Abstract
The aim of this work was to compare the multiple alignment methods MAHDS, T-Coffee, MUSCLE, Clustal Omega, Kalign, MAFFT, and PRANK in their ability to align highly divergent amino acid sequences. To accomplish this, we created test amino acid sequences with an average number of substitutions per amino acid (x) from 0.6 to 5.6, a total of 81 sets. Comparison of the performance of sequence alignments constructed by MAHDS and previously developed algorithms using the CS and Z score criteria and the benchmark alignment database (BAliBASE) indicated that, although the quality of the alignments built with MAHDS was somewhat lower than that of the other algorithms, it was compensated by greater statistical significance. MAHDS could construct statistically significant alignments of artificial sequences with x ≤ 4.8, whereas the other algorithms (T-Coffee, MUSCLE, Clustal Omega, Kalign, MAFFT, and PRANK) could not perform that at x > 2.4. The application of MAHDS to align 21 families of highly diverged proteins (identity < 20%) from Pfam and HOMSTRAD databases showed that it could calculate statistically significant alignments in cases when the other methods failed. Thus, MAHDS could be used to construct statistically significant multiple alignments of highly divergent protein sequences, which accumulated multiple mutations during evolution.
1. Introduction
Algorithms for multiple sequence alignment (MSA) are widely used in modern molecular biology for numerous purposes, including evolutionary studies and molecular modeling. Thus, the publication describing the ClustalW multiple alignment algorithm [1] is among the top 10 most cited publications [2]. MSA is indispensable when it is necessary to determine the relatedness of polypeptide sequences containing substitutions and/or insertions/deletions (indels) of amino acid residues, and MSA algorithms are fundamental tools for the construction of phylogenetic trees, calculation of profiles for hidden Markov models (HMMs), and search for common motifs. An important application of MSA methods is the prediction of secondary and tertiary structures of polypeptides and RNA, as well as their molecular functions and interactions [3,4].
In general, the construction of an MSA is an NP-complete problem, for which no direct solution by dynamic programming methods is possible in a reasonable time, even using modern supercomputers [5]. Therefore, many heuristic methods for performing MSA in a relatively short computational time have been developed. Among them, one of the most popular is the progressive procedure based on the construction of a binary (guiding) tree in which sequences are presented as leaves [6]. Then, MSA can be built through a series of pairwise alignments added according to the branching order in the path tree. Methods that employ the progressive approach include ClustalW [1] and its variant Clustal Omega, based on HMM for protein alignments [7], MAFFT [8], and T-Coffee [9]; the latter uses a paired alignment library, which allows for the construction of more accurate multiple alignments than those produced by ClustalW or Clustal Omega.
Iterative methods such as PRRN/PRRP [10] and MUSCLE [11] have been developed with the aim to reduce errors in constructing progressive alignments, which is achieved by repeatedly rebuilding the generated MSAs. Iterative methods revert to pairwise alignments, which can optimize the objective function and improve the quality of MSA.
Some MSA algorithms such as SAM [12] and HMMER [13] use HMM. Compared to progressive methods, HMM-based methods often calculate more significant alignments, which is achieved by rebuilding the alignment each time a new sequence is added.
There are also other methods for calculating MSA, such as genetic algorithms [14], annealing modeling [15], and phylogeny-aware gap placement [16,17]. Recently, we have developed an algorithm for multiple alignments of highly diverged sequences (MAHDS), which is based on the optimization of random position weight matrix (PWM) images of MSAs [18,19,20]. To optimize random patterns in MSA calculation, the MAHDS method uses a genetic algorithm and two-dimensional dynamic programming rather than pairwise equalization; as a result, multiple alignments for highly divergent nucleotide sequences can be obtained. The application of MAHDS to random nucleotide sequences shows that it can calculate statistically significant multiple alignments of sequences, for which the number of random base substitutions per nucleotide (x) ranges from 0 to 4.4. At the same time, the other methods such as ClustalW [21], Clustal Omega [7], MAFFT [22], T-Coffee [9], Muscle [11], and Kalign [23] could do it for x from 0 to 2.4 [18], indicating that for highly divergent nucleotide sequences (x = 2.4–4.4), the MAHDS algorithm is the only method capable of building statistically significant alignments. Previously, MAHDS has been used to construct MSAs of highly diverged promoter sequences (x = 3.6) from different genomes [19,24,25], which made it possible to identify a large number of potential promoters in rice genes [25].
In this study, we used the MAHDS method for the calculation of MSAs of protein sequences. The application of MAHDS to the alignment of random amino acid sequences with different x showed that the method could produce statistically significant MSAs for sequences with x ranging from 0 to 3.6. The alignments constructed by MAHDS and the other methods mentioned above were performed using the column score (CS) test [26,27,28], Z-score, and the benchmark alignment database (BAliBASE) (reference set 10) containing 218 protein families. Although, according to the CS criterion, the quality of the alignments built by the MAHDS method was somewhat lower than that of the other algorithms, it was compensated by greater statistical significance. Furthermore, in the alignment of 21 highly diverged protein families from the Pfam and HOMSTRAD databases, MAHDS could calculate statistically significant MSAs of more families than the other methods. Our results demonstrate that the developed MAHDS algorithm can be applied to analyze the phylogenetic relationships of evolutionary distant proteins.
