.E.; Brandberg, G.; Zander, C.S.; Thuresson, A.-C.; et al. Mutations in HECW2 are associated with intellectual disability and epilepsy. J. Med. Genet. 2016, 53, 697–704. [Google Scholar] [CrossRef]
- Ullman, N.L.; Smith-Hicks, C.L.; Desai, S.; Stafstrom, C.E. De Novo HECW2 Mutation Associated With Epilepsy, Developmental Decline, and Intellectual Disability: Case Report and Review of Literature. Pediatr. Neurol. 2018, 85, 76–78. [Google Scholar] [CrossRef]
- Drinjakovic, J.; Jung, H.; Campbell, D.S.; Strochlic, L.; Dwivedy, A.; Holt, C.E. E3 Ligase Nedd4 Promotes Axon Branching by Downregulating PTEN. Neuron 2010, 65, 341–357. [Google Scholar] [CrossRef]
- Lin, A.; Hou, Q.; Jarzylo, L.; Amato, S.; Gilbert, J.; Shang, F.; Man, H.-Y. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J. Neurochem. 2011, 119, 27–39. [Google Scholar] [CrossRef]
- Han, C.; Cui, K.; Bi, X.; Wang, L.; Sun, M.; Yang, L.; Liu, L. Association between polymorphism of the NEDD4 gene and cognitive dysfunction of schizophrenia patients in Chinese Han population. BMC Psychiatry 2019, 19, 405. [Google Scholar] [CrossRef]
- Ekberg, J.A.; Boase, N.A.; Rychkov, G.; Manning, J.; Poronnik, P.; Kumar, S. Nedd4-2 (NEDD4L) controls intracellular Na+-mediated activity of voltage-gated sodium channels in primary cortical neurons. Biochem. J. 2014, 457, 27–31. [Google Scholar] [CrossRef]
- Dibbens, L.M.; Ekberg, J.; Taylor, I.; Hodgson, B.L.; Conroy, S.-J.; Lensink, I.L.; Kumar, S.; Zielinski, M.A.; Harkin, L.A.; Sutherland, G.R.; et al. NEDD4-2 as a potential candidate susceptibility gene for epileptic photosensitivity. Genes Brain Behav. 2007, 6, 750–755. [Google Scholar] [CrossRef]
- Moortgat, S.; Berland, S.; Aukrust, I.; Maystadt, I.; Baker, L.; Benoit, V.; Caro-Llopis, A.; Cooper, N.S.; Debray, F.G.; Faivre, L.; et al. HUWE1 variants cause dominant X-linked intellectual disability: A clinical study of 21 patients. Eur. J. Hum. Genet. 2018, 26, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.W.K.; Zhang, Z.N.; Kim, T.Y. Volunteering and health benefits in general adults: Cumulative effects and forms. BMC Public Health 2017, 18, 8. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Lanznaster, D.; Hergesheimer, R.; Vourc’H, P.; Corcia, P.; Blasco, H. TDP43 aggregates: The ‘Schrödinger’s cat’ in amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 2021, 22, 514. [Google Scholar] [CrossRef] [PubMed]
- Sap, K.A.; Reits, E.A. Strategies to Investigate Ubiquitination in Huntington’s Disease. Front. Chem. 2020, 8, 485. [Google Scholar] [CrossRef]
- Sugeno, N.; Hasegawa, T.; Tanaka, N.; Fukuda, M.; Wakabayashi, K.; Oshima, R.; Konno, M.; Miura, E.; Kikuchi, A.; Baba, T.; et al. Lys-63-linked Ubiquitination by E3 Ubiquitin Ligase Nedd4-1 Facilitates Endosomal Sequestration of Internalized α-Synuclein. J. Biol. Chem. 2014, 289, 18137–18151. [Google Scholar] [CrossRef]
- Tofaris, G.K.; Kim, H.T.; Hourez, R.; Jung, J.-W.; Kim, K.P.; Goldberg, A.L. Ubiquitin ligase Nedd4 promotes α-synuclein degradation by the endosomal–lysosomal pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 17004–17009. [Google Scholar] [CrossRef]
- Davies, S.E.; Hallett, P.; Moens, T.; Smith, G.; Mangano, E.; Kim, H.T.; Goldberg, A.L.; Liu, J.-L.; Isacson, O.; Tofaris, G.K. Enhanced ubiquitin-dependent degradation by Nedd4 protects against α-synuclein accumulation and toxicity in animal models of Parkinson’s disease. Neurobiol. Dis. 2014, 64, 79–87. [Google Scholar] [CrossRef]
- Chai, A.B.; Callaghan, R.; Gelissen, I.C. The Ubiquitin E3 Ligase Nedd4 Regulates the Expression and Amyloid-β Peptide Export Activity of P-Glycoprotein. Int. J. Mol. Sci. 2022, 23, 1019. [Google Scholar] [CrossRef]
