Isoprene Emission Influences the Proteomic Profile of Arabidopsis Plants under Well-Watered and Drought-Stress Conditions
Abstract
1. Introduction
2. Results
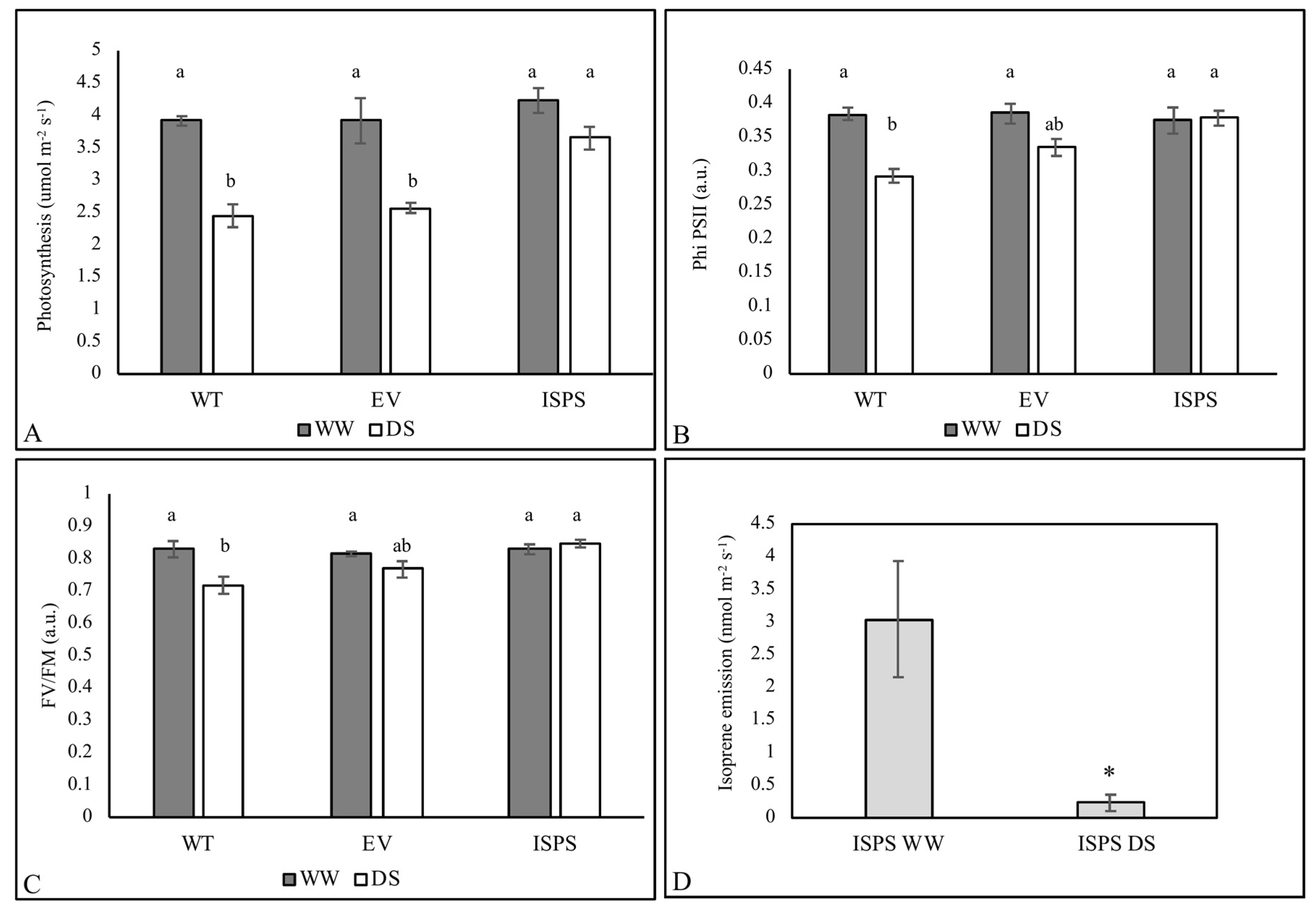
2.1. Physiological Data
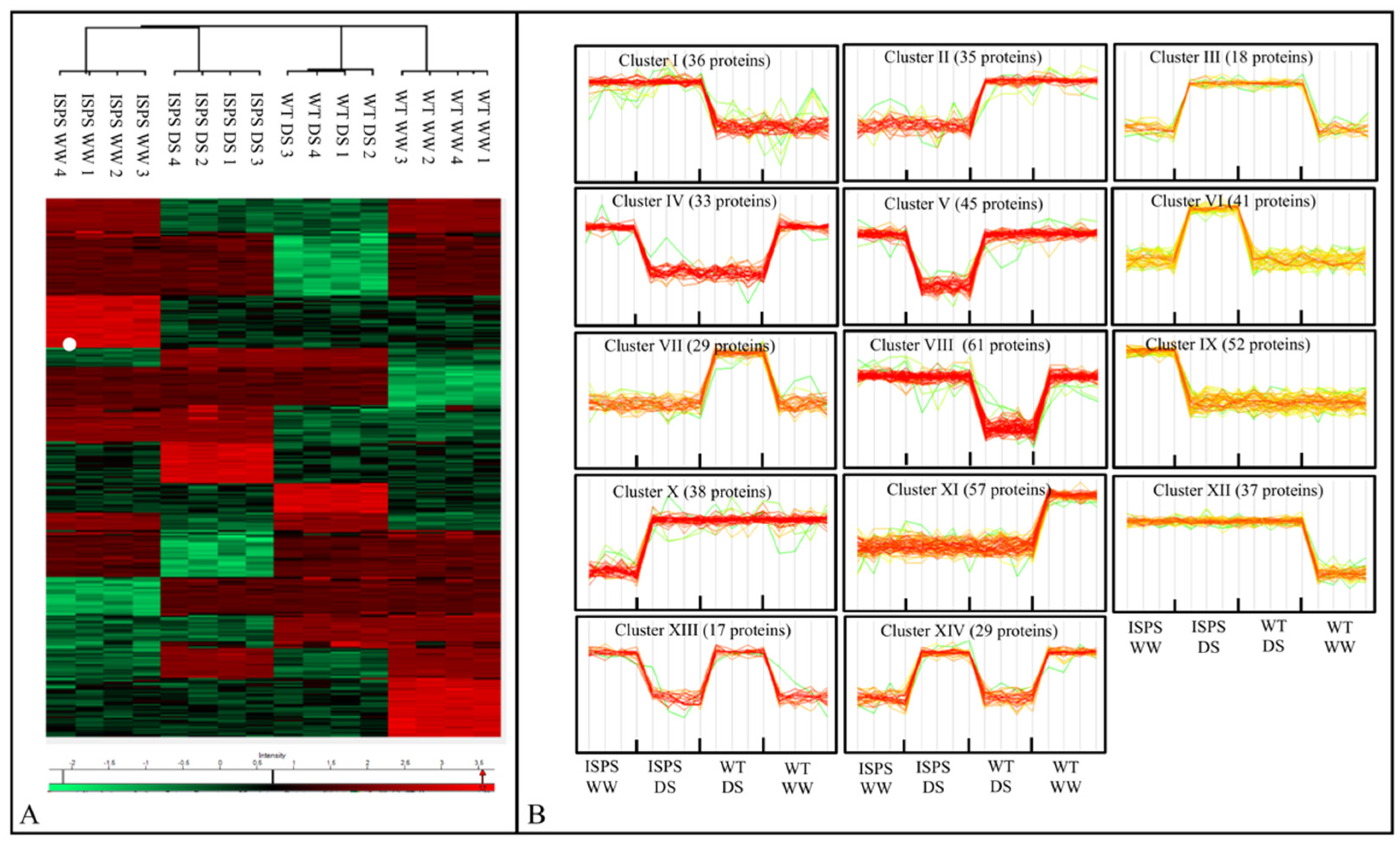
2.2. Overview of Proteomic Profiles
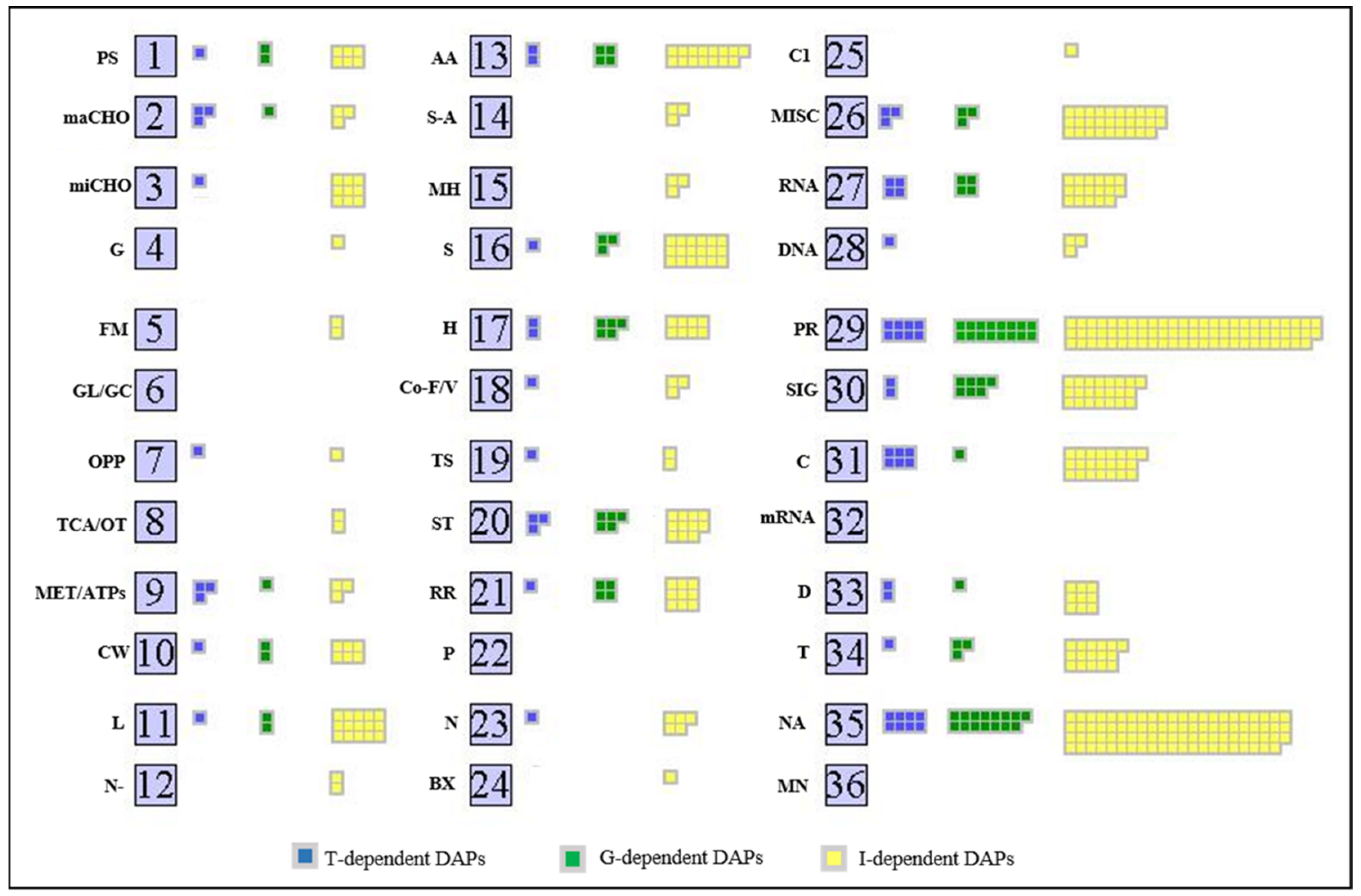
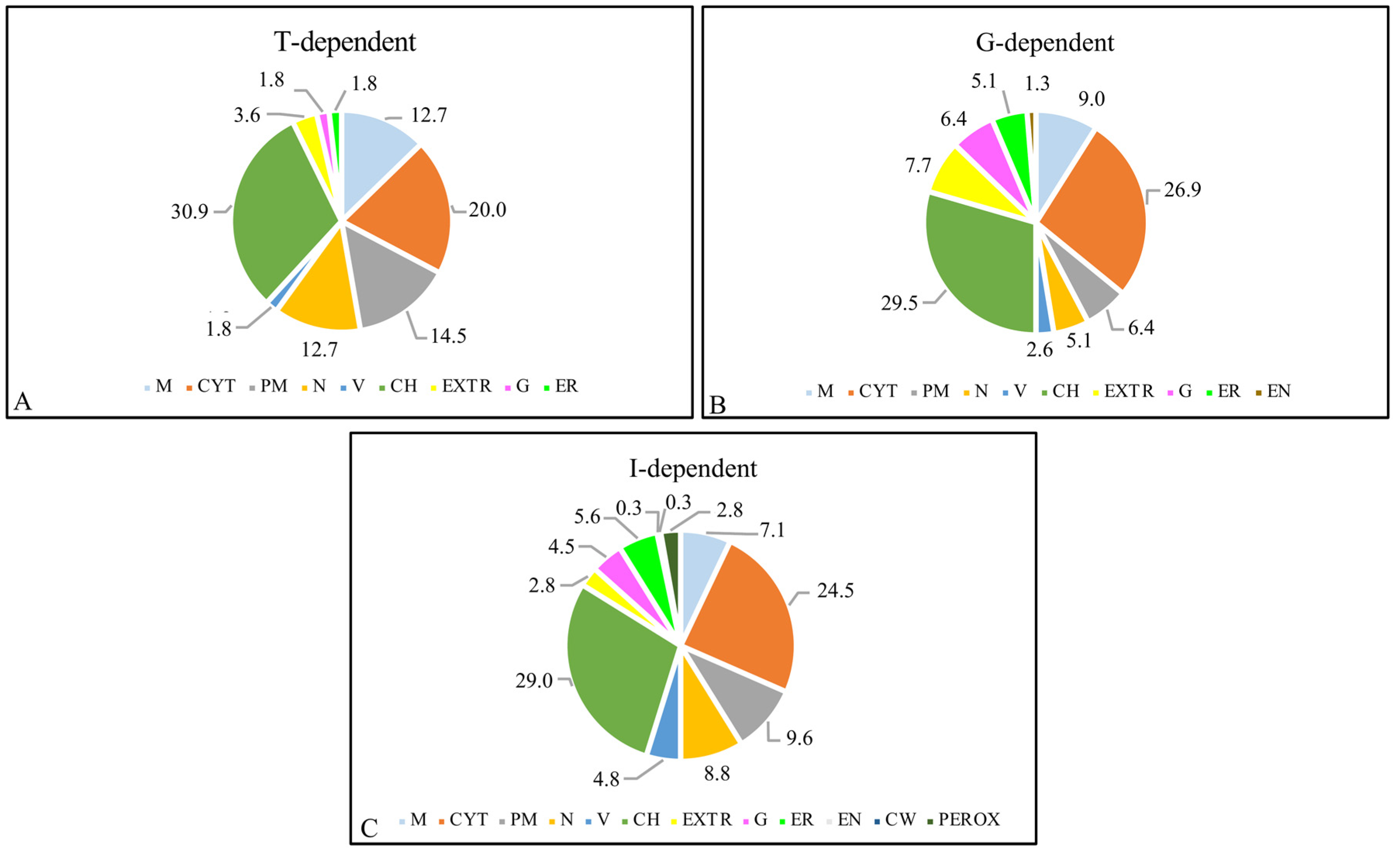
2.3. Analysis of Proteomic Differences
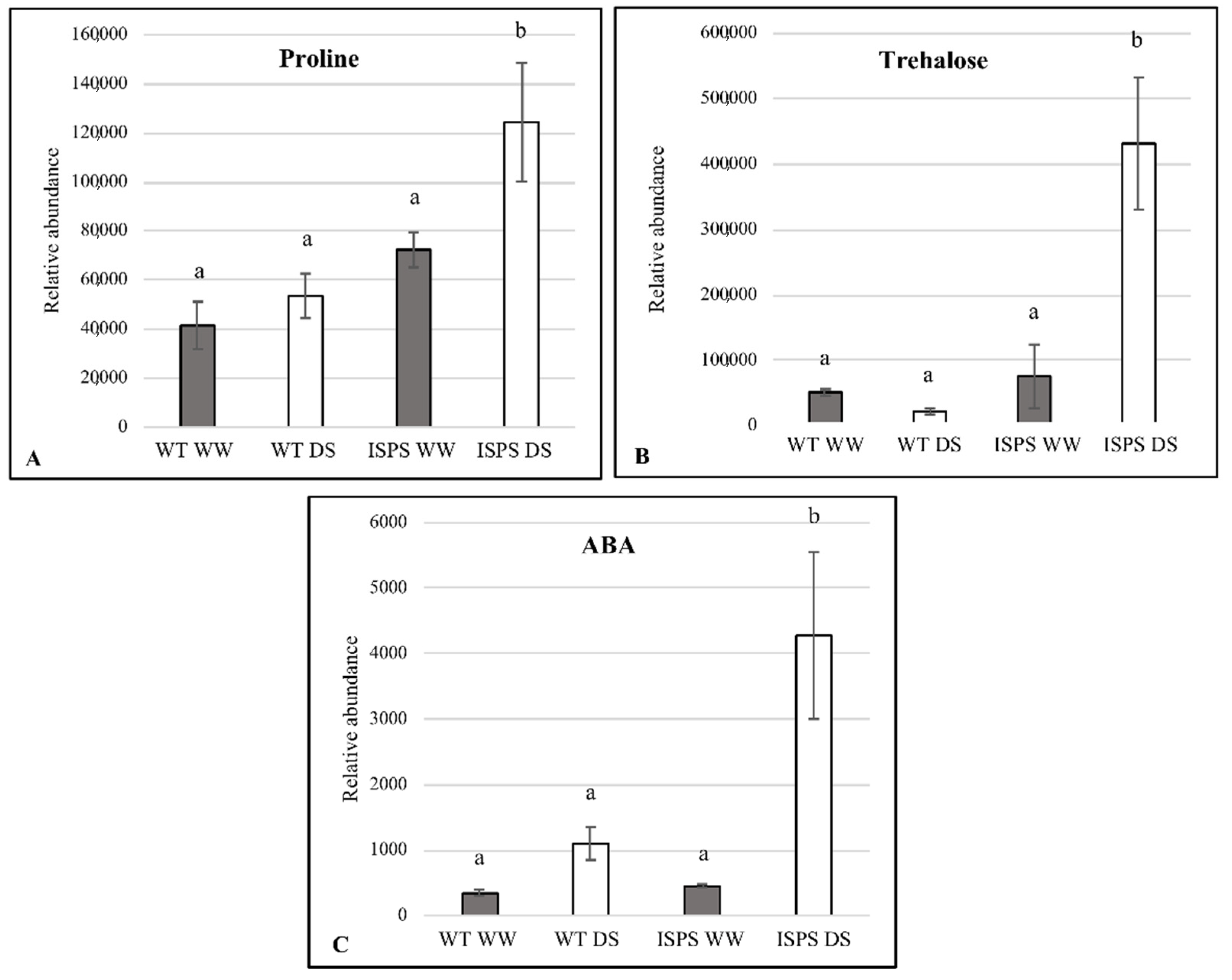
2.4. Free Proline, Trehalose, and ABA Accumulation
3. Discussion
3.1. Genotype-Dependent Daps and Underlying Processes
3.2. Treatment-Dependent Daps and Underlying Processes
3.3. Genotype–Treatment Interaction-Dependent Daps and Underlying Processes
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Drought-Stress Treatment
4.2. Leaf Gas–Exchange and Chlorophyll Fluorescence Measurements
4.3. Biomass and Leaf Relative Water Content and Measurements
4.4. Protein Extraction
4.5. Trypsin Digestion
4.6. LC–MS/MS Analysis and Elaboration of Raw Data
4.7. Quantification of Free-Proline Content
4.8. Determination of ABA Content
4.9. Trehalose Determination
4.10. Statistical Analysis
4.11. Downstream Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, M.; He, C.-Q.; Ding, N.-Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef]
- Loreto, F.; Schnitzler, J.-P. Abiotic stresses and induced BVOCs. Trends Plant Sci. 2010, 15, 154–166. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Schwender, J.; Disch, A.; Rohmer, M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997, 400, 271–274. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Yeh, S. Isoprene emission from plants. Annu. Rev. Plant Biol. 2001, 52, 407–436. [Google Scholar] [CrossRef]
- Loreto, F.; Fineschi, S. Reconciling functions and evolution of isoprene emission in higher plants. New Phytol. 2015, 206, 578–582. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Chen, X.; Yeh, S. Isoprene increases thermotolerance of fosmidomycin-fed leaves. Plant Physiol. 2001, 125, 2001–2006. [Google Scholar] [CrossRef]
- Loivamäki, M.; Louis, S.; Cinege, G.; Zimmer, I.; Fischbach, R.J.; Schnitzler, J.-P. Circadian rhythms of isoprene biosynthesis in grey poplar leaves. Plant Physiol. 2007, 143, 540–551. [Google Scholar] [CrossRef]
- Sasaki, K.; Saito, T.; Lämsä, M.; Oksman-Caldentey, K.-M.; Suzuki, M.; Ohyama, K.; Muranaka, T.; Ohara, K.; Yazaki, K. Plants utilize isoprene emission as a thermotolerance mechanism. Plant Cell Physiol. 2007, 48, 1254–1262. [Google Scholar] [CrossRef]
- Velikova, V.; Loreto, F. On the relationship between isoprene emission and thermotolerance in Phragmites australis leaves exposed to high temperatures and during the recovery from a heat stress. Plant Cell Environ. 2005, 28, 318–327. [Google Scholar] [CrossRef]
- Tattini, M.; Loreto, F.; Fini, A.; Guidi, L.; Brunetti, C.; Velikova, V.; Gori, A.; Ferrini, F. Isoprenoids and phenylpropanoids are part of the antioxidant defense orchestrated daily by drought-stressed P latanus× acerifolia plants during Mediterranean summers. New Phytol. 2015, 207, 613–626. [Google Scholar] [CrossRef]
- Vickers, C.E.; Gershenzon, J.; Lerdau, M.T.; Loreto, F. A unified mechanism of action for volatile isoprenoids in plant abiotic stress. Nat. Chem. Biol. 2009, 5, 283–291. [Google Scholar] [CrossRef]