2. Results
The performance of the MAHDS algorithm was compared with that of the MSA methods provided by EMBL-EBI resources, including T-Coffee, MUSCLE, PRANK, Clustal Omega, Kalign, and MAFFT, for which the web application programming interface is available. The following test data were used for analysis: benchmark alignment database (BAliBASE) [27,28,29], artificial sequences with certain properties, and protein families with low average sequence identity in full alignment.
The quality of alignments was evaluated using the CS [26,27,28] and Z-score criteria. In the calculation of the CS, the columns of the alignment are considered. If all remainders in the i-th column are aligned as in the reference alignment, then Ci = 1; otherwise, Ci = 0. The formula for calculating the CS is:
where L is the number of columns in the evaluated alignment. As in order to use the CS, it is necessary to have reference alignments, the CS was applied only to evaluate the BAliBASE using its tools bali score and bali_score_reliable; the former considers all sequences, whereas the latter ignores sequences with discrepancies (annotated with SEQERR features [30]). The MAHDS algorithm uses the following parameters: gap opening penalty (d), gap extension penalty (e), RL, and Kd. RL is the multiplier parameter, which can be used to scale R2 (Formula (2)), and Kd is the equivalent of an expected E score value features [31], which defines the accuracy of determining the start and end of the local alignment (Formula (3)). Therefore, the primary task in testing MAHDS was to find a biologically relevant combination of d, e, RL, and Kd.
2.1. Testing MAHDS Performance Using the BAliBASE
We used BAliBASE reference set 10, comprising 218 protein families. However, because of restraints set on the file size and the number of sequences by EMBL-EBI services, it was necessary to exclude five families (BBA0039.fasta, BBA0101.fasta, BBA0134.fasta, BBA0190.fasta, and BBA0213.fasta) for which alignment could not be performed by at least one of the compared methods. As a result, we considered the remaining 213 protein families and calculated the average CS for the MSAs as an indicator of the biological correctness, which should be maximized when choosing the parameters. At the same time, we tried to ensure that Z (Methods, Formula (7)) was close to the maximum.
We carried out a series of experiments for different values of parameters d, e, and Kd at fixed RL = 5.0, which can be chosen arbitrarily, as d, e, and Kd would then be adjusted accordingly. Initially, we considered e = 0.25 d as the optimal ratio, which was previously established for the application of MAHDS to DNA sequences [18], and estimated Z and CS scores for the sets of d values {8, 10, 12, 16, 20, 24, 28, 32, 36, 40, 44, 48, 52, 56} and Kd values {−0.1, −1, −2}. However, it appeared that with e = 0.25 d there was a discrepancy between the peaks in the CS and Z; therefore, an additional series of experiments was conducted with varying e-to-d ratios (Table 1).

Table 1.
Mean Z and CS values for BAliBASE protein families’ alignments produced by MAHDS with different Kd, d, and e parameters.
The results indicated that the combination d = 40, e = 1, and Kd = −1.0 was the optimal for protein sequence alignment and Z estimation with MAHDS. Then, we aligned the 213 protein families by the other methods and estimated the average Z (calculated as described in Section 4.6.1) and CS (Table 2). The results showed that the MAHDS algorithm was superior to the compared methods in terms of statistical significance (Z) but inferior in terms of CS.

Table 2.
Mean Z and CS values for BAliBASE protein families’ alignments produced by different methods.
2.2. Determining the Significance Threshold for Z
As a rule of thumb, paired alignment is considered statistically significant at Z ≥ 8 [32]. For MAHDS, the threshold value was 6 [18] or 10 [33], depending on the nature of the sequences; however, it has been determined for the alphabet of four nucleotides and may not be relevant for protein sequences composed of 20 amino acids.
To determine the threshold of statistical significance Zt, we created random sequences. To ensure that the number of aligned sequences and their average length did not affect Z, two datasets were generated: H1 containing 100 subsets of 100 random sequences, each 600 amino acids long, and H2 containing 100 subsets, including 20, 40, and 40 random sequences of 600, 900, and 300 amino acids, respectively. For each H1 and H2 subset, we built sequence alignments using the MAHDS algorithm and calculated Z. For set H1, = 4.147 and 2.11, whereas for set H2, = 4.04 and 1.78. These results showed that the variations in sequence lengths did not affect Z, indicating that the same Zt could be used for sequences of different lengths. Therefore, we chose Zt = 10.0, which was approximately equal to the average value of .
2.3. Testing MAHDS Performance Using Artificial Sequences
To compare methods for constructing multiple alignments, we created 81 sets, each containing 100 artificial sequences Des(i) generated from ancestor sequence Anc of length (L) = 600 symbols (Methods, Section 4.7). To create Des(i), the following random changes were made to Anc: the numbers of insertions or deletions were 2, 5, or 10, the indel length was 1, 5, or 20, and the number of substitutions per amino acid was 0.3, 0.6, 0.9, 1.2, 1.5, 1.8, 2.1, 2.4, or 2.7. In accordance with Formula (11), the average number of substitutions per symbol between sequences in sets Des was 0.6, 1.2, 1.8, 2.4, 3.0, 3.6, 4.2, 4.8, or 5.4.
We aligned each of the 81 artificial Des sequence sets using MAHDS, T-Coffee, MUSCLE, Clustal Omega, Kalign, MAFFT, and PRANK and calculated Z values (Table 3, Table 4, Table 5 and Table 6; only x values for which Z > 0.0 are shown).

Table 3.
Z values for multiple alignments of 81 Des sets built using MAHDS (Z values > 10.0 are in bold).

