Identification of Tumor-Suppressive miR-30e-3p Targets: Involvement of SERPINE1 in the Molecular Pathogenesis of Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Expression Levels of miR-30e-3p in Clinical Specimens
2.2. Effects of Ectopic Expression of miR-30e-3p on HNSCC Cell Lines
2.3. Identification of Putative Oncogenic Targets of miR-30e-3p in HNSCC Cells
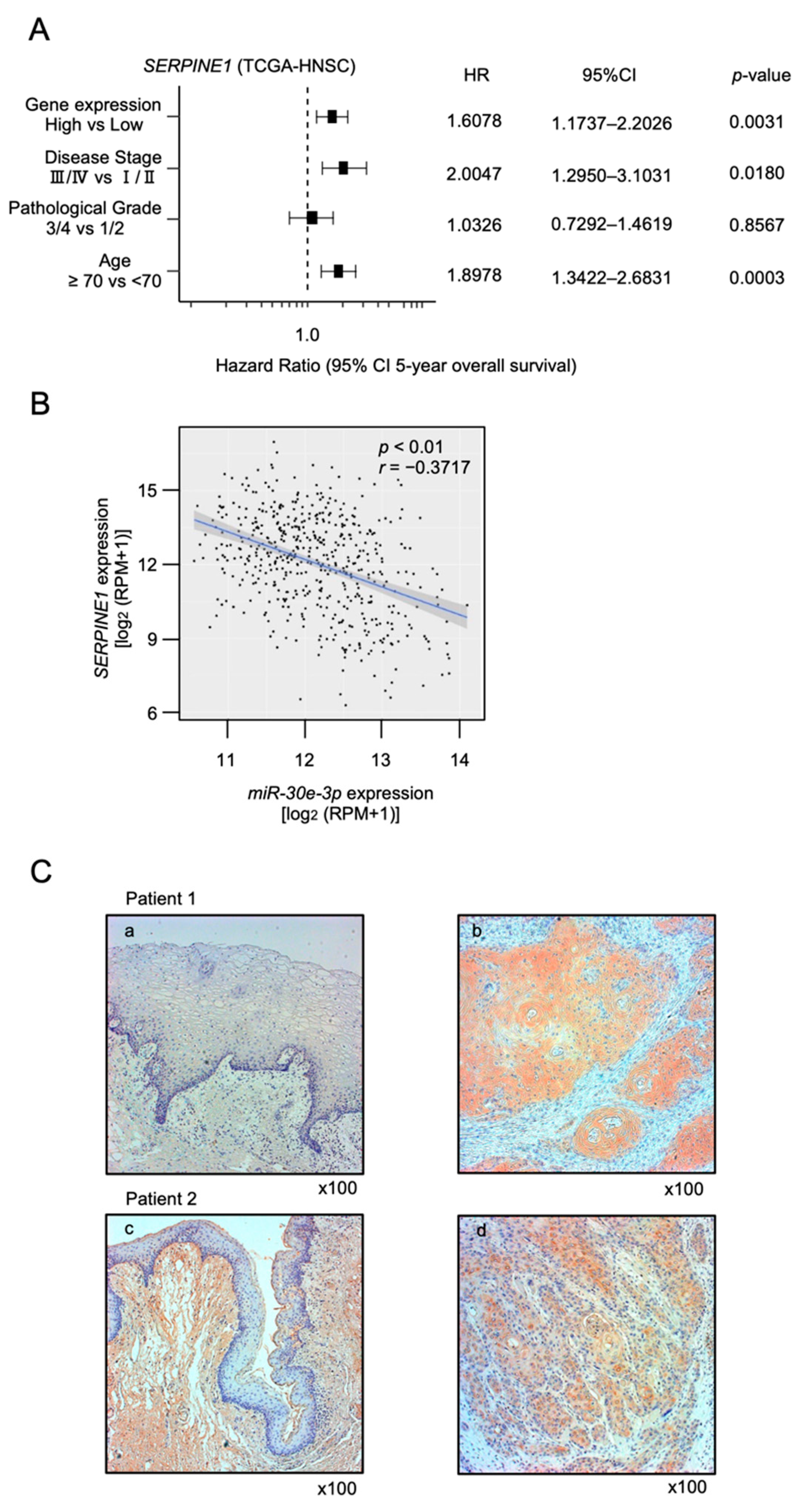
2.4. Clinical Significance of miR-30e-3p Target Genes Determined by TCGA Analysis
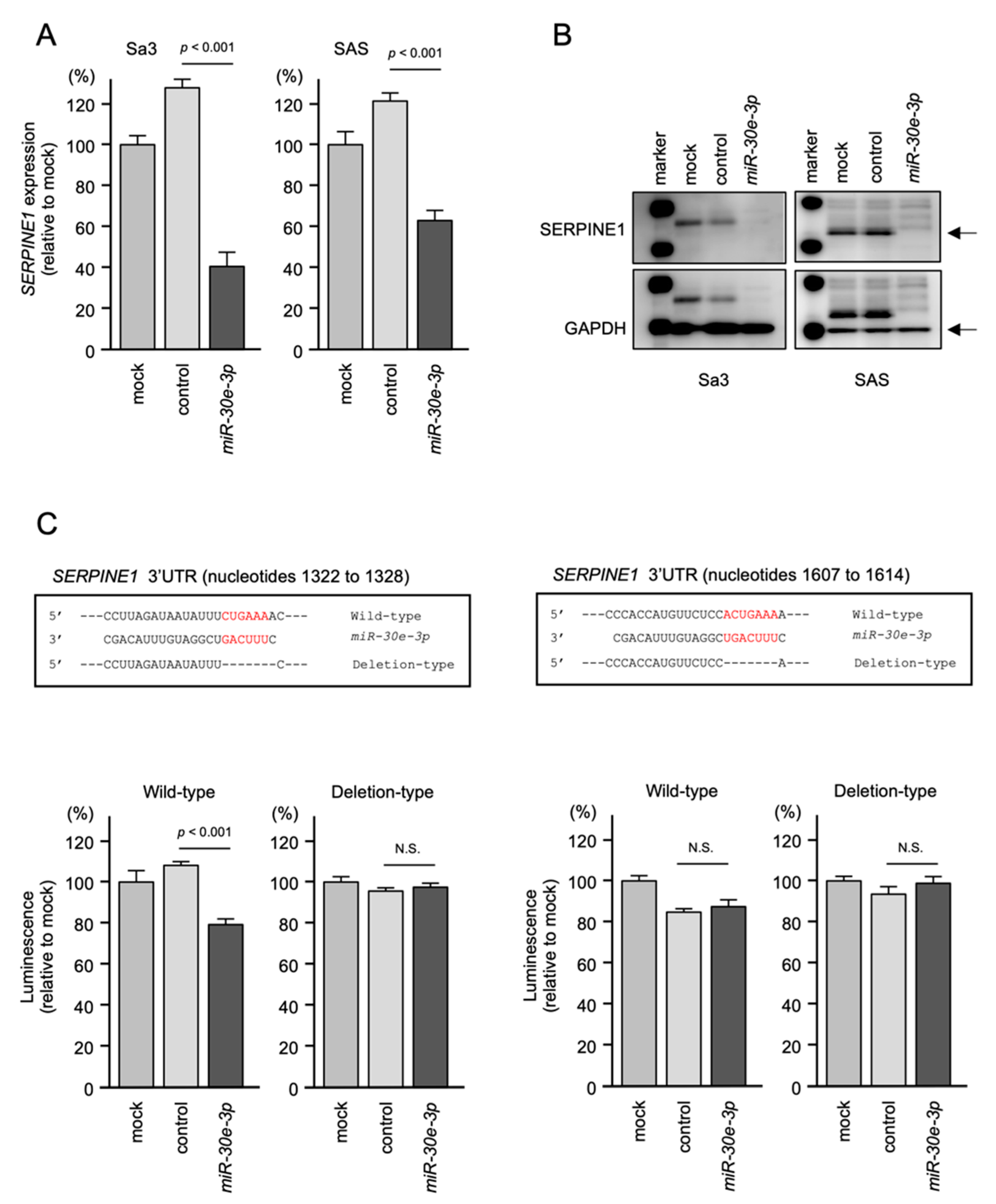
2.5. Regulation of SERPINE1 Expression by miR-30e-3p in HNSCC Cells
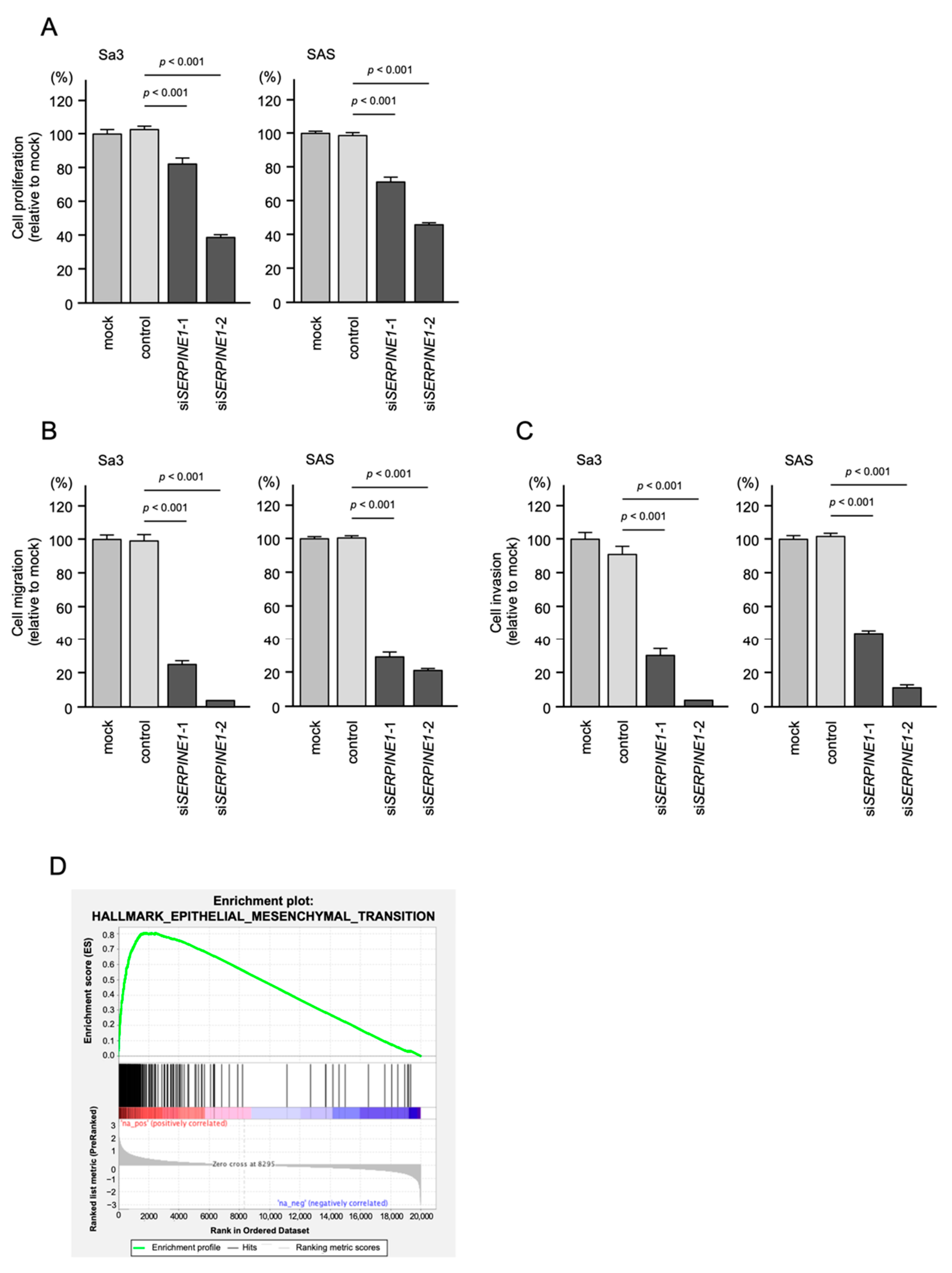
2.6. Effects of SERPINE1 Knockdown on the Proliferation, Migration, and Invasion Assays
3. Discussion
4. Materials and Methods
4.1. Transfection of Mature miRNAs and Small-Interfering RNAs (siRNAs) into HNSCC Cells
4.2. Functional Assays of miR-30e-3p and SERPINE1 on HNSCC Cells
4.3. RNA Extraction and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.4. Western Blotting and Immunohistochemistry
4.5. Dual Luciferase Reporter Assays
4.6. Identification of miR-30e-3p Targets in HNSCC
4.7. Analysis of miRNAs and miRNA Target Genes in HNSCC Patients
4.8. Gene Set Enrichment Analysis (GSEA)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Cramer, J.D.; Burtness, B.; Le, Q.T.; Ferris, R.L. The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 2019, 16, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Perry, C.; Jovanovic, L.; Nelson, C.; Punyadeera, C. Circulating tumour cells in metastatic head and neck cancers. Int. J. Cancer 2015, 136, 2515–2523. [Google Scholar] [CrossRef]
- Tran, G.; Zafar, S.Y. Financial toxicity and implications for cancer care in the era of molecular and immune therapies. Ann. Transl. Med. 2018, 6, 166. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-coding RNA regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Carron, J.; Torricelli, C.; Silva, J.K.; Queiroz, G.S.R.; Ortega, M.M.; Lima, C.S.P.; Lourenço, G.J. microRNAs deregulation in head and neck squamous cell carcinoma. Head Neck 2021, 43, 645–667. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Tanigawa, K.; Misono, S.; Suetsugu, T.; Sanada, H.; Uchida, A.; Kawano, M.; Machida, K.; Asai, S.; Moriya, S.; et al. Regulation of Oncogenic Targets by Tumor-Suppressive miR-150-3p in Lung Squamous Cell Carcinoma. Biomedicines 2021, 9, 1883. [Google Scholar] [CrossRef]