- Monson, R.K.; Winkler, B.; Rosenstiel, T.N.; Block, K.; Merl-Pham, J.; Strauss, S.H.; Ault, K.; Maxfield, J.; Moore, D.J.; Trahan, N.A. High productivity in hybrid-poplar plantations without isoprene emission to the atmosphere. Proc. Natl. Acad. Sci. USA 2020, 117, 1596–1605. [Google Scholar] [CrossRef]
- Velikova, V.; Várkonyi, Z.; Szabó, M.; Maslenkova, L.; Nogues, I.; Kovács, L.; Peeva, V.; Busheva, M.; Garab, G.; Sharkey, T.D. Increased thermostability of thylakoid membranes in isoprene-emitting leaves probed with three biophysical techniques. Plant Physiol. 2011, 157, 905–916. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Singsaas, E.L. Why plants emit isoprene. Nature 1995, 374, 769. [Google Scholar] [CrossRef]
- Pollastri, S.; Jorba, I.; Hawkins, T.J.; Llusià, J.; Michelozzi, M.; Navajas, D.; Peñuelas, J.; Hussey, P.J.; Knight, M.R.; Loreto, F. Leaves of isoprene-emitting tobacco plants maintain PSII stability at high temperatures. New Phytol. 2019, 223, 1307–1318. [Google Scholar] [CrossRef]
- Pollastri, S.; Tsonev, T.; Loreto, F. Isoprene improves photochemical efficiency and enhances heat dissipation in plants at physiological temperatures. J. Exp. Bot. 2014, 65, 1565–1570. [Google Scholar] [CrossRef]
- Harvey, C.M.; Sharkey, T.D. Exogenous isoprene modulates gene expression in unstressed Arabidopsis thaliana plants. Plant Cell Environ. 2016, 39, 1251–1263. [Google Scholar] [CrossRef]
- Zuo, Z.; Weraduwage, S.M.; Lantz, A.T.; Sanchez, L.M.; Weise, S.E.; Wang, J.; Childs, K.L.; Sharkey, T.D. Isoprene Acts as a Signaling Molecule in Gene Networks Important for Stress Responses and Plant Growth. Plant Physiol 2019, 180, 124–152. [Google Scholar] [CrossRef]
- Vanzo, E.; Merl-Pham, J.; Velikova, V.; Ghirardo, A.; Lindermayr, C.; Hauck, S.M.; Bernhardt, J.; Riedel, K.; Durner, J.; Schnitzler, J.-P. Modulation of protein S-nitrosylation by isoprene emission in poplar. Plant Physiol. 2016, 170, 1945–1961. [Google Scholar] [CrossRef]
- Velikova, V.; Ghirardo, A.; Vanzo, E.; Merl, J.; Hauck, S.M.; Schnitzler, J.-P. Genetic manipulation of isoprene emissions in poplar plants remodels the chloroplast proteome. J. Proteome Res. 2014, 13, 2005. [Google Scholar] [CrossRef]
- Behnke, K.; Kaiser, A.; Zimmer, I.; Brüggemann, N.; Janz, D.; Polle, A.; Hampp, R.; Hänsch, R.; Popko, J.; Schmitt-Kopplin, P. RNAi-mediated suppression of isoprene emission in poplar transiently impacts phenolic metabolism under high temperature and high light intensities: A transcriptomic and metabolomic analysis. Plant Mol. Biol. 2010, 74, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Ghirardo, A.; Wright, L.P.; Bi, Z.; Rosenkranz, M.; Pulido, P.; Rodríguez-Concepción, M.; Niinemets, Ü.; Brüggemann, N.; Gershenzon, J.; Schnitzler, J.-P. Metabolic flux analysis of plastidic isoprenoid biosynthesis in poplar leaves emitting and nonemitting isoprene. Plant Physiol. 2014, 165, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Way, D.A.; Ghirardo, A.; Kanawati, B.; Esperschütz, J.; Monson, R.K.; Jackson, R.B.; Schmitt-Kopplin, P.; Schnitzler, J.P. Increasing atmospheric CO 2 reduces metabolic and physiological differences between isoprene-and non-isoprene-emitting poplars. New Phytol. 