Table 4.
Z values calculated for multiple alignments of 81 Des sets constructed using Clustal Omega and Kalign (Z values > 10.0 are in bold).

Table 5.
Z values calculated for multiple alignments of 81 Des sets constructed using MAFFT and MUSCLE (Z values > 10.0 are in bold).

Table 6.
Z values calculated for multiple alignments of 81 Des sets built using PRANK and T-Coffee (Z > 10.0 are in bold).
The results indicated that MAHDS could produce statistically significant alignments even if the evolutionary divergence between the aligned sequences (x) was 4.8 substitutions per amino acid; however, it applied to sequences with relatively small numbers of short indels. In other cases, MAHDS can build multiple alignments for x ≤ 3.6 (Table 3). At the same time, most of the other tested methods could not produce statistically significant results for sequences with x > 1.2 (Table 4, Table 5 and Table 6). MUSCLE showed the best performance among the other tested methods, being able to build statistically significant alignments for sequences with x ≤ 2.4 and small numbers of short indels; however, with five indels of 20 residues each, the x value dropped to ≤ 1.2 (Table 5).
2.4. Testing MAHDS Performance on Protein Families with Low Identity
As shown above, MAHDS could construct statistically significant MSAs for artificial sequences with x ≤ 4.8, whereas the other tested algorithms could not produce statistically significant alignments at x > 2.4. In the next experiment, we verified the efficiency of MAHDS in aligning divergent sequences with a high x value using protein families from Pfam and HOMSTRAD databases with a low percentage of identity.
In total, 21 families with an identity of less than 20% were aligned using the MAHDS algorithm, and the constructed MSAs were compared with those built using T-Coffee and MUSCLE chosen because they could produce statistically significant alignments for artificial sequences with the largest x among the other tested methods. These 21 families are shown in Table 7, and the names of protein families received from Pfam are shown in Table 8.

Table 7.
Performance of MAHDS, T-Coffee, and MUSCLE in building MSAs of low identity protein families (Z values > 10.0 are in bold). The first two families are taken from https://mizuguchilab.org/cgi-bin/homstrad/browse.cgi (accessed on 5 June 2021) The remaining 19 were obtained from https://pfam.xfam.org/ (accessed on 7 June 2021). The protein family names obtained from the Pfam database are shown in Table 8.

Table 8.
Protein family names from the Pfam database whose accession number is used in Table 7 are shown in this table.
The performance of MAHDS, T-Coffee, and MUSCLE was evaluated based on statistical significance (Z), the number of gap openings, and the total number of gaps in alignments. The results shown in Table 7 indicated that, for most analyzed families, MAHDS could build alignments of higher statistical significance than the other two methods, with the exception of PF02950, PF09624, PF09987, and PF19443 families, for which the MSAs constructed by MUSCLE had greater Z (however, it should be noted that for PF02950, both MAHDS and MUSCLE showed Z < 10.0). At the same time, only MAHDS was able to construct statistically significant alignments for PF00915, PF10846, PF10895, and PF13944 families. Furthermore, there was a tendency for MAHDS to use fewer gap openings, although the series of gaps tended to be longer than for the other two methods.
3. Discussion
Here, we described the application of our MAHDS method, which was previously used for the alignment of DNA sequences [18,19,20], to the construction of statistically significant alignments of highly divergent protein sequences that have accumulated multiple amino acid substitutions and/or indels. Comparison with the currently available MSA algorithms revealed that MAHDS could produce significant alignments of model sequences with low identity (x up to 4.8), whereas the other methods (T-Coffee, MUSCLE, Clustal Omega, Kalign, MAFFT, and PRANK) could only carry this out for less diverged sequences (x < 2.4). The superior performance of the MAHDS method is due to the use of the progressive algorithm instead of pairwise alignment and calculation of MSA based on patterns of random alignments. In this case, it is possible to replace N-dimensional dynamic programming, which requires significant computational resources, with two-dimensional dynamic programming. Such an approach enabled the construction of more statistically significant MSAs for 21 protein families with a low percentage of identity than could be performed by the other methods.
We made an assessment of the speed of the MAHDS. For 100 amino acid sequences with a length of 100, 400, and 700 amino acids, the running time of the algorithm was 58, 851, and 2370 s, respectively. This suggests that the calculation time grows approximately in a quadratic dependence on the length of the sequences. For sets of amino acid sequences containing 100, 400, and 700 sequences having a length of 100 amino acids, the multiple alignment building times were 58, 333, and 629 s. These results show that there is an approximately linear relationship between the time required to build an alignment and the number of sequences. For calculations, two computing nodes were used. Each computing node includes two Intel Xeon Gold 6240 CPUs. Each processor has nine physical cores, i.e., we used 36 cores.
The developed algorithm made it possible to calculate the statistical significance of any alignment and compare the main methods used in terms of the statistical significance of the alignments that they can construct for protein families. Statistical significance in our work is estimated by the Monte Carlo method based on the generation of random multiple alignments. Using such a Z-score allows us to make direct probabilistic estimates based on which we can filter out statistically insignificant multiple alignments. Such probabilistic estimates are difficult to make using currently used object functions. Such functions include sum of pairs, minimum entropy, or NorMD [34,35].