- Hozaka, Y.; Kita, Y.; Yasudome, R.; Tanaka, T.; Wada, M.; Idichi, T.; Tanabe, K.; Asai, S.; Moriya, S.; Toda, H.; et al. RNA-Sequencing Based microRNA Expression Signature of Colorectal Cancer: The Impact of Oncogenic Targets Regulated by miR-490-3p. Int. J. Mol. Sci. 2021, 22, 9876. [Google Scholar] [CrossRef]
- Oshima, S.; Asai, S.; Seki, N.; Minemura, C.; Kinoshita, T.; Goto, Y.; Kikkawa, N.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Identification of Tumor Suppressive Genes Regulated by miR-31-5p and miR-31-3p in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 6199. [Google Scholar] [CrossRef]
- Shinden, Y.; Hirashima, T.; Nohata, N.; Toda, H.; Okada, R.; Asai, S.; Tanaka, T.; Hozaka, Y.; Ohtsuka, T.; Kijima, Y.; et al. Molecular pathogenesis of breast cancer: Impact of miR-99a-5p and miR-99a-3p regulation on oncogenic genes. J. Hum. Genet. 2021, 66, 519–534. [Google Scholar] [CrossRef]
- Tanaka, N.; Minemura, C.; Asai, S.; Kikkawa, N.; Kinoshita, T.; Oshima, S.; Koma, A.; Kasamatsu, A.; Hanazawa, T.; Uzawa, K.; et al. Identification of miR-199-5p and miR-199-3p Target Genes: Paxillin Facilities Cancer Cell Aggressiveness in Head and Neck Squamous Cell Carcinoma. Genes 2021, 12, 1910. [Google Scholar] [CrossRef]
- Okada, R.; Goto, Y.; Yamada, Y.; Kato, M.; Asai, S.; Moriya, S.; Ichikawa, T.; Seki, N. Regulation of Oncogenic Targets by the Tumor-Suppressive miR-139 Duplex (miR-139-5p and miR-139-3p) in Renal Cell Carcinoma. Biomedicines 2020, 8, 599. [Google Scholar] [CrossRef] [PubMed]
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y.; Nohata, N.; Moriya, S.; Idichi, T.; Maemura, K.; Fujii, T.; et al. RNA-sequence-based microRNA expression signature in breast cancer: Tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol. Oncol. 2020, 14, 426–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, M.; Goto, Y.; Tanaka, T.; Okada, R.; Moriya, S.; Idichi, T.; Noda, M.; Sasaki, K.; Kita, Y.; Kurahara, H.; et al. RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: Oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J. Hum. Genet. 2020, 65, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, K.; Hanazawa, T.; Arai, T.; Okato, A.; Kikkawa, N.; Seki, N. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastasis Rev. 2017, 36, 525–545. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Okada, R.; Hozaka, Y.; Wada, M.; Moriya, S.; Satake, S.; Idichi, T.; Kurahara, H.; Ohtsuka, T.; Seki, N. Molecular Pathogenesis of Pancreatic Ductal Adenocarcinoma: Impact of miR-30c-5p and miR-30c-2-3p Regulation on Oncogenic Genes. Cancers 2020, 12, 2731. [Google Scholar] [CrossRef]