2013, 200, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Velikova, V.; Vickers, C.; Brunetti, C.; Di Ferdinando, M.; Trivellini, A.; Fineschi, S.; Agati, G.; Ferrini, F.; Loreto, F. Isoprene production in transgenic tobacco alters isoprenoid, non-structural carbohydrate and phenylpropanoid metabolism, and protects photosynthesis from drought stress. Plant Cell Environ. 2014, 37, 1950–1964. [Google Scholar] [CrossRef]
- Lantz, A.T.; Allman, J.; Weraduwage, S.M.; Sharkey, T.D. Isoprene: New insights into the control of emission and mediation of stress tolerance by gene expression. Plant Cell Environ. 2019, 42, 2808–2826. [Google Scholar] [CrossRef]
- Pollastri, S.; Baccelli, I.; Loreto, F. Isoprene: An Antioxidant Itself or a Molecule with Multiple Regulatory Functions in Plants? Antioxidants 2021, 10, 684. [Google Scholar] [CrossRef]
- Schneider, A.; Steinberger, I.; Strissel, H.; Kunz, H.H.; Manavski, N.; Meurer, J.; Burkhard, G.; Jarzombski, S.; Schünemann, D.; Geimer, S. The Arabidopsis Tellurite resistance C protein together with ALB 3 is involved in photosystem II protein synthesis. Plant J. 2014, 78, 344–356. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Ovečka, M.; Basheer, J.; Zapletalová, V.; Šamaj, J.; Takáč, T. FSD1: Developmentally-regulated plastidial, nuclear and cytoplasmic enzyme with anti-oxidative and osmoprotective role. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Essentials of proteolytic machineries in chloroplasts. Mol. Plant 2017, 10, 4–19. [Google Scholar] [CrossRef]
- Harris, A.; Owen, S.M.; Sleep, D.; Pereira, M.G. Constitutive changes in pigment concentrations: Implications for estimating isoprene emissions using the photochemical reflectance index. Physiol. Plant. 2016, 156, 190–200. [Google Scholar] [CrossRef]
- Behnke, K.; Ehlting, B.; Teuber, M.; Bauerfeind, M.; Louis, S.; Hänsch, R.; Polle, A.; Bohlmann, J.; Schnitzler, J.P. Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J. 2007, 51, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Monson, R.K.; Weraduwage, S.M.; Rosenkranz, M.; Schnitzler, J.-P.; Sharkey, T.D. Leaf isoprene emission as a trait that mediates the growth-defense tradeoff in the face of climate stress. Oecologia 2021, 197, 885–902. [Google Scholar] [CrossRef] [PubMed]
- Frank, L.; Wenig, M.; Ghirardo, A.; van der Krol, A.; Vlot, A.C.; Schnitzler, J.P.; Rosenkranz, M. Isoprene and β-caryophyllene confer plant resistance via different plant internal signalling pathways. Plant Cell Environ. 2021, 44, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Srikanta Dani, K.G.; Pollastri, S.; Pinosio, S.; Reichelt, M.; Sharkey, T.D.; Schnitzler, J.P.; Loreto, F. Isoprene enhances leaf cytokinin metabolism and induces early senescence. New Phytol. 2021. [Google Scholar] [CrossRef]
- Ding, Z.; Galván-Ampudia, C.S.; Demarsy, E.; Łangowski, Ł.; Kleine-Vehn, J.; Fan, Y.; Morita, M.T.; Tasaka, M.; Fankhauser, C.; Offringa, R. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 2011, 13, 447–452. [Google Scholar] [CrossRef]
- Rakusová, H.; Abbas, M.; Han, H.; Song, S.; Robert, H.S.; Friml, J. Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr. Biol. 2016, 26, 3026–3032. [Google Scholar] [CrossRef]