A question arises regarding when the developed MAHDS method for MSA of amino acid sequences should be applied and what results could be expected. Obviously, MAHDS would be most useful for analysis of those amino acid sequences that have accumulated multiple mutations (x > 2.4), thus facilitating the prediction of the biological role for the amino acid sequences of unknown function [36]. For this, MSA of the protein family in question could be calculated using MAHDS and then analyzed by similarity search using the profile method [37] or HMM [38]. However, the quality of the MSA can strongly influence the search for statistically significant similarities [39]. Therefore, it is important to use MAHDS to compare highly divergent sequences and calculate MSA. Probably, it could be possible to identify residues, for which the biological function is either unknown or the reliability of the annotation is questionable.
The MAHDS method could also be used to reconstruct the evolutionary history of sequences. In this respect, MAHDS can provide an advantage by constructing statistically significant alignments and tracing similarity between evolutionary distant sequences with x = 2.4–4.8, thus resolving the problem of MSA uncertainty [40] and revealing previously unknown evolutionary relationships between different protein families.
By disclosing similarities among highly diverged proteins, MAHDS can also be applied to predict various parameters of protein conformation, including fold recognition and structure-based functional activity [41,42,43], which can help in the construction of three-dimensional models for various proteins [44].
We have developed a site, http://victoria.biengi.ac.ru/mahds/main (3 June 2020), where the user can build multiple alignments for any set of amino acid sequences using MAHDS. Our work was performed using resources of the NRNU MEPhI high-performance computing center.
4. Materials and Methods
4.1. Algorithm to Calculate Multiple Alignment
The MAHDS method was developed early for the multi alignment of promoter sequences and DNA sequences with weak similarity [18,19,20]. This method uses mathematical approaches and programs that were previously created to search for tandem repeats in various sequences [33,45,46,47,48,49]. In the present work, the site http://victoria.biengi.ac.ru/mahds/main (3 June 2020) was used to construct a multiple alignment by the MAHDS method for various amino acid sequences.
Let us briefly consider the mathematical algorithm that underlies the MAHDS method. This algorithm is described in detail in [18]. Let us calculate multiple alignment for a set of N sequences denoted as SI. The optimal multiple alignment (MA) for this set is characterized by a maximum of some function Ψ (maxΨ), which is calculated from MA. Then, we define the image of multiple alignment (IMA) as a function of MA: IMA = f(MA) and also consider an inverse function to f: f−1(IMA) = MA, which allows us to construct MA from IMA and the SI sequences. Specifying functions f and f−1 makes it possible to unambiguously convert MA to its image (IMA) and vice versa, i.e., to establish one-to-one correspondence between MA and IMA.
Several existing algorithms, including progressive alignment [6], HMM [7], and others [3,4] can produce alignment MA’, which is close to the optimal MA. In these methods, the construction of multiple alignment is defined by function ζ: MA’ = ζ(SI). The main disadvantage of such an approach is that the alignment is based on pairwise comparisons of sequences from set SI, which precludes building of statistically significant MA’ when the number of nucleotide substitutions x is greater than 2.4 [18]. A statistically significant MA’ has a probability P(Ψ > Ψ0) < α, where Ψ0 is the value of function Ψ for MA’. This probability is calculated by aligning a large number of sets SR, each of which comprising a series of randomly shuffled sequences of set SI and determining the distribution of function Ψ for this alignment. The value of α can be chosen as 0.05, if the alignment is built once.
However, there is another way of constructing MA’, which is not based on pairwise alignments and direct calculation of MA’ = ζ(SI) but uses patterns (images) of random multiple alignments (IMAR) [18]. These images can be subjected to optimization using genetic algorithms in order to create from a random image IMAR such an image IMAm that would be the closest to IMA = f(MA). The degree of closeness between IMAm and IMA cannot be assessed, since MA and its image IMA = f(MA) are unknown at x > 2.4 for most protein families. However, we can arrive as close as possible to IMA by increasing the similarity measure between each sequence from set SI and IMAm., which can be carried out using global two-dimensional alignment. Each such comparison would produce a maximum value of similarity function Fmax(i) (i = 1, 2, ..., N), and the sum would be used as the measure of similarity between IMAm and IMA. The greater mF is obtained with the same global alignment parameters, the more similarity there is between IMAm and each sequence from set SI and the closer IMAm is to IMA. We will also use mF as an objective function when running a genetic algorithm to calculate IMAm, which then would be used to define multiple alignment: MAm = f−1(IMAm).
In this approach to calculate multiple alignment, we only need to define functions f and f−1. As function f, we can take the algorithm for creating a position weight matrix (PWM), which would be the image of multiple alignment, i.e., IMA = PWM and IMAm = PWMm (described below in Section 4.2). As function f−1, we take the algorithm for the global alignment of PWMm and each sequence from set SI (described earlier [50] and in Section 4.3 and Section 4.4). As a result, it is possible to build multiple alignment with good accuracy using PWMm. Since the sequences in set SI can have different lengths, we calculate the average length of the sequence in the set and create a PWM in the range from to .
Compared to the method described in [18], here we did not combine all sequences from set SI into one large sequence to calculate global alignment with the PWM but instead aligned the PWM with each SI sequence separately, which significantly improved the quality of MSA. Furthermore, the alphabet of SI sequences (Ain) was expanded from 4 to 20 symbols to correspond to the number of amino acids so that protein sequences could be aligned. The scheme of the algorithm is shown in Figure 1.