- Nepal, P.; Hozaka, Y.; Tanaka, T.; Wada, M.; Asai, S.; Minemura, C.; Idichi, T.; Arigami, T.; Kurahara, H.; Seki, N.; et al. Impact of Oncogenic Targets Controlled by Tumor-Suppressive miR-30a-5p in Pancreatic Ductal Adenocarcinoma. Anticancer Res. 2021, 41, 4821–4836. [Google Scholar] [CrossRef]
- Yang, S.J.; Yang, S.Y.; Wang, D.D.; Chen, X.; Shen, H.Y.; Zhang, X.H.; Zhong, S.L.; Tang, J.H.; Zhao, J.H. The miR-30 family: Versatile players in breast cancer. Tumour Biol 2017, 39, 1010428317692204. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Li, G.; Liu, C.; Lu, S.; Jing, Q.; Chen, X.; Zheng, H.; Ma, H.; Zhang, D.; Ren, S.; et al. miR-30e-5p represses angiogenesis and metastasis by directly targeting AEG-1 in squamous cell carcinoma of the head and neck. Cancer Sci. 2020, 111, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.D.; Cheng, H.; Martin, S.E.; Si, H.; Ormanoglu, P.; Carlson, S.; Clavijo, P.E.; Yang, X.; Das, R.; Cornelius, S.; et al. Integrated Genomic and Functional microRNA Analysis Identifies miR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin. Cancer Res. 2019, 25, 2860–2873. [Google Scholar] [CrossRef] [Green Version]
- Mitra, R.; Adams, C.M.; Jiang, W.; Greenawalt, E.; Eischen, C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020, 11, 968. [Google Scholar] [CrossRef] [Green Version]
- Shimomura, H.; Okada, R.; Tanaka, T.; Hozaka, Y.; Wada, M.; Moriya, S.; Idichi, T.; Kita, Y.; Kurahara, H.; Ohtsuka, T.; et al. Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis. Int. J. Mol. Sci. 2020, 21, 6459. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Okato, A.; Idichi, T.; Arai, T.; Sugawara, S.; Katada, K.; Okamoto, Y.; et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int. J. Oncol. 2018, 52, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Koma, A.; Asai, S.; Minemura, C.; Oshima, S.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 9947. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhu, C.; Zhang, Y.; Zheng, Y.; Ma, F.; Su, L.; Shao, G. MicroRNA-30e-3p inhibits cell invasion and migration in clear cell renal cell carcinoma by targeting Snail1. Oncol. Lett. 2017, 13, 2053–2058. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Yan, C.; Chen, Y.; Wang, Z.; Guo, J.; Pei, H. Propofol inhibits the proliferation, migration, invasion and epithelial to mesenchymal transition of renal cell carcinoma cells by regulating microRNA-363/Snail1. Am. J. Transl. Res. 2021, 13, 2256–2269. [Google Scholar]