- Schröder, F.; Lisso, J.; Müssig, C. EXORDIUM-LIKE1 promotes growth during low carbon availability in Arabidopsis. Plant Physiol. 2011, 156, 1620–1630. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, M.; Gao, C.; Zeng, Y.; Cui, Y.; Shen, W.; Jiang, L. The roles of endomembrane trafficking in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 55–69. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef]
- Schulz, P.; Piepenburg, K.; Lintermann, R.; Herde, M.; Schöttler, M.A.; Schmidt, L.K.; Ruf, S.; Kudla, J.; Romeis, T.; Bock, R. Improving plant drought tolerance and growth under water limitation through combinatorial engineering of signalling networks. Plant Biotechnol. J. 2021, 19, 74–86. [Google Scholar] [CrossRef]
- Chmielewska, K.; Rodziewicz, P.; Swarcewicz, B.; Sawikowska, A.; Krajewski, P.; Marczak, Ł.; Ciesiołka, D.; Kuczyńska, A.; Mikołajczak, K.; Ogrodowicz, P. Analysis of drought-induced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Front. Plant Sci. 2016, 7, 1108. [Google Scholar] [CrossRef] [PubMed]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef]
- Ghirardo, A.; Nosenko, T.; Kreuzwieser, J.; Winkler, J.B.; Kruse, J.; Albert, A.; Merl-Pham, J.; Lux, T.; Ache, P.; Zimmer, I. Protein expression plasticity contributes to heat and drought tolerance of date palm. Oecologia 2021, 197, 903–919. [Google Scholar] [CrossRef]
- Jia, Q.; Kong, D.; Li, Q.; Sun, S.; Song, J.; Zhu, Y.; Liang, K.; Ke, Q.; Lin, W.; Huang, J. The function of inositol phosphatases in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2019, 20, 3999. [Google Scholar] [CrossRef]
- Wu, X.; Qiao, Z.; Liu, H.; Acharya, B.R.; Li, C.; Zhang, W. CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front. Plant Sci. 2017, 8, 824. [Google Scholar] [CrossRef]
- Wang, S.; Kurepa, J.; Smalle, J.A. The Arabidopsis 26S proteasome subunit RPN1a is required for optimal plant growth and stress responses. Plant Cell Physiol. 2009, 50, 1721–1725. [Google Scholar] [CrossRef] [PubMed]
- Meiri, D.; Tazat, K.; Cohen-Peer, R.; Farchi-Pisanty, O.; Aviezer-Hagai, K.; Avni, A.; Breiman, A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 2010, 72, 191. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Fujioka, S.; Takatsuto, S.; Yoshida, S. An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 2002, 130, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, F.; Xie, Q. Balancing growth and adaptation to stress: Crosstalk between brassinosteroid and abscisic acid signaling. Plant Cell Environ. 2020, 43, 2325–2335. [Google Scholar] [CrossRef]
- Hardtke, C.S.; Dorcey, E.; Osmont, K.S.; Sibout, R. Phytohormone collaboration: Zooming in on auxin–brassinosteroid interactions. Trends Cell Biol. 2007, 17, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Halliday, K.J. Plant hormones: The interplay of brassinosteroids and auxin. Curr. Biol. 2004, 14, R1008–R1010. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-W.; Zang, B.-S.; Deng, X.-W.; Wang, X.-P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar] [CrossRef]
- Qamar, A.; Mysore, K.; Senthil-Kumar, M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 2015, 6, 503. [Google Scholar] [CrossRef]