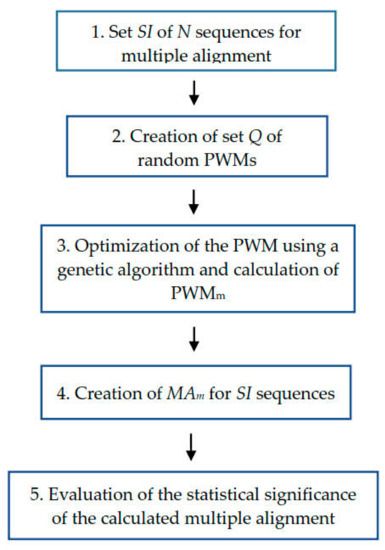
Figure 1.
Diagram of the algorithm for multiple alignment of amino acid sequences shown in Section 4.1. Set of N sequences for multiple alignment is denoted as SI. Q is set of random PWMs (see Section 4.2). PWMm is matrix that has the maximum value of the similarity function when aligning sequences from the set SI (see Section 4.3 and Formula (4)). MAm is a multiple alignment built for sequences from SI set using PWMm (see Section 4.5 and Formula (5)).
At the beginning, we have SI sequences whose number is N (step 1). In step 2, we created a set (Q) of random PWMs, each of which served as a random image IMAR. In step 3, we used a genetic algorithm to optimize each PWM from set Q and determine PWMm = IMAm. In step 4, we created multiple alignment MAm for sequences from set SI, which corresponded to the found PWMm. Finally, in step 5 we evaluated the statistical significance of the Monte Carlo alignment.
4.2. Creation of Set Q of Random PWMs
To create set Q of random PWMs, PWM rows were represented by 20 amino acids and columns by integers from 1 to L; thus, the PWM dimensions were 20 rows and L columns. A random PWM was obtained from a random amino acid sequence S1 of length Lk (k = 103), in which the frequencies of amino acids corresponded with those in the sequences of set SI. Then, a sequence containing integers from 1 to L was generated, copied k times, and joined in tandem to yield sequence S2.
Next, we filled in frequency matrix M(s1(i), s2(i)) = M(s1(i), s2(i)) + 1 for all i from 1 to Lxk, determined sums and , and estimated probability . After this, each PWM element was calculated as . The procedure was repeated 500 times, yielding set Q (step 2, Figure 1).
Then, we performed transformation of the matrices from set Q, which was necessary to ensure that the distribution function of different Fmax(i) (i = 1, 2, ..., N) was similar in the alignment of sequences from set SI with each random PWM. For normalization purposes, two restrictions were imposed on PWMs:
where p1(i) and p2(i) are probabilities of amino acids in sequence S1 and of symbols in sequence S2., respectively. Upon transformation, any PWM used in the algorithm must be reduced to the given R2 and Kd. The matrix transformation procedure is described in detail in [33].
However, it should be noted that R2 cannot be set constant, since in this case the number of cells in the PWM would increase and the cell values would not remain in approximately the same order (as needed) but would rather tend to approach zero in order to correspond to Equation (1). Therefore, R2 was specified as a function of the period and cardinality of the SI sequence alphabet: R2 = RLAinL, where RL is the multiplier parameter, which can be used to scale R2.
As the sequences in set SI may have different lengths, we created sets of matrices Q(L) with length L ranging from to (see Section 4.1). Each Q(L) set contained 500 PWMs.
4.3. Using a Genetic Algorithm to Optimize PWMs
Then, we calculated PWMm using a genetic algorithm described in detail in [18,33] (Figure 1, step 3). Briefly, PWMs of sets Q(L) with the size |Q| = 500 PWMs (Section 4.2) were considered as organisms. To calculate the objective function for each PWM from set Q(L), global alignment with each sequence from set SI was carried out. We calculated similarity function Fmax(l) at point (L1(l), L), where l is the sequence number in set SI, L is the number of columns in the PWM, and L1 is the length of the l-th sequence from set SI. Then, the mF value was calculated and used as an objective function for PWM matrices from set Q(L):
where k is the index of the matrix from set Q(L).
The PWMs were ranked according to mF in the descending order, and the matrix with the largest mF value, mF(1), was saved, whereas two PWMs with the smallest mF values were excluded from set Q(L). Then, we created two children from the remaining PWMs; as before [18,33], the offspring were generated by gluing of two parent matrices, which were chosen randomly. However, the probability of selecting a particular matrix increased with its mF value. In addition, random mutations were introduced into 50 randomly selected PWMs (except children created at this step) by replacing one random element by a random value evenly distributed in the interval from −10 to 10. After these changes, a new Q(L) set was generated, its matrices compared with the sequences from set SI, and a new vector mF(k) (k = 1, 2, ..., 500) was obtained. The procedure of modifying set Q(L) was iterated until mF(1) ceased to increase during the last 10 cycles. As a result of the genetic algorithm for set Q(L), mF(1) (denoted as mFL(1)) and the corresponding PWM were obtained.
The iterative procedure was performed in each Q(L) set for L from to . Finally, we chose the L value for which mFL(1) was maximal; it was denoted as Lm and the corresponding matrix as PWMm.
4.4. Global Alignment of PWMs from Set Q and Sequences from Set SI
Next, sequence SI(l) of length L1(l) was aligned with a PWM of number k from set Q(L). Sequence SI(l) was denoted as S3 and its elements as s3(i), where i = 1, 2, ..., L1(l). We also used sequence S4 with elements s4(j), where j = 1, 2, ..., L.
