Profiles of Cytokinins Metabolic Genes and Endogenous Cytokinins Dynamics during Shoot Multiplication In Vitro of Phalaenopsis
Abstract
:1. Introduction
2. Results
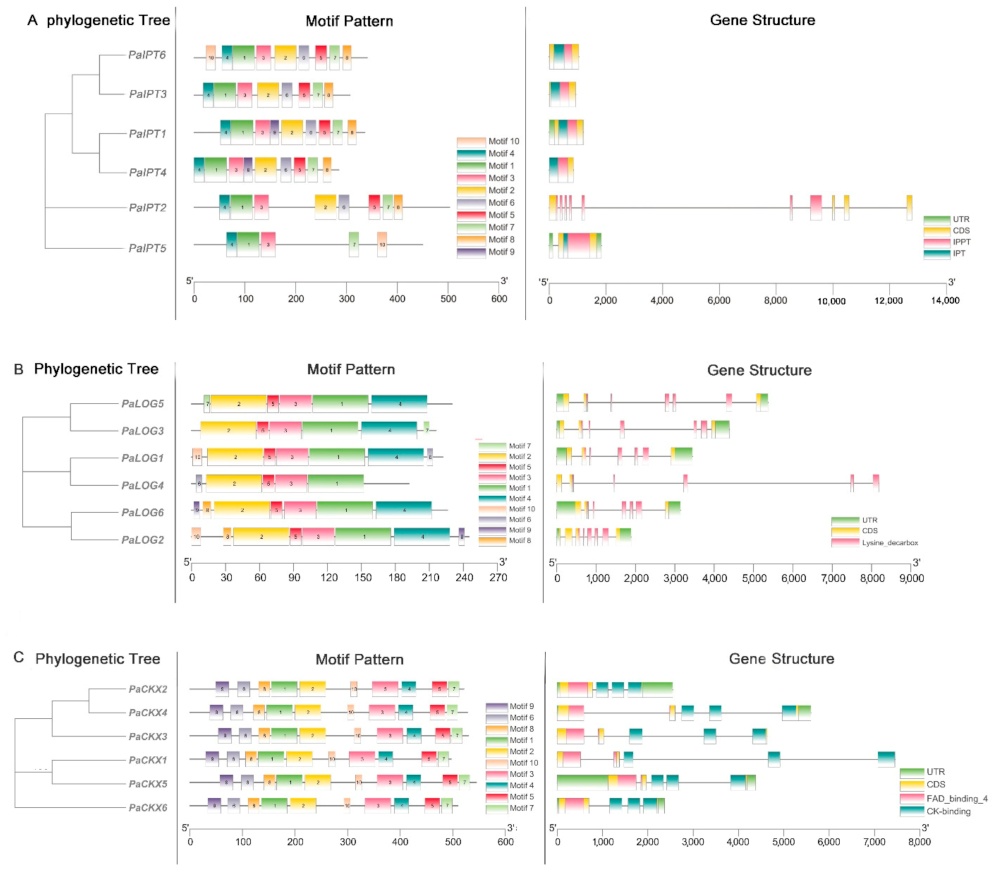
2.1. Identification of IPT, LOG, and CKX Proteins in Phalaenopsis
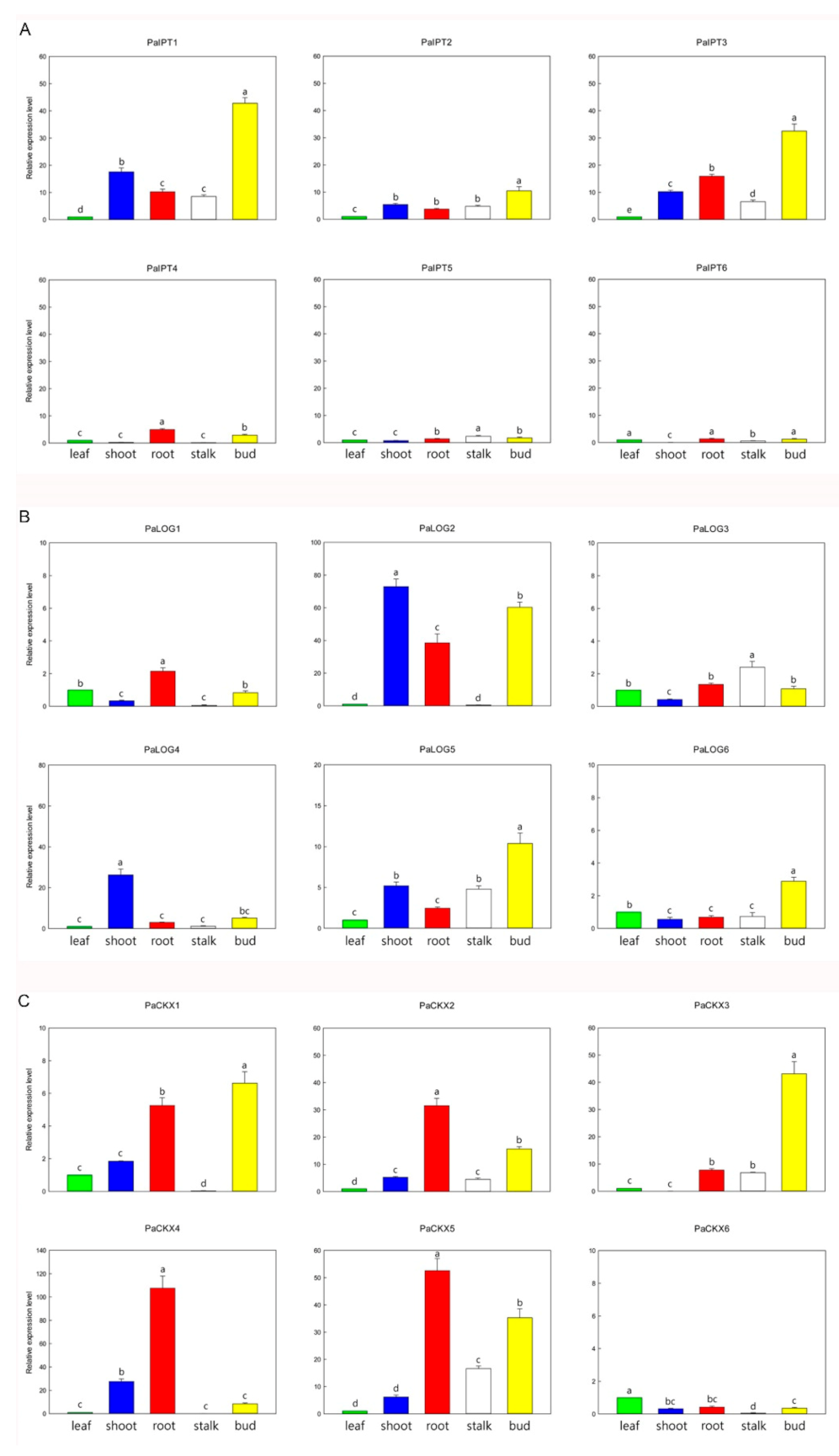
2.2. Expression of IPT, LOG, and CKX Genes in Different Organs
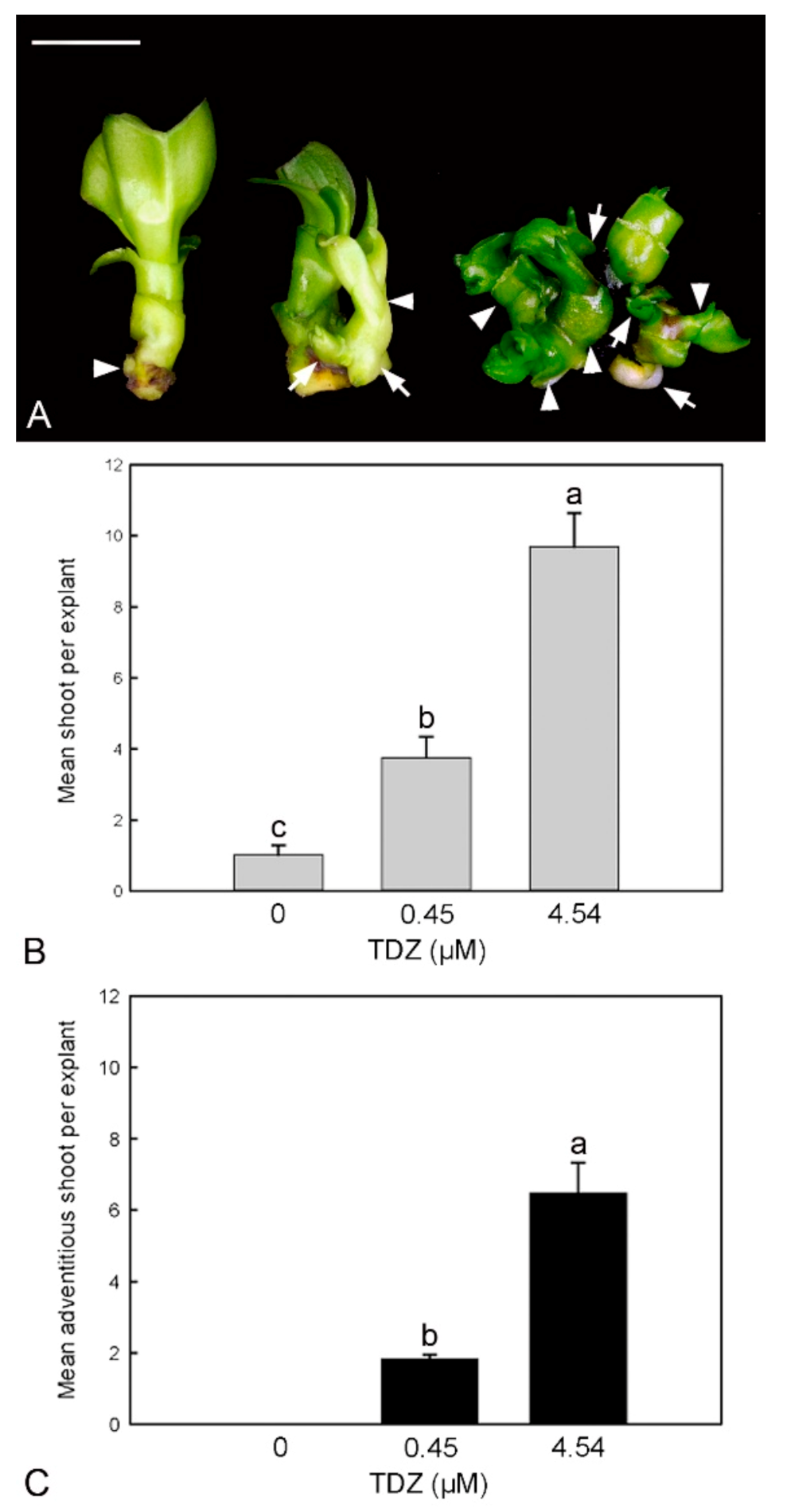