- Barta, C.; Loreto, F. The relationship between the methyl-erythritol phosphate pathway leading to emission of volatile isoprenoids and abscisic acid content in leaves. Plant Physiol. 2006, 141, 1676–1683. [Google Scholar] [CrossRef][Green Version]
- Dani, K.G.S.; Fineschi, S.; Michelozzi, M.; Loreto, F. Do cytokinins, volatile isoprenoids and carotenoids synergically delay leaf senescence? Plant Cell Environ. 2016, 39, 1103–1111. [Google Scholar] [CrossRef]
- Jones, R.J.; Mansfield, T. Suppression of stomatal opening in leaves treated with abscisic acid. J. Exp. Bot. 1970, 21, 714–719. [Google Scholar] [CrossRef]
- Wilkinson, S.; Davies, W.J. ABA-based chemical signalling: The co-ordination of responses to stress in plants. Plant Cell Environ. 2002, 25, 195–210. [Google Scholar] [CrossRef]
- Dietz, K.J.; Sauter, A.; Wichert, K.; Messdaghi, D.; Hartung, W. Extracellular β-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 2000, 51, 937–944. [Google Scholar] [CrossRef]
- Schurr, U.; Schulze, E.D. The concentration of xylem sap constituents in root exudate, and in sap from intact, transpiring castor bean plants (Ricinus communis L.). Plant Cell Environ. 1995, 18, 409–420. [Google Scholar] [CrossRef]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Merilo, E.; Yarmolinsky, D.; Jalakas, P.; Parik, H.; Tulva, I.; Rasulov, B.; Kilk, K.; Kollist, H. Stomatal VPD response: There is more to the story than ABA. Plant Physiol. 2018, 176, 851–864. [Google Scholar] [CrossRef]
- Loreto, F.; Velikova, V. Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 2001, 127, 1781–1787. [Google Scholar] [CrossRef] [PubMed]
- Velikova, V.; Edreva, A.; Loreto, F. Endogenous isoprene protects Phragmites australis leaves against singlet oxygen. Physiol. Plant. 2004, 122, 219–225. [Google Scholar] [CrossRef]
- Khafif, M.; Balagué, C.; Huard-Chauveau, C.; Roby, D. An essential role for the VASt domain of the Arabidopsis VAD1 protein in the regulation of defense and cell death in response to pathogens. PLoS ONE 2017, 12, e0179782. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.; Goring, D.R. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 2002, 50, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Keichinger, C. Manipulating cellular factors to combat viruses: A case study from the plant eukaryotic translation initiation factors eIF4. Front. Microbiol. 2019, 10, 17. [Google Scholar] [CrossRef]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta (BBA) Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Smart, R.E.; Bingham, G.E. Rapid estimates of relative water content. Plant Physiol. 1974, 53, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xiong, E.; Wang, W.; Scali, M.; Cresti, M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 2014, 9, 362. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Seco, D.; Chiapello, M.; Bracale, M.; Pesce, C.; Bagnaresi, P.; Dubois, E.; Moulin, L.; Vannini, C.; Koebnik, R. Transcriptome and proteome analysis reveal new insight into proximal and distal responses of wheat to foliar infection by Xanthomonas translucens. Sci. Rep. 2017, 7, 10157. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