To construct the alignment of sequences s3(i) and s4(j), we used the global alignment algorithm [51] with an affine gap penalty function and PWM values from set Q(L) [33] instead of substitution matrices. Then, similarity function F was calculated as:
where d and e are gap opening and gap extension penalties, respectively. Fmax(l) = F(L1(l),L) (Section 4.2) was used as a measure of similarity between sequences S3 and S4. We also filled in matrix F’, where in each cell (i, j) we remembered the number of the cell from which we arrived at this cell using Formula (4). As a measure of similarity between set SI(l) and the PWM from set Q(L), we have chosen:
The PWMm of length Lm (Section 4.3) was aligned with set SI(l) using matrix F’, and the results were applied to calculate multiple alignments of sequences SI(l) (l = 1, 2, …, N), which were evaluated according to mF(PWMm) calculated with Formula (6).
4.5. Algorithm for Constructing Multiple Alignment
To obtain MAm for the SI sequences (Figure 1, step 4), we used PWMm and a set of global alignments of the SI(l) sequences (l = 1, 2, ..., N,) with PWMm. For this, sequence S4 with elements s4(j) (j = 1, 2, ..., Lm) was arranged horizontally, and under it the alignments of SI(l) sequences (l = 1, 2, ..., N) generated in Section 4.4 were written. If any of the SI(l) sequences had an insertion of k amino acids compared to sequence S4, then additional k columns were created for these amino acids in the s4(j) sequence (if they were not created for any other SI(l) previously). As a result, MAm was constructed.
4.6. Estimating the Statistical Significance of Multiple Alignments
4.6.1. Assessing the Statistical Significance of MAm
To evaluate the statistical significance of the constructed multiple alignment MAm obtained with PWMm (Figure 1, step 5), we used the Monte Carlo method. First, we generated 300 sets of random sequences Qr through random shuffling of residues in the SI sequences. For each Qr we calculated mF(PWMm) using Formula (6), its arithmetic mean and dispersion D(mF(PWMm)), and, finally, Z:
A Z value greater than threshold Z0 indicated that the alignment obtained with PWMm was statistically significant.
4.6.2. Estimating the Statistical Significance of an Arbitrary MA
The algorithm described above can be applied to evaluate the statistical significance of any alignment. Let us take an MA with length K containing α sequences, each denoted as and its elements as (where k = 1, 2, ..., α and j = 1, 2, ..., K). Let us also introduce sequence S6 with elements s6(j), j = 1, 2, ..., K representing consecutive numbers from 1 to K; S6 would be used as column numbers for MA. First, we transform MA to remove those columns where the number of amino acids is less than α/2 and that of deletions is more than α/2; as a result, multiple alignment MA’ of length K’ is obtained. At the same time, sequence S6 is transformed by eliminating the numbers equal to those of the columns to be removed. As a result, we obtain sequence with length K’. Then, we calculate the amino acid frequency matrix for MA’ denoted as V(i,j), where i = 1, 2, …, 20 and j = 1, 2, …, K’; each element shows the number of type i residues at position j of MA’. Next, we calculate the PWM based on matrix V(i,j) as: , where U is the total number of amino acids in MA’, , X(i) is the number of type i residues, and Y(j) is the total number of residues in the j-th column of MA’. Then, we transform PWM(i,j) according to Formulas (2) and (3).
After calculating PWM′(i,j), the weight of MA can be determined. First, we compute the sum for all k = 1, 2, ..., α and j = 1, 2, ..., K: . Next, we obtain the Del(i) vector, which shows the number of deletions with size i in MA. Finally, we calculate , which is considered as the MA weight. Then, statistical significance should be evaluated by estimating mean and variance D(FMA). For this, symbols characterizing deletions are removed from sequences, which are then randomly shuffled to yield sequences, where k = 1, 2, ..., α, and the statistical significance is determined for matrix PWM′(i,j) and sequences according to Formula (7), assuming that , N = K, and PWMm = PWM′(i,j). Thus, we calculate the Z value for MA, which enables comparison of different MAs by their statistical significance.
4.7. Creation of Artificial Sequences to Compare Different Methods of Constructing MAs
To compare the performance of MA methods T-Coffee, MUSCLE, Clustal Omega, Kalign, MAFFT, and PRANK with that of the MAHDS algorithm, we used artificial sequences, which can be created with a given number of amino acid substitutions and indels for convenience of analysis. To generate an artificial sequence, we first created a random ancestor sequence Anc of length L and then a set of descendant sequences Des(i) (i = 1, 2, …, 100) by adding a given number of random substitutions and/or indels to Anc in a random order. In case of random substitutions without indels, the lengths of child sequences were the same and equal to L, and the number of substitutions in each Des(i) sequence with respect to Anc was denoted as s1. Let us show that the presence of s1 random substitutions in each Des(i) sequence leads to 2s1 random substitutions between any two Des sequences.
When one random substitution is made in sequence Anc, the probability that in sequence Des(1) with the number of substitutions s1 a residue at position j will be changed is 1/L. Then, the probability that after s1 replacements residue j will not be replaced is:
We assume that amino acid substitutions during the creation of the Des(1) sequence occur with equal probability. For the rest of amino acids, it is likely that during the last replacement in a given position, the original amino acid will appear as substitution. The probability of falling into this category is:
where is 20. Thus, the probability that residue j in sequence Des(1) will match the residue at Anc is:
The events described in Formulas (9) and (10) are applicable to all situations, when a residue at position j remains unchanged after random substitutions.