2.3. Effect of TDZ Treatments on Shoot Multiplication In Vitro
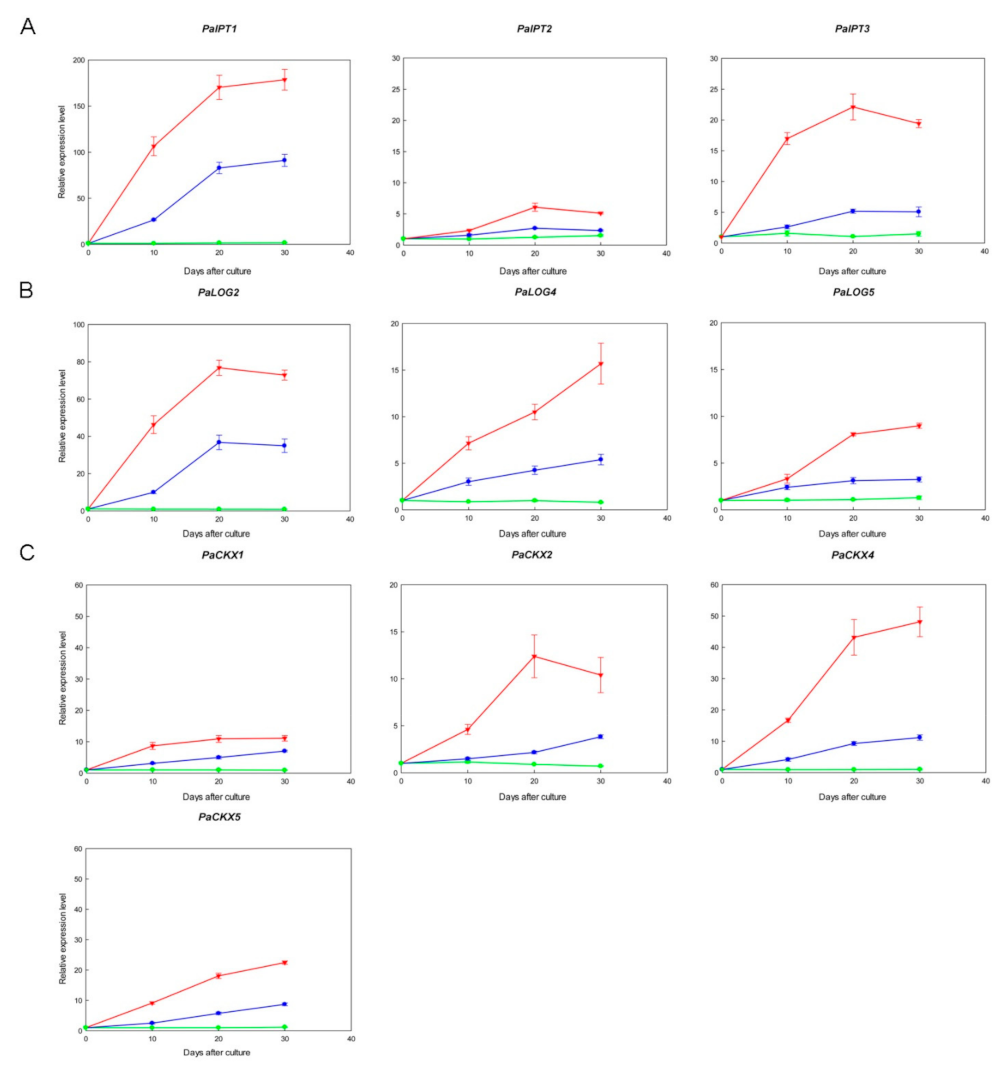
2.4. Expression of IPT, LOG, and CKX Genes in Micropropagated Shoots
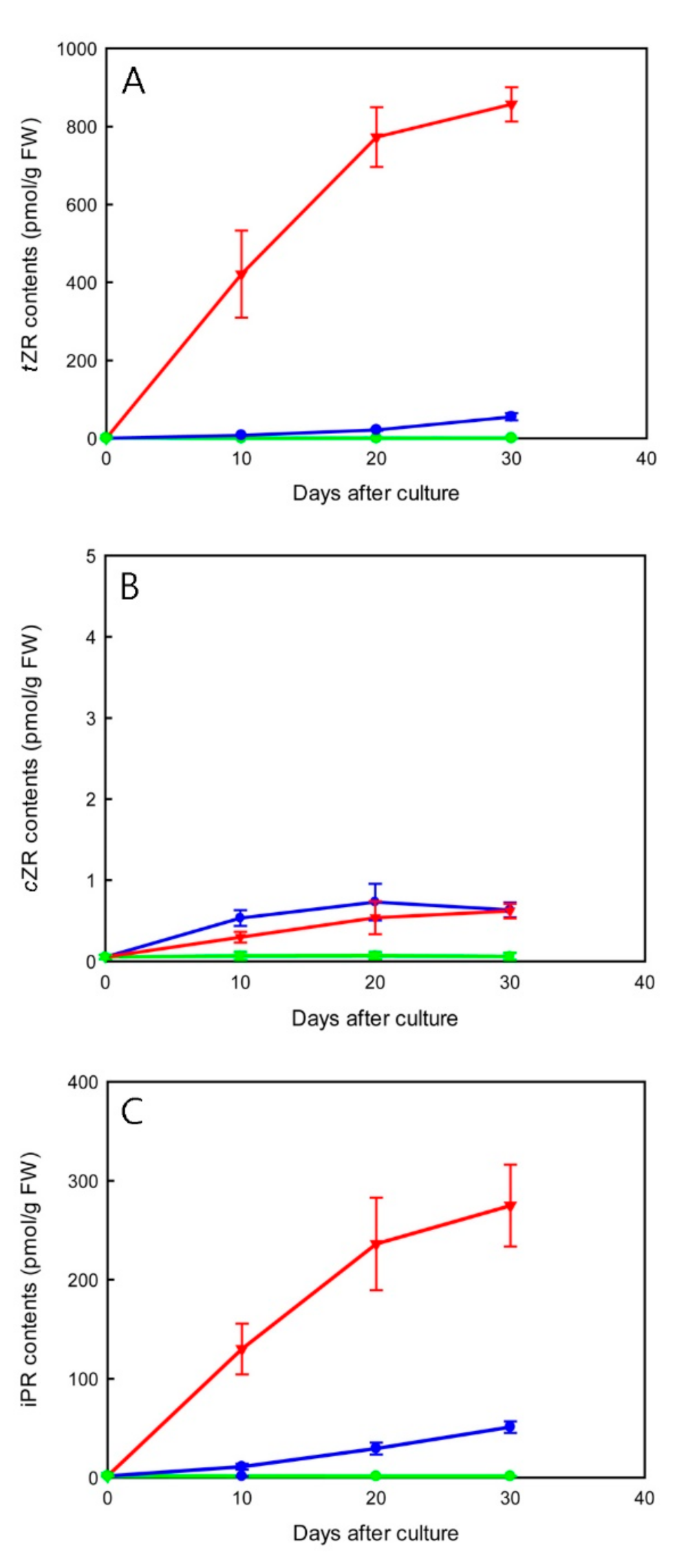
2.5. Effect of TDZ Treatments on Endogenous Cytokinin-Ribosides Levels
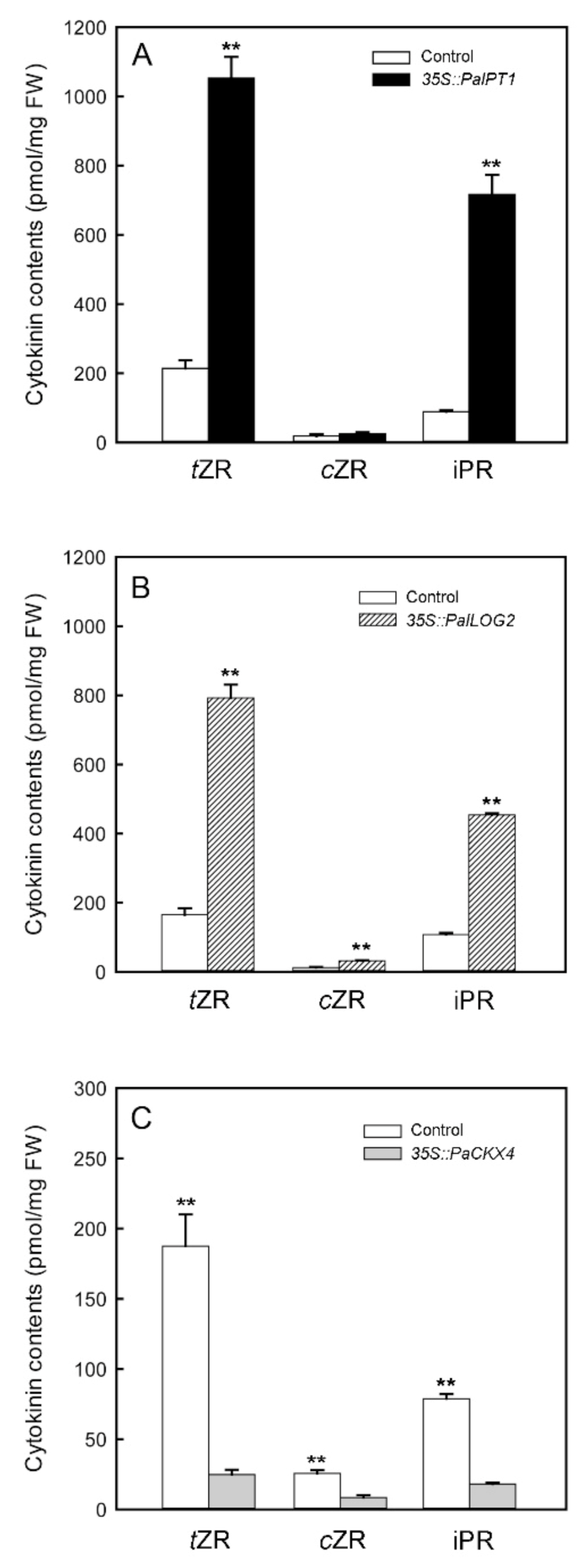
2.6. Transient Overexpression of PaIPT1, PaLOG2, and PaCKX4 in Tobacco Leaves