- Ciucanu, I.; Kerek, F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984, 131, 209–217. [Google Scholar] [CrossRef]
- Ciucanu, I.; Costello, C.E. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J. Am. Chem. Soc. 2003, 125, 16213–16219. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef] [PubMed]
| ID TAIR | Protein Description | WT DS vs. WT WW | ISPS WW vs. WT WW | ISPS DS vs. WT WW | ISPS WW vs. WT DS | ISPS WW vs. ISPS DS | ISPS DS vs. WT DS | Suba CC |
|---|---|---|---|---|---|---|---|---|
| GENOTYPE-DEPENDENT DAPS | ||||||||
| CHLOROPLAST | ||||||||
| AT5G10470.1 | KCA1 | −0.08 | 5.98 | 5.97 | 6.07 | 0.02 | 6.05 | CYT |
| AT1G31800.1 | CYP97A3, LUT5 | 0.31 | 6.26 | 6.34 | 5.95 | −0.07 | 6.02 | CH |
| AT1G53520.1 | Chalcone-flavanone isomerase | 0.28 | 4.75 | 4.55 | 4.46 | 0.20 | 4.26 | CH |
| AT4G25100.5 | FSD1 | −0.07 | 4.09 | 5.79 | 4.16 | −1.70 | 5.86 | CH |
| AT5G53170.1 | FTSH11 | −0.03 | 4.40 | 4.22 | 4.43 | 0.18 | 4.25 | CH |
| AT1G12270.1 | Hop1 | −0.08 | 4.39 | 4.56 | 4.47 | −0.17 | 4.64 | CYT |
| ATCG00190.1 | RPOB | −0.68 | 4.08 | 4.55 | 4.77 | −0.47 | 5.23 | CH |
| ENDOMEMBRANE TRAFFICKING | ||||||||
| AT1G01910.4 | GET3A | −0.39 | 7.36 | 7.17 | 7.75 | 0.20 | 7.56 | CYT |
| AT5G16300.3 | COG1 | 0.67 | 3.31 | 3.14 | 2.64 | 0.18 | 2.46 | CYT,G |
| AT4G29160.3 | SNF7.1 | −0.84 | 5.28 | 5.64 | 6.12 | −0.35 | 6.48 | ENDO |
| AT5G54440.1 | CLUB/TRS130 | 0.13 | 4.63 | 4.36 | 4.50 | 0.27 | 4.23 | G |
| AT5G58030.1 | TRAPPC5 | 0.62 | 6.19 | 5.97 | 5.57 | 0.22 | 5.35 | G |
| CELLULAR SIGNALLING | ||||||||
| AT5G07300.1 | BON2 | 0.34 | 5.04 | 4.69 | 4.70 | 0.35 | 4.35 | PM |
| AT1G70940.1 | PIN3 | −0.82 | 4.99 | 4.54 | 5.81 | 0.45 | 5.36 | PM |
| AT3G21220.1 | MKK5 | −0.30 | 2.83 | 3.05 | 3.13 | −0.22 | 3.35 | M |
| AT5G05590.1 | PAI2 | −0.27 | 5.45 | 5.67 | 5.72 | −0.22 | 5.94 | CH |
| AT5G09440.1 | EXL4 | −0.22 | −5.45 | −5.56 | −5.23 | 0.11 | −5.35 | EXTR |
| TREATMENT-DEPENDENT DAPS | ||||||||
| AT5G23040.1 | CDF1 | −4.46 | 0.15 | −4.99 | 4.61 | 5.14 | −0.53 | CH |
| AT4G04850.2 | KEA3 K+ efflux antiporter 3 | −5.20 | −0.13 | −5.56 | 5.07 | 5.43 | −0.36 | CH |
| AT5G48570.1 | FKBP65 | −10.96 | −0.32 | −10.19 | 10.64 | 9.87 | 0.61 | CYT |
| AT5G16010.1 | 3-oxo-5-alpha-steroid 4-dehydrogenase family protein | −6.99 | −0.10 | −6.80 | 6.89 | 6.70 | 0.41 | CH |
| AT3G02260.1 | auxin transport protein (BIG) | −3.90 | 0.07 | −3.39 | 3.97 | 3.46 | −0.07 | CYT |
| AT1G58290.1 | HEMA1 | 4.50 | 0.65 | 5.11 | −3.85 | −4.46 | −0.01 | CH |
| AT3G53560.1 | Tetratricopeptide repeat (TPR)-like superfamily protein | 4.86 | −0.16 | 5.26 | −5.01 | −5.42 | 0.52 | CH |
| AT4G05090.1 | Inositol monophosphatase family protein | 5.81 | 0.48 | 5.74 | −5.33 | −5.26 | 0.77 | CH |
| AT3G50360.1 | CEN2 | 4.07 | 0.21 | 4.06 | −3.87 | −3.86 | 0.19 | CYT |
| AT4G28470.1 | RPN1B | 3.69 | 0.16 | 4.21 | −3.53 | −4.05 | 0.51 | CYT |
| INTERACTION-DEPENDENT DAPS | ||||||||
| ISOPRENE-SPECIFIC RESPONSE TO DROUGHT STRESS | ||||||||
| OSMOPROTECTANS AND REDOX HOMESOTASIS | ||||||||
| AT1G06410.1 | trehalose-phosphatase/synthase 7 | 1.08 | 0.90 | 5.36 | −0.18 | −4.45 | 4.27 | CYT |
| AT3G55610.1 | P5CS2 | 0.52 | −0.02 | 4.45 | −0.54 | −4.47 | 3.93 | CH |