- Gramantieri, L.; Pollutri, D.; Gagliardi, M.; Giovannini, C.; Quarta, S.; Ferracin, M.; Casadei-Gardini, A.; Callegari, E.; De Carolis, S.; Marinelli, S.; et al. MiR-30e-3p Influences Tumor Phenotype through MDM2/TP53 Axis and Predicts Sorafenib Resistance in Hepatocellular Carcinoma. Cancer Res. 2020, 80, 1720–1734. [Google Scholar] [CrossRef]
- Kurashige, J.; Yokobori, T.; Mima, K.; Sawada, G.; Takahashi, Y.; Ueo, H.; Takano, Y.; Matsumura, T.; Uchi, R.; Eguchi, H.; et al. Plastin3 is associated with epithelial-mesenchymal transition and poor prognosis in gastric cancer. Oncol. Lett. 2019, 17, 2393–2399. [Google Scholar] [CrossRef]
- Wolff, L.; Strathmann, E.A.; Müller, I.; Mählich, D.; Veltman, C.; Niehoff, A.; Wirth, B. Plastin 3 in health and disease: A matter of balance. Cell Mol. Life Sci. 2021, 78, 5275–5301. [Google Scholar] [CrossRef]
- Sugimachi, K.; Yokobori, T.; Iinuma, H.; Ueda, M.; Ueo, H.; Shinden, Y.; Eguchi, H.; Sudo, T.; Suzuki, A.; Maehara, Y.; et al. Aberrant expression of plastin-3 via copy number gain induces the epithelial-mesenchymal transition in circulating colorectal cancer cells. Ann. Surg. Oncol. 2014, 21, 3680–3690. [Google Scholar] [CrossRef]
- Kuriyama, K.; Yokobori, T.; Sohda, M.; Nakazawa, N.; Yajima, T.; Naruse, I.; Kuwano, H.; Shirabe, K.; Kaira, K.; Saeki, H. Plasma plastin-3: A tumor marker in patients with non-small-cell lung cancer treated with nivolumab. Oncol. Lett. 2021, 21, 11. [Google Scholar] [CrossRef]
- Hori, T.; Kato, S.; Saeki, M.; DeMartino, G.N.; Slaughter, C.A.; Takeuchi, J.; Toh-e, A.; Tanaka, K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene 1998, 216, 113–122. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Zhen, C.; Yang, B.; Feng, L. Gankyrin as a potential target for tumor therapy: Evidence and perspectives. Am. J. Transl. Res. 2018, 10, 1949–1960. [Google Scholar] [PubMed]
- Li, J.; Knobloch, T.J.; Kresty, L.A.; Zhang, Z.; Lang, J.C.; Schuller, D.E.; Weghorst, C.M. Gankyrin, a biomarker for epithelial carcinogenesis, is overexpressed in human oral cancer. Anticancer Res. 2011, 31, 2683–2692. [Google Scholar] [PubMed]
- Yan, C.; Theodorescu, D. RAL GTPases: Biology and Potential as Therapeutic Targets in Cancer. Pharm. Rev. 2018, 70, 1–11. [Google Scholar] [CrossRef]
- Chardin, P.; Tavitian, A. Coding sequences of human ralA and ralB cDNAs. Nucleic Acids Res. 1989, 17, 4380. [Google Scholar] [CrossRef] [Green Version]