- Chhangani, D.; Upadhyay, A.; Amanullah, A.; Joshi, V.; Mishra, A. Ubiquitin ligase ITCH recruitment suppresses the aggregation and cellular toxicity of cytoplasmic misfolded proteins. Sci. Rep. 2014, 4, 5077. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Z.; Alimandi, M.; Chen, C. WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer. Oncogene 2009, 28, 2948–2958. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Fukuda-Yuzawa, Y.; Yoshimura, J.; Toyoda, A.; Kurppa, K.; Moritoyo, H.; Belzil, V.V.; Dion, P.A.; Higasa, K.; Doi, K.; et al. ERBB4 Mutations that Disrupt the Neuregulin-ErbB4 Pathway Cause Amyotrophic Lateral Sclerosis Type 19. Am. J. Hum. Genet. 2013, 93, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kaneko, S.; Wate, R.; Asayama, S.; Nakamura, Y.; Fujita, K.; Ito, H.; Kusaka, H. Regionally different immunoreactivity for Smurf2 and pSmad2/3 in TDP-43-positive inclusions of amyotrophic lateral sclerosis. Neuropathol. Appl. Neurobiol. 2013, 39, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Haraguchi, S.; Koda, T.; Hashimoto, K.; Nakagawara, A. Muscle Atrophy and Motor Neuron Degeneration in Human NEDL1 Transgenic Mice. J. Biomed. Biotechnol. 2011, 2011, 831092. [Google Scholar] [CrossRef]
- Bianchi, V.E.; Locatelli, V.; Rizzi, L. Neurotrophic and Neuroregenerative Effects of GH/IGF1. Int. J. Mol. Sci. 2017, 18, 2441. [Google Scholar] [CrossRef]
- Kwak, Y.-D.; Wang, B.; Li, J.J.; Wang, R.; Deng, Q.; Diao, S.; Chen, Y.; Xu, R.; Masliah, E.; Xu, H.; et al. Upregulation of the E3 ligase NEDD4-1 by Oxidative Stress Degrades IGF-1 Receptor Protein in Neurodegeneration. J. Neurosci. 2012, 32, 10971–10981. [Google Scholar] [CrossRef]
- Kim, E.; Wang, B.; Sastry, N.; Masliah, E.; Nelson, P.T.; Cai, H.; Liao, F.-F. NEDD4-mediated HSF1 degradation underlies α-synucleinopathy. Hum. Mol. Genet. 2015, 25, 211–222. [Google Scholar] [CrossRef]
- Xing, L.; Guo, H.; Wang, D.; Sun, L.; Pan, S. Protective effects and mechanisms of Ndfipl on SH-SY5Y cell apoptosis in an in vitro Parkinson’s disease model. Genet. Mol. Res. 2016, 15, gmr.15026963. [Google Scholar] [CrossRef]
- Canal, M.; Martín-Flores, N.; Pérez-Sisqués, L.; Romaní-Aumedes, J.; Altas, B.; Man, H.-Y.; Kawabe, H.; Alberch, J.; Malagelada, C. Loss of NEDD4 contributes to RTP801 elevation and neuron toxicity: Implications for Parkinson’s disease. Oncotarget 2016, 7, 58813–58831. [Google Scholar] [CrossRef]
- Makioka, K.; Yamazaki, T.; Takatama, M.; Ikeda, M.; Okamoto, K. Immunolocalization of Smurf1 in Hirano bodies. J. Neurol. Sci. 2014, 336, 24–28. [Google Scholar] [CrossRef]
- Shao, L.; Liu, X.; Zhu, S.; Liu, C.; Gao, Y.; Xu, X. The Role of Smurf1 in Neuronal Necroptosis after Lipopolysaccharide-Induced Neuroinflammation. Cell. Mol. Neurobiol. 2018, 38, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Seth, B.; Yadav, A.; Agarwal, S.; Tiwari, S.K.; Chaturvedi, R.K. Inhibition of the transforming growth factor-β/SMAD cascade mitigates the anti-neurogenic effects of the carbamate pesticide carbofuran. J. Biol. Chem. 2017, 292, 19423–19440. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, Y.; Cao, Z.; Liu, Q.; Cheng, Y. Cerebrospinal Fluid Inflammatory Cytokine Aberrations in Alzheimer’s Disease, Parkinson’s Disease and Amyotrophic Lateral Sclerosis: A Systematic Review and Meta-Analysis. Front. Immunol. 2018, 9, 2122. [Google Scholar] [CrossRef]