| AT2G26230.1 | urate oxidase | 0.16 | 0.09 | 3.99 | −0.06 | −3.89 | 3.83 | PEROX |
| AT1G75270.1 | DHAR2 | 0.03 | 0.33 | 4.25 | 0.30 | −3.92 | 4.23 | CYT |
| AT5G18100.2 | copper/zinc superoxide dismutase 3 | −0.61 | 0.25 | 4.30 | 0.87 | −4.05 | 4.92 | PEROX |
| AT2G22500.1 | uncoupling protein 5 | −0.12 | 0.06 | 4.35 | 0.17 | −4.30 | 4.47 | M |
| AT5G54430.1 | PHOS32 | 0.46 | 0.43 | 6.17 | −0.04 | −5.74 | 5.71 | CYT |
| AT3G13470.1 | TCP-1/cpn60 chaperonin | 0.03 | 0.24 | 5.11 | 0.21 | −4.87 | 5.08 | CH |
| AT1G04980.1 | PDIL2-2 | −0.07 | −0.03 | 4.08 | 0.03 | −4.11 | 4.14 | ER |
| AT1G12010.1 | ACO3 | −0.06 | 0.28 | 5.18 | 0.34 | −4.90 | 5.24 | CYT |
| AT3G10130.1 | | SOUL heme-binding | −0.15 | −0.03 | −2.60 | 0.12 | 2.57 | −2.45 | CH |
| AT1G21350.3 | Thioredoxin | 0.14 | −0.03 | −5.49 | −0.17 | 5.46 | −5.63 | CH |
| DEFENSE | ||||||||
| AT1G02120.1 | VAD1 | −0.16 | 0.72 | 4.03 | 0.89 | −3.31 | 4.20 | ER |
| AT3G24550.1 | PERK1 | −0.78 | −0.45 | 3.42 | 0.33 | −3.87 | 4.21 | PM |
| AT2G44490.1 | PEN2 | 0.03 | −0.20 | −6.26 | −0.23 | 6.06 | −6.29 | PEROX |
| AT5G35620.1 | Eukaryotic initiation factor 4E protein | −0.29 | 0.12 | −3.26 | 0.40 | 3.37 | −2.97 | N |
| AT1G55210.2 | Disease resistance-responsive (dirigent-like protein) | 0.44 | −0.34 | −7.38 | −0.78 | 7.03 | −7.82 | EXTR |
| AT4G33500.1 | Protein phosphatase 2C | 0.62 | −4.35 | −4.30 | −4.97 | −0.05 | −4.92 | CH |
| AT3G18165.1 | MOS4 | 0.17 | 3.92 | 4.06 | 3.75 | −0.15 | 3.90 | N |
| AT4G16990.2 | RLM3 | 0.12 | 4.83 | 4.94 | 4.71 | −0.11 | 4.82 | CYT |
| DAPS SIMILARLY ALTERED IN WT STRESSED AND ISOPRENE-EMITTING UNSTRESSED PLANTS | ||||||||
| CHLOROPLAST | ||||||||
| AT5G48790.1 | low accumulation PSII protein | 6.06 | 5.66 | 5.53 | −0.40 | 0.13 | −0.53 | CH |
| AT1G32550.1 | FdC2 | 5.13 | 5.64 | 5.97 | 0.51 | −0.33 | 0.84 | CH |
| ATCG00150.1 | ATPI | 7.42 | 7.17 | 7.06 | −0.25 | 0.11 | −0.36 | CH |
| ATCG00170.1 | RPOC2 | 5.06 | 5.74 | 5.25 | 0.68 | 0.49 | 0.19 | CH |
| CELLULAR SIGNALLING | ||||||||
| AT4G35860.1 | RABB1B | 5.81 | 5.52 | 5.69 | −0.29 | −0.17 | −0.12 | CH |
| AT3G44340.2 | CEF | 5.31 | 5.53 | 5.03 | 0.22 | 0.49 | −0.28 | CH |
| AT5G07340.1 | Calreticulin | 3.52 | 3.52 | 3.58 | 0.00 | −0.05 | 0.05 | ER |
| AT4G29810.1 | MKK2 | 4.99 | 5.00 | 5.16 | 0.00 | −0.16 | 0.16 | PM |
| AT3G01090.3 | KIN10 | 6.74 | 6.81 | 6.75 | 0.07 | 0.06 | 0.01 | CYT |
| AT3G11660.1 | NDR1/HIN1-like 1 | 5.51 | 5.33 | 5.24 | −0.18 | 0.09 | −0.27 | PM |
| AT2G21600.1 | RER1B | −3.34 | −3.86 | −3.85 | −0.51 | −0.01 | −0.50 | ER |
| AT5G65950.1 | TRAPPC11 | −6.27 | −6.16 | −6.31 | 0.11 | 0.14 | −0.03 | G |
| AT1G18210.2 | Calcium-binding EF-hand family protein | −5.46 | −6.30 | −6.00 | −0.84 | −0.30 | −0.54 | CYT+ |
| AT3G22370.1 | alternative oxidase 1° | −5.33 | −4.53 | −5.13 | 0.79 | 0.59 | 0.20 | M |
| AT4G23150.1 | CRK7 | −5.31 | −4.86 | −5.69 | 0.45 | 0.83 | −0.38 | PM |
| AT1G49340.2 | ATPI4K ALPHA | −5.83 | −5.15 | −5.50 | 0.68 | 0.35 | 0.33 | PM |
| AT1G76040.2 | CPK29 | −4.13 | −4.27 | −3.81 | −0.14 | −0.46 | 0.31 | CYT |
| DAPS ALTERED BY DROUGHT STRESS IN WT BUT NOT IN TRANSGENIC ARABIDOPSIS | ||||||||
| CHLOROPLAST | ||||||||
| ATCG00520.1 | YCF4 | −5.40 | 0.05 | 0.33 | 5.45 | −0.28 | 5.73 | CH |
| AT3G48870.1 | CLPC | −4.06 | 0.27 | 0.41 | 4.33 | −0.14 | 4.47 | CH |