- Chien, Y.; White, M.A. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003, 4, 800–806. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.H.; O’Hayer, K.; Adam, S.J.; Kendall, S.D.; Campbell, P.M.; Der, C.J.; Counter, C.M. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 2006, 16, 2385–2394. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.K.; Vaughan, D.E. PAI-1 in tissue fibrosis. J. Cell Physiol. 2012, 227, 493–507. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wei, X.; He, J.; Tian, X.; Yuan, S.; Sun, L. Plasminogen activator inhibitor-1 in cancer research. Biomed. Pharm. 2018, 105, 83–94. [Google Scholar] [CrossRef]
- Kubala, M.H.; DeClerck, Y.A. The plasminogen activator inhibitor-1 paradox in cancer: A mechanistic understanding. Cancer Metastasis Rev. 2019, 38, 483–492. [Google Scholar] [CrossRef]
- Pavón, M.A.; Arroyo-Solera, I.; Céspedes, M.V.; Casanova, I.; León, X.; Mangues, R. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget 2016, 7, 57351–57366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavón, M.A.; Arroyo-Solera, I.; Téllez-Gabriel, M.; León, X.; Virós, D.; López, M.; Gallardo, A.; Céspedes, M.V.; Casanova, I.; López-Pousa, A.; et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015, 6, 29016–29033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo-Solera, I.; Pavón, M.; León, X.; López, M.; Gallardo, A.; Céspedes, M.V.; Casanova, I.; Pallarès, V.; López-Pousa, A.; Mangues, M.A.; et al. Effect of serpinE1 overexpression on the primary tumor and lymph node, and lung metastases in head and neck squamous cell carcinoma. Head Neck 2019, 41, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Liu, Z. MicroRNA 617 Targeting SERPINE1 Inhibited the Progression of Oral Squamous Cell Carcinoma. Mol. Cell Biol. 2021, 41, e0056520. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, P.; Tang, Y.; Fan, Y.; Liu, Y.; Fang, X.; Wang, W.; Zhao, S. Down-Regulation of MiR-181c-5p Promotes Epithelial-to-Mesenchymal Transition in Laryngeal Squamous Cell Carcinoma via Targeting SERPINE1. Front. Oncol. 2020, 10, 544476. [Google Scholar] [CrossRef]
- Mizuno, K.; Tanigawa, K.; Nohata, N.; Misono, S.; Okada, R.; Asai, S.; Moriya, S.; Suetsugu, T.; Inoue, H.; Seki, N. FAM64A: A Novel Oncogenic Target of Lung Adenocarcinoma Regulated by Both Strands of miR-99a (miR-99a-5p and miR-99a-3p). Cells 2020, 9, 2083. [Google Scholar] [CrossRef]
- Kase-Kato, I.; Asai, S.; Minemura, C.; Tsuneizumi, K.; Oshima, S.; Koma, A.; Kasamatsu, A.; Hanazawa, T.; Uzawa, K.; Seki, N. Molecular Pathogenesis of the Coronin Family: CORO2A Facilitates Migration and Invasion Abilities in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 2684. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anaya, J. Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J. Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]