- Li, Y.; Ozaki, T.; Kikuchi, H.; Yamamoto, H.; Ohira, M.; Nakagawara, A. A novel HECT-type E3 ubiquitin protein ligase NEDL1 enhances the p53-mediated apoptotic cell death in its catalytic activity-independent manner. Oncogene 2008, 27, 3700–3709. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, G.; Holtzman, D.M. Amyloid-β and Tau at the Crossroads of Alzheimer’s Disease. Adv. Exp. Med. Biol. 2019, 1184, 187–203. [Google Scholar] [CrossRef]
- Rodrigues, E.M.; Scudder, S.L.; Goo, M.S.; Patrick, G.N. Aβ-Induced Synaptic Alterations Require the E3 Ubiquitin Ligase Nedd4-1. J. Neurosci. 2016, 36, 1590–1595. [Google Scholar] [CrossRef]
- Park, M.; Jung, H.-G.; Kweon, H.-J.; Kim, Y.-E.; Park, J.-Y.; Hwang, E.M. The E3 ubiquitin ligase, NEDD4L (NEDD4-2) regulates bestrophin-1 (BEST1) by ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2019, 514, 344–350. [Google Scholar] [CrossRef]
- Lau, A.; Tymianski, M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflüg. Arch. Eur. J. Physiol. 2010, 460, 525–542. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Meng, X.; Wu, X.; Tong, H.; Zhang, X.; Qu, S. Regulation of glutamate transporter trafficking by Nedd4-2 in a Parkinson’s disease model. Cell Death Dis. 2017, 8, e2574. [Google Scholar] [CrossRef]
- García-Tardón, N.; González-González, I.M.; Martínez-Villarreal, J.; Fernández-Sánchez, E.; Gimenez, C.; Zafra, F. Protein Kinase C (PKC)-promoted Endocytosis of Glutamate Transporter GLT-1 Requires Ubiquitin Ligase Nedd4-2-dependent Ubiquitination but Not Phosphorylation. J. Biol. Chem. 2012, 287, 19177–19187. [Google Scholar] [CrossRef]
- Weber, J.; Polo, S.; Maspero, E. HECT E3 Ligases: A Tale With Multiple Facets. Front. Physiol. 2019, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Dudeck, M.A.; Horan, T.C.; Peterson, K.D.; Allen-Bridson, K.; Morrell, G.; Pollock, D.A.; Edwards, J.R. National Healthcare Safety Network (NHSN) Report, data summary for 2010, device-associated module. Am. J. Infect. Control. 2011, 39, 798–816. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gu, H.; Zhang, E.; Chen, Q.; Cao, W.; Yan, H.; Chen, J.; Yang, L.; Lv, N.; He, J.; et al. The NEDD4-1 E3 ubiquitin ligase: A potential molecular target for bortezomib sensitivity in multiple myeloma. Int. J. Cancer 2020, 146, 1963–1978. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Rotblat, B.; Ansell, K.; Amelio, I.; Caraglia, M.; Misso, G.; Bernassola, F.; Cavasotto, C.N.; Knight, R.A.; Ciechanover, A.; et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis. 2014, 5, e1203. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haouari, S.; Vourc’h, P.; Jeanne, M.; Marouillat, S.; Veyrat-Durebex, C.; Lanznaster, D.; Laumonnier, F.; Corcia, P.; Blasco, H.; Andres, C.R. The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 3882. https://doi.org/10.3390/ijms23073882
Haouari S, Vourc’h P, Jeanne M, Marouillat S, Veyrat-Durebex C, Lanznaster D, Laumonnier F, Corcia P, Blasco H, Andres CR. The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. International Journal of Molecular Sciences. 2022; 23(7):3882. https://doi.org/10.3390/ijms23073882
Chicago/Turabian StyleHaouari, Shanez, Patrick Vourc’h, Médéric Jeanne, Sylviane Marouillat, Charlotte Veyrat-Durebex, Débora Lanznaster, Frédéric Laumonnier, Philippe Corcia, Hélène Blasco, and Christian R. Andres. 2022. "The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration" International Journal of Molecular Sciences 23, no. 7: 3882. https://doi.org/10.3390/ijms23073882
APA StyleHaouari, S., Vourc’h, P., Jeanne, M., Marouillat, S., Veyrat-Durebex, C., Lanznaster, D., Laumonnier, F., Corcia, P., Blasco, H., & Andres, C. R. (2022). The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. International Journal of Molecular Sciences, 23(7), 3882. https://doi.org/10.3390/ijms23073882