| AT2G32480.1 | ARASP | −5.14 | 0.12 | 0.06 | 5.26 | 0.06 | 5.19 | CH |
| AT1G06820.1 | CCR2 carotenoid isomerase | −4.43 | −0.11 | 0.03 | 4.32 | −0.14 | 4.45 | CH |
| AT2G26540.1 | UROS uroporphyrinogen-III synthase | −4.81 | 0.13 | −0.13 | 4.94 | 0.26 | 4.68 | CH |
| AT1G09940.1 | HEMA2 | 7.41 | 0.00 | −0.32 | −7.42 | 0.32 | −7.74 | CH |
| AT1G17050.1 | SPS2 solanesyl diphosphate synthase 2 | 5.37 | −0.24 | 0.09 | −5.61 | −0.33 | −5.28 | CH |
| ATCG00360.1 | YCF3 | 7.02 | 0.56 | 0.64 | −6.46 | −0.08 | −6.38 | CH |
| CELLULAR TRAFFICKING | ||||||||
| AT1G09210.1 | CRT1b | 5.54 | −0.46 | −0.77 | −6.00 | 0.31 | −6.31 | ER |
| AT1G71270.1 | POK Vps52 / Sac2 family | 6.43 | −0.19 | −0.91 | −6.62 | 0.71 | −5.72 | G |
| AT5G54750.1 | Transport protein particle (TRAPP) | 5.77 | 0.05 | 0.05 | −5.72 | 0.00 | 5.42 | G |
| AT1G15130.1 | Endosomal targeting BRO1-like domain-containing protein | −5.47 | 0.07 | −0.05 | 5.54 | 0.12 | 5.92 | ENDO |
| AT3G25220.1 | FKBP15-1 | −5.59 | 0.33 | 0.33 | 5.92 | 0.00 | 5.78 | ER |
| AT1G31730.1 | Adaptin family protein | −5.46 | 0.02 | 0.32 | 5.48 | −0.30 | 3.52 | CYT,G |
| AT1G67930.1 | Golgi transport complex protein-related | −4.00 | 0.02 | −0.48 | 4.02 | 0.50 | 3.53 | G |
| AT4G02350.1 | SEC15B | −3.72 | 0.31 | −0.19 | 4.03 | 0.50 | 5.40 | CYT |
| AT3G46830.1 | ATRABA2C | −5.58 | −0.14 | −0.17 | 5.44 | 0.03 | 4.87 | G-ENDO |
| AT5G35160.1 | Endomembrane protein 70 protein | −4.70 | 0.17 | 0.17 | 4.87 | 0.00 | 3.13 | G |
| AT5G15970.1 | KIN2 | −2.89 | −0.01 | 0.24 | 2.88 | −0.25 | 6.24 | CYT |
| AT3G48170.1 | ALDH10A9 | −5.35 | 0.86 | 0.89 | 6.22 | −0.03 | 5.67 | PEROX |
| AT4G01850.2 | SAM2 | −5.04 | 0.34 | 0.64 | 5.38 | −0.29 | 11.74 | CYT |
| AT1G35670.1 | CDPK2 calcium-dependent protein kinase 2 | −11.73 | 0.06 | 0.01 | 11.79 | 0.05 | 6.02 | CYT |
| AT5G13710.2 | SMT1 | −5.89 | 0.33 | 0.14 | 6.22 | 0.20 | 5.68 | V |
| AT1G20050.1 | HYD1 | −5.94 | 0.22 | −0.26 | 6.16 | 0.48 | −7.34 | PM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancini, I.; Domingo, G.; Bracale, M.; Loreto, F.; Pollastri, S. Isoprene Emission Influences the Proteomic Profile of Arabidopsis Plants under Well-Watered and Drought-Stress Conditions. Int. J. Mol. Sci. 2022, 23, 3836. https://doi.org/10.3390/ijms23073836
Mancini I, Domingo G, Bracale M, Loreto F, Pollastri S. Isoprene Emission Influences the Proteomic Profile of Arabidopsis Plants under Well-Watered and Drought-Stress Conditions. International Journal of Molecular Sciences. 2022; 23(7):3836. https://doi.org/10.3390/ijms23073836
Chicago/Turabian StyleMancini, Ilaria, Guido Domingo, Marcella Bracale, Francesco Loreto, and Susanna Pollastri. 2022. "Isoprene Emission Influences the Proteomic Profile of Arabidopsis Plants under Well-Watered and Drought-Stress Conditions" International Journal of Molecular Sciences 23, no. 7: 3836. https://doi.org/10.3390/ijms23073836
APA StyleMancini, I., Domingo, G., Bracale, M., Loreto, F., & Pollastri, S. (2022). Isoprene Emission Influences the Proteomic Profile of Arabidopsis Plants under Well-Watered and Drought-Stress Conditions. International Journal of Molecular Sciences, 23(7), 3836. https://doi.org/10.3390/ijms23073836