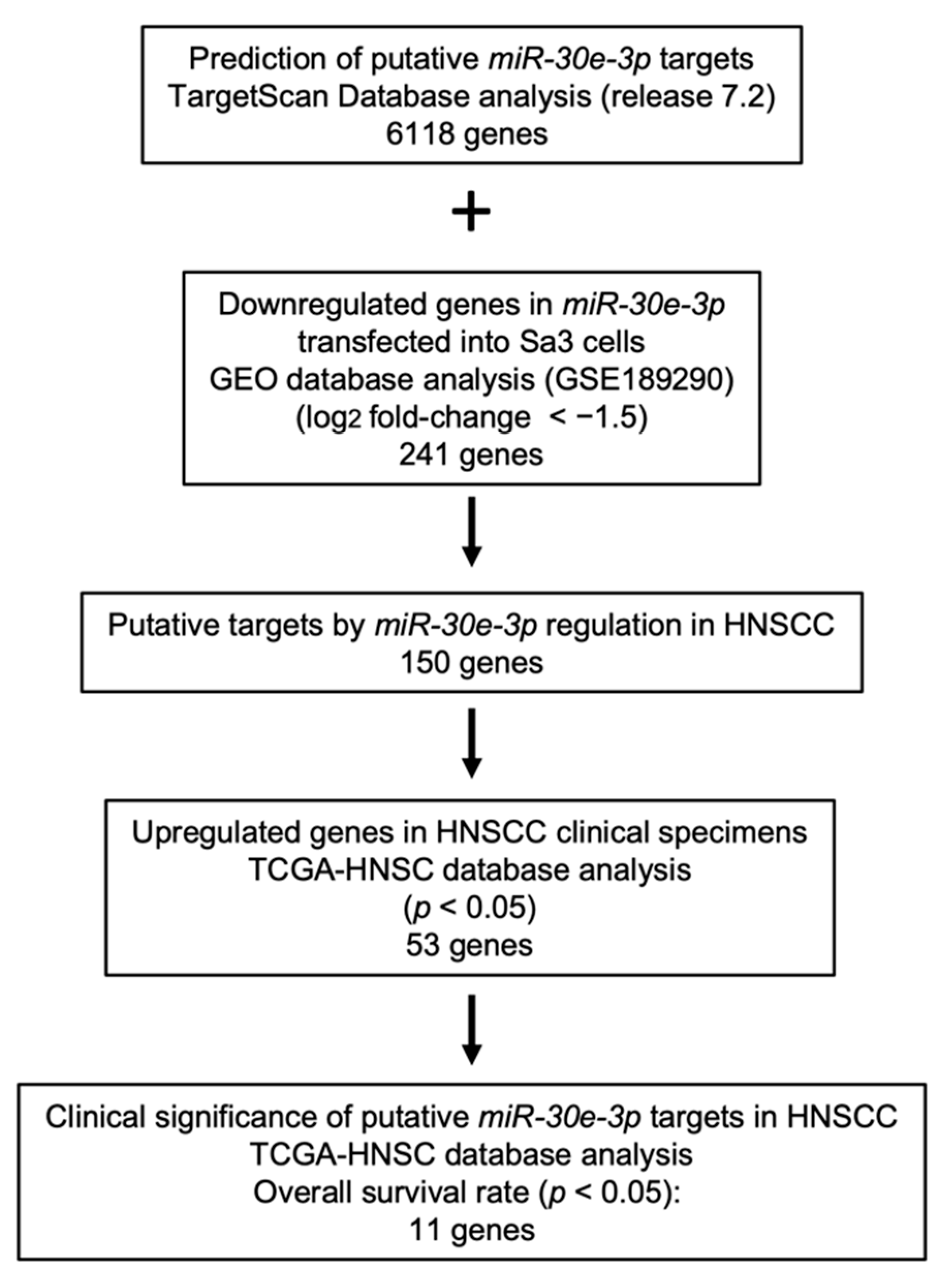
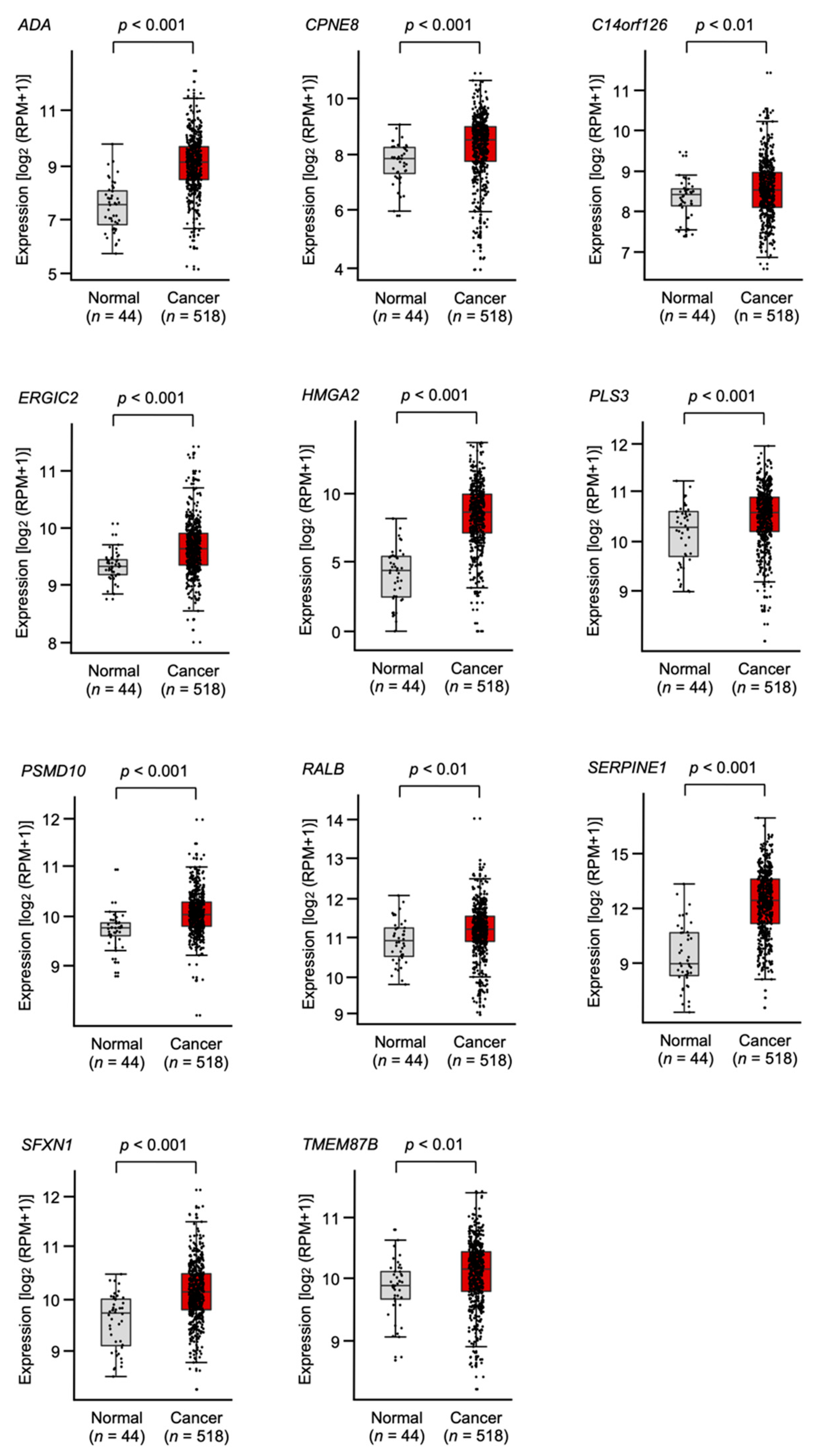
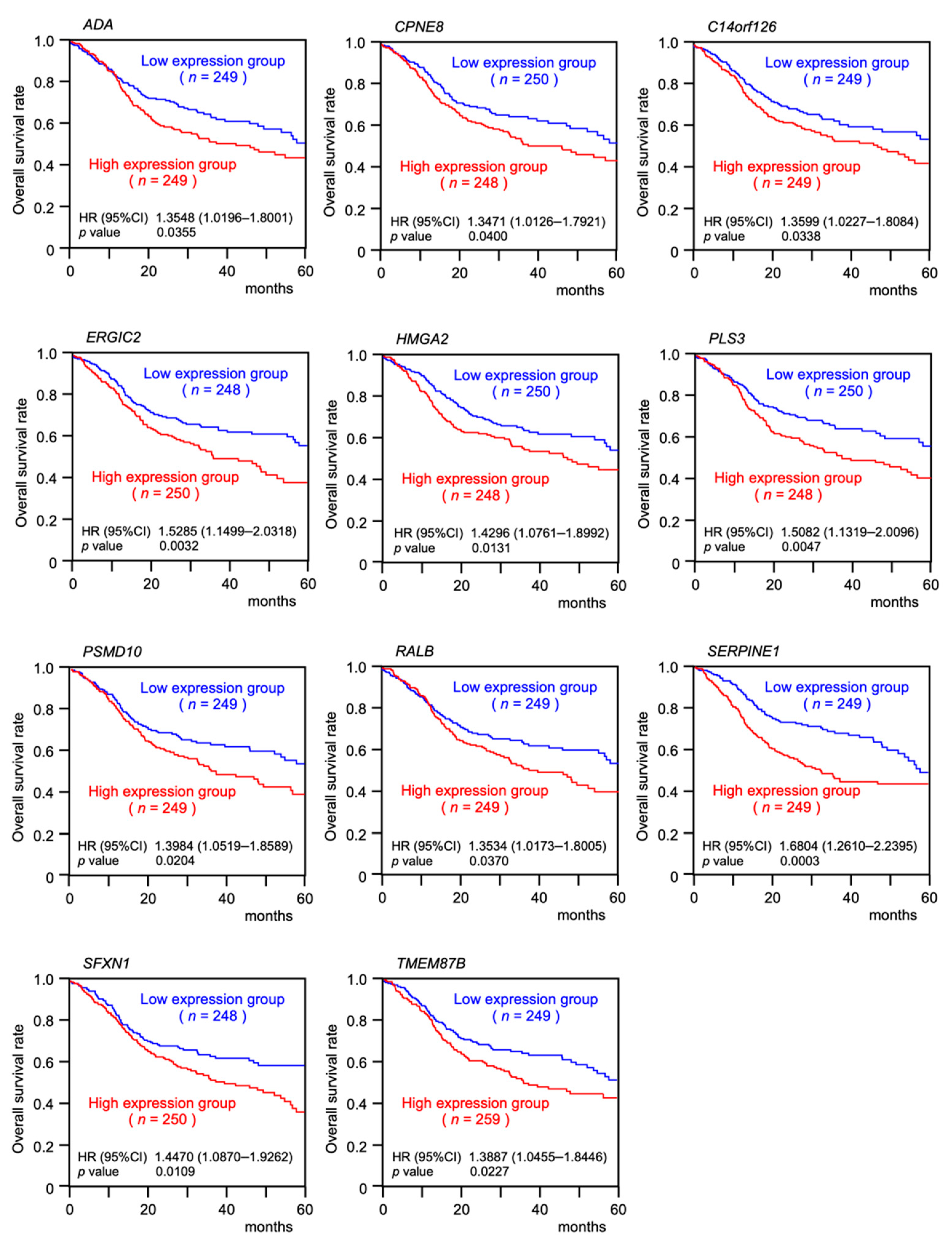
| Entrez Gene ID | Gene Symbol | Gene Name | Total Binding Sites | GEO log2 FC 1 | 5y OS 2 p-Value | FDR 3 |
|---|---|---|---|---|---|---|
| 5054 | SERPINE1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 2 | −1.98 | 0.0003 | 0.0209 |
| 51290 | ERGIC2 | ERGIC and golgi 2 | 2 | −1.68 | 0.0032 | 0.0730 |
| 5358 | PLS3 | plastin 3 | 2 | −1.53 | 0.0047 | 0.0898 |
| 94081 | SFXN1 | sideroflexin 1 | 2 | −1.92 | 0.0109 | 0.1423 |
| 8091 | HMGA2 | high mobility group AT-hook 2 | 1 | −2.65 | 0.0131 | 0.1575 |
| 5716 | PSMD10 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | 1 | −1.52 | 0.0204 | 0.2014 |
| 84910 | TMEM87B | transmembrane protein 87B | 1 | −1.64 | 0.0227 | 0.2137 |
| 112487 | C14orf126 | D-tyrosyl-tRNA deacylase 2 (putative) | 1 | −1.58 | 0.0338 | 0.2663 |
| 100 | ADA | adenosine deaminase | 1 | −1.64 | 0.0355 | 0.2736 |
| 5899 | RALB | v-ral simian leukemia viral oncogene homolog B | 2 | −1.56 | 0.0370 | 0.2799 |
| 144402 | CPNE8 | copine VIII | 1 | −1.74 | 0.0400 | 0.2921 |
| 10923 | SUB1 | SUB1 homolog (S. cerevisiae) | 3 | −2.53 | 0.0933 | 0.4584 |
| 6566 | SLC16A1 | solute carrier family 16 (monocarboxylate transporter), member 1 | 4 | −1.75 | 0.0955 | 0.4640 |
| 3336 | HSPE1 | heat shock 10kDa protein 1 (chaperonin 10) | 1 | −1.53 | 0.0960 | 0.4652 |
| 55156 | ARMC1 | armadillo repeat containing 1 | 4 | −1.91 | 0.0997 | 0.4743 |
| 79624 | C6orf211 | chromosome 6 open reading frame 211 | 4 | −1.50 | 0.1251 | 0.5316 |
| 528 | ATP6V1C1 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 2 | −2.04 | 0.1309 | 0.5436 |
| 64841 | GNPNAT1 | glucosamine-phosphate N-acetyltransferase 1 | 3 | −1.73 | 0.1564 | 0.5927 |
| 10552 | ARPC1A | actin related protein 2/3 complex, subunit 1A, 41 kDa | 1 | −1.97 | 0.1871 | 0.6449 |
| 6780 | STAU1 | staufen double-stranded RNA binding protein 1 | 3 | −1.77 | 0.1883 | 0.6468 |
| 9265 | CYTH3 | cytohesin 3 | 2 | −1.52 | 0.2113 | 0.6817 |
| 90874 | ZNF697 | zinc finger protein 697 | 3 | −1.58 | 0.2182 | 0.6915 |
| 5923 | RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 | 2 | −1.51 | 0.2359 | 0.7157 |
| 136 | ADORA2B | adenosine A2b receptor | 1 | −2.09 | 0.2367 | 0.7168 |
| 81539 | SLC38A1 | solute carrier family 38, member 1 | 5 | −2.00 | 0.2493 | 0.7330 |
| 10473 | HMGN4 | high-mobility group nucleosomal binding domain 4 | 2 | −1.84 | 0.2678 | 0.7555 |