Now, let us consider the case when sequences Des(2) and Des(3) are independently generated from sequence Anc by making s2 random substitutions. Then, for both sequences, Formulas (8)–(10) can be presented as: , , . Let us take Pm3 as a probability that in Des(2) and Des(3) two residues could match. If Pm3 = Pm, then the evolutionary distance between Anc and Des(1) is equal to that between Des(2) and Des(3), which means that, on average, the same number of amino acids coincide between Anc and Des(1) and between Des(2) and Des(3). In Des(2) and Des(3), the proportion of residues unchanged compared to Anc is P02; then, the proportion of residues preserved at the same positions of child sequences is , since it is the probability for a residue to remain in both Des(2) and Des(3). Other amino acids matching in Des(2) and Des(3) include those that have been substituted with the same residues in both sequences, and the probability of such events is .
Finally, there is a probability that a residue that remained from Anc in Des(2) could match the one randomly replaced in Des(3) and vice versa. The sum of the two probabilities is: . Then, = and . If Pm3 = Pm, then , i.e., . If the values for P02 and P0 are substituted, then:
Formula (11) means that the number of s2 random substitutions introduced in Des(i) sequences is equivalent to twice the number of s2 mutations in pairwise comparison of Des(i) sequences relative to Anc, which happens under the condition that all child sequences in set Des(i) (i = 1, 2, ..., 100) are generated independently of each other. We used this property in the calculation of the average number of mutations between Des(i) sequences.
Programs developed in paragraphs 4.6.2 and 4.7 can be obtained from the site https://github.com/katri2/MAHDS_addition (21 March 2022).
Author Contributions
Conceptualization, E.V.K.; methodology, E.V.K. and D.O.K.; software, D.O.K.; validation, E.V.K. and D.O.K.; formal analysis, E.V.K. and D.O.K.; investigation, D.O.K.; resources, D.O.K.; data curation, E.V.K. and D.O.K.; writing—original draft preparation, D.O.K.; writing—review and editing, E.V.K.; supervision, E.V.K.; project administration, E.V.K.; funding acquisition, E.V.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly funded by the RFBR grant N.20-016-00057A.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Noorden, R.; Maher, B.; Nuzzo, R. The top 100 papers. Nature 2014, 514, 550–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Linard, B.; Lecompte, O.; Poch, O. A comprehensive benchmark study of multiple sequence alignment methods: Current challenges and future perspectives. PLoS ONE 2011, 6, e18093. [Google Scholar] [CrossRef] [PubMed]
- Kemena, C.; Notredame, C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics 2009, 25, 2455–2465. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Jiang, T. On the Complexity of Multiple Sequence Alignment. J. Comput. Biol. 1994, 1, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Hogeweg, P.; Hesper, B. The alignment of sets of sequences and the construction of phyletic trees: An integrated method. J. Mol. Evol. 1984, 20, 175–186. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2014, 7, 539. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, O. Multiple sequence alignment: Algorithms and applications. Adv. Biophys. 1999, 36, 159–206. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- UCSC-CRL-96-22: SAM: Sequence Alignment and Modeling Software System | Technical Reports. Available online: https://tr.soe.ucsc.edu/research/technical-reports/UCSC-CRL-96-22 (accessed on 19 November 2021).
- Analysis, B.S. Multiple Sequence Alignment using Profile HMM. Nucleic Acids Res. 1998, 26, 320–322. [Google Scholar]
- Notredame, C.; O’Brien, E.A.; Higgins, D.G. RAGA: RNA sequence alignment by genetic algorithm. Nucleic Acids Res. 1997, 25, 4570–4580. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Pramanik, S.; Chung, M.J. Multiple sequence alignment using simulated annealing. Comput. Appl. Biosci. 1994, 10, 419–426. [Google Scholar] [CrossRef]
- Löytynoja, A.; Goldman, N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 2008, 320, 1632–1635. [Google Scholar] [CrossRef] [Green Version]
- Szalkowski, A.M. Fast and robust multiple sequence alignment with phylogeny-aware gap placement. BMC Bioinf. 2012, 13, 129. [Google Scholar] [CrossRef] [Green Version]
- Korotkov, E.V.; Suvorova, Y.M.; Kostenko, D.O.; Korotkova, M.A. Multiple alignment of promoter sequences from the arabidopsis thaliana l. Genome. Genes 2021, 12, 135. [Google Scholar] [CrossRef]
- Kamionskaya, A.M.; Korotkova, M.A. Multiple Alignment of Promoter Sequences from the Human Genome. Biotekhnologiya 2020, 36, 7–14. [Google Scholar] [CrossRef]
- Korotkov, E.V.; Korotkova, M.A. Developing mathematical method for multi alignment of DNA sequences with weak similarity. J. Phys. Conf. Ser. 2019, 1205, 12025. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Frith, M.C. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 2012, 28, 3144–3146. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Sonnhammer, E. Kalign—An accurate and fast multiple sequence alignment algorithm. BMC Bioinf. 2005, 6, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korotkova, M.A.; Kamionskya, A.M.; Korotkov, E.V. A mathematical method for the classification of promoter sequences from the A.thaliana genome. J. Phys. Conf. Ser. 2020, 1686, 012031. [Google Scholar] [CrossRef]