| 51762 | RAB8B | RAB8B, member RAS oncogene family | 4 | −1.59 | 0.2683 | 0.7561 |
| 54165 | DCUN1D1 | DCN1, defective in cullin neddylation 1, domain containing 1 | 5 | −1.52 | 0.3327 | 0.8236 |
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 | 2 | −1.55 | 0.3663 | 0.8530 |
| 3556 | IL1RAP | interleukin 1 receptor accessory protein | 1 | −3.20 | 0.3916 | 0.8729 |
| 112399 | EGLN3 | egl-9 family hypoxia-inducible factor 3 | 1 | −2.10 | 0.4389 | 0.9051 |
| 1021 | CDK6 | cyclin-dependent kinase 6 | 3 | −1.82 | 0.4733 | 0.9249 |
| 54108 | CHRAC1 | chromatin accessibility complex 1 | 1 | −1.65 | 0.4733 | 0.9249 |
| 7172 | TPMT | thiopurine S-methyltransferase | 1 | −1.66 | 0.4754 | 0.9260 |
| 2113 | ETS1 | v-ets avian erythroblastosis virus E26 oncogene homolog 1 | 3 | −1.89 | 0.5119 | 0.9436 |
| 51199 | NIN | ninein (GSK3B interacting protein) | 2 | −1.58 | 0.5582 | 0.9616 |
| 8862 | APLN | apelin | 1 | −2.05 | 0.5596 | 0.9621 |
| 5597 | MAPK6 | mitogen-activated protein kinase 6 | 2 | −2.10 | 0.5818 | 0.9690 |
| 57045 | TWSG1 | twisted gastrulation BMP signaling modulator 1 | 1 | −1.69 | 0.5857 | 0.9701 |
| 55142 | HAUS2 | HAUS augmin-like complex, subunit 2 | 4 | −1.98 | 0.5862 | 0.9702 |
| 4015 | LOX | lysyl oxidase | 1 | −1.55 | 0.6133 | 0.9769 |
| 4678 | NASP | nuclear autoantigenic sperm protein (histone-binding) | 2 | −1.74 | 0.6811 | 0.9864 |
| 55824 | PAG1 | phosphoprotein associated with glycosphingolipid microdomains 1 | 5 | −2.10 | 0.7043 | 0.9871 |
| 3553 | IL1B | interleukin 1, beta | 1 | −2.15 | 0.7316 | 0.9871 |
| 65062 | ALS2CR4 | transmembrane protein 237 | 1 | −1.86 | 0.7378 | 0.9871 |
| 51715 | RAB23 | RAB23, member RAS oncogene family | 2 | −1.70 | 0.7820 | 0.9871 |
| 4603 | MYBL1 | v-myb avian myeloblastosis viral oncogene homolog-like 1 | 1 | −1.55 | 0.7897 | 0.9871 |
| 4893 | NRAS | neuroblastoma RAS viral (v-ras) oncogene homolog | 3 | −2.53 | 0.7935 | 0.9871 |
| 84668 | FAM126A | family with sequence similarity 126, member A | 2 | −1.61 | 0.8471 | 0.9871 |
| 51633 | OTUD6B | OTU domain containing 6B | 2 | −1.63 | 0.9276 | 0.9871 |
| 3930 | LBR | lamin B receptor | 1 | −2.40 | 0.9492 | 0.9871 |
| 8869 | ST3GAL5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | 1 | −2.38 | 0.9686 | 0.9871 |
| 1719 | DHFR | dihydrofolate reductase | 4 | −1.96 | 0.9722 | 0.9871 |
| Name | Normalized Enrichment Score | FDR q-Value |
|---|---|---|
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 3.149 | q < 0.001 |
| HALLMARK_MYOGENESIS | 2.866 | q < 0.001 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 2.489 | q < 0.001 |
| HALLMARK_ANGIOGENESIS | 2.349 | q < 0.001 |
| HALLMARK_KRAS_SIGNALING_UP | 2.289 | q < 0.001 |
| HALLMARK_COAGULATION | 2.285 | q < 0.001 |
| HALLMARK_HYPOXIA | 2.282 | q < 0.001 |
| HALLMARK_APICAL_JUNCTION | 2.247 | q < 0.001 |
| HALLMARK_UV_RESPONSE_DN | 2.189 | q < 0.001 |
| HALLMARK_INFLAMMATORY_RESPONSE | 2.156 | q < 0.001 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 2.054 | q < 0.001 |
| HALLMARK_TGF_BETA_SIGNALING | 2.027 | q < 0.001 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.896 | q < 0.001 |
| HALLMARK_COMPLEMENT | 1.846 | q < 0.001 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.836 | q < 0.001 |
| HALLMARK_NOTCH_SIGNALING | 1.793 | 0.001 |
| HALLMARK_APOPTOSIS | 1.716 | 0.002 |
| HALLMARK_HEDGEHOG_SIGNALING | 1.692 | 0.002 |
| HALLMARK_IL2_STAT5_SIGNALING | 1.618 | 0.004 |
| HALLMARK_GLYCOLYSIS | 1.593 | 0.005 |
| HALLMARK_P53_PATHWAY | 1.396 | 0.038 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.376 | 0.043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minemura, C.; Asai, S.; Koma, A.; Kase-Kato, I.; Tanaka, N.; Kikkawa, N.; Kasamatsu, A.; Yokoe, H.; Hanazawa, T.; Uzawa, K.; et al. Identification of Tumor-Suppressive miR-30e-3p Targets: Involvement of SERPINE1 in the Molecular Pathogenesis of Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3808. https://doi.org/10.3390/ijms23073808
Minemura C, Asai S, Koma A, Kase-Kato I, Tanaka N, Kikkawa N, Kasamatsu A, Yokoe H, Hanazawa T, Uzawa K, et al. Identification of Tumor-Suppressive miR-30e-3p Targets: Involvement of SERPINE1 in the Molecular Pathogenesis of Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2022; 23(7):3808. https://doi.org/10.3390/ijms23073808
Chicago/Turabian StyleMinemura, Chikashi, Shunichi Asai, Ayaka Koma, Ikuko Kase-Kato, Nozomi Tanaka, Naoko Kikkawa, Atsushi Kasamatsu, Hidetaka Yokoe, Toyoyuki Hanazawa, Katsuhiro Uzawa, and et al. 2022. "Identification of Tumor-Suppressive miR-30e-3p Targets: Involvement of SERPINE1 in the Molecular Pathogenesis of Head and Neck Squamous Cell Carcinoma" International Journal of Molecular Sciences 23, no. 7: 3808. https://doi.org/10.3390/ijms23073808
APA StyleMinemura, C., Asai, S., Koma, A., Kase-Kato, I., Tanaka, N., Kikkawa, N., Kasamatsu, A., Yokoe, H., Hanazawa, T., Uzawa, K., & Seki, N. (2022). Identification of Tumor-Suppressive miR-30e-3p Targets: Involvement of SERPINE1 in the Molecular Pathogenesis of Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences, 23(7), 3808. https://doi.org/10.3390/ijms23073808