- Korotkov, E.V.; Suvorova, Y.M.; Nezhdanova, A.V.; Gaidukova, S.E.; Yakovleva, I.V.; Kamionskaya, A.M.; Korotkova, M.A. Mathematical Algorithm for Identification of Eukaryotic Promoter Sequences. Symmetry 2021, 13, 917. [Google Scholar] [CrossRef]
- Thompson, J.D.; Plewniak, F.; Poch, O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999, 27, 2682–2690. [Google Scholar] [CrossRef]
- Thompson, J.D.; Koehl, P.; Ripp, R.; Poch, O. BAliBASE 3.0: Latest developments of the multiple sequence alignment benchmark. Proteins Struct. Funct. Genet. 2005, 61, 127–136. [Google Scholar] [CrossRef]
- Pais, F.S.M.; Ruy, P.C.; Oliveira, G.; Coimbra, R.S. Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol. Biol. 2014, 9, 4. [Google Scholar] [CrossRef] [Green Version]
- Bahr, A.; Thompson, J.D.; Thierry, J.-C.; Poch, O. BAliBASE (Benchmark Alignment dataBASE): Enhancements for repeats, transmembrane sequences and circular permutations. Nucleic Acid Res. 2001, 29, 323–326. [Google Scholar] [CrossRef]
- Browning, B.L.; Browning, S.R. Detecting Identity by Descent and Estimating Genotype Error Rates in Sequence Data. Am. J. Hum. Genet. 2013, 93, 840–851. [Google Scholar] [CrossRef] [Green Version]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [Green Version]
- Bastien, O.; Aude, J.-C.; Roy, S.; Maréchal, E. Fundamentals of massive automatic pairwise alignments of protein sequences: Theoretical significance of Z-value statistics. Bioinformatics 2004, 20, 534–537. [Google Scholar] [CrossRef]
- Pugacheva, V.; Korotkov, A.; Korotkov, E. Search of latent periodicity in amino acid sequences by means of genetic algorithm and dynamic programming. Stat. Appl. Genet. Mol. Biol. 2016, 15, 381–400. [Google Scholar] [CrossRef]
- Heringa, J.; Argos, P. A method to recognize distant repeats in protein sequences. Proteins 1993, 17, 391–411. [Google Scholar] [CrossRef]
- Russell, D.J. Multiple Sequence Alignment Methods—Edited by David J. Russell. Syst. Biol. 2015, 64, 690–692. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef]
- Haritha, P.; Shanmugavadivu, P.; Dhamodharan, S. A Comprehensive Review on Protein Sequence Analysis Techniques. Int. J. Comput. Sci. Eng. 2018, 6, 1433–1442. [Google Scholar] [CrossRef]
- Yoon, B.-J. Hidden Markov Models and their Applications in Biological Sequence Analysis. Curr. Genom. 2009, 10, 402–415. [Google Scholar] [CrossRef] [Green Version]
- Gil, N.; Fiser, A. The choice of sequence homologs included in multiple sequence alignments has a dramatic impact on evolutionary conservation analysis. Bioinformatics 2019, 35, 12–19. [Google Scholar] [CrossRef]
- Spielman, S.J.; Miraglia, M.L. Relative model selection of evolutionary substitution models can be sensitive to multiple sequence alignment uncertainty. BMC Ecol. Evol. 2021, 21, 214. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, Y. ANGLOR: A composite machine-learning algorithm for protein backbone torsion angle prediction. PLoS ONE 2008, 3, e3400. [Google Scholar] [CrossRef]
- Hanson, J.; Paliwal, K.; Litfin, T.; Yang, Y.; Zhou, Y. Accurate prediction of protein contact maps by coupling residual two-dimensional bidirectional long short-term memory with convolutional neural networks. Bioinformatics 2018, 34, 4039–4045. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Wuyun, Q.; Pearce, R.; Li, Y.; Zhang, Y. LOMETS2: Improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins. Nucleic Acids Res. 2019, 47, W429–W436. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Mortuza, S.M.; He, B.; Wang, Y.; Zhang, Y. Template-based and free modeling of I-TASSER and QUARK pipelines using predicted contact maps in CASP12. Proteins Struct. Funct. Bioinform. 2018, 86 (Suppl. 1), 136–151. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, V.; Korotkov, E. Search for Highly Divergent Tandem Repeats in Amino Acid Sequences. Int. J. Mol. Sci. 2021, 22, 7096. [Google Scholar] [CrossRef]
- Korotkov, E.V.; Kamionskya, A.M.; Korotkova, M.A. Detection of highly divergent tandem repeats in the rice genome. Genes 2021, 12, 473. [Google Scholar] [CrossRef] [PubMed]
- Nor, A.; Korotkov, E. Search of Fuzzy Periods in the Works of Poetry of Different Authors. Adv. Fuzzy Syst. 2018, 2018, 4028417. [Google Scholar] [CrossRef]
- Korotkov, E.V.; Korotkova, M.A. Search for regions with periodicity using the random position weight matrices in the C. elegans genome. Int. J. Data Min. Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef]
- Korotkov, E.; Korotkova, M. Study of the periodicity in Euro-US Dollar exchange rates using local alignment and random matrixes. Procedia Comput. Sci. 2017, 108, 1344–1353. [Google Scholar] [CrossRef]
- Laskin, A.A.; Korotkov, E.V.; Chalei, M.B.; Kudryashov, N.A. The locally optimal method of cyclic alignment to reveal latent periodicities in genetic texts. The NAD-binding protein sites. Mol. Biol. 2003, 37, 663–673. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).