3. Discussion
4. Materials and Methods
4.1. Identification of Phalaenopsis IPT, LOG, and CKX Genes
4.2. Plant Materials and Tissue Culture Experiments
4.3. Quantitative RT-PCR (RT-qPCR)
4.4. Transient Overexpression of PaIPT, PaLOG, and PaCKX Proteins in Tobacco Leaves
4.5. Quantification of Endogenous CK-Ribosides
4.6. Experimental Design and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yam, T.W.; Arditti, J. Micropropagation of Orchids; John Wiley and Sons, Inc.: Chichester, UK, 2017. [Google Scholar]
- Tokuhara, K.; Mii, M. Micropropagation of Phalaenopsis and Doritaenopsis by shoot tips of flower stalk buds. Plant Cell Rep. 1993, 13, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Tokuhara, K.; Mii, M. Induction of embryogenic callus and cell suspension culture from shoot tips excised from flower stalk buds of Phalaenopsis (Orchidaceae). In Vitro Cell. Dev. Biol. Plant 2001, 37, 457–461. [Google Scholar] [CrossRef]
- Mok, D.W.S.; Mok, M.C. Cytokinin metabolism and action. Ann. Rev. Plant Physiol. Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, W.M.; Kerbauy, G.B.; Kraus, J.E.; Pescador, R.; Suzuki, R.M. Thidiazuron influences the endogenous levels of cytokinins and IAA during the flowering of isolated shoots of Dendrobium. J. Plant Physiol. 2006, 163, 1126–1134. [Google Scholar] [CrossRef]
- Podwyszynska, M.; Novak, O.; Dolezal, K.; Strnad, M. Endogenous cytokinin dynamics in micropropagated tulips during bulb formation process influenced by TDZ and iP pretreatment. Plant Cell Tiss. Organ. Cult. 2014, 119, 331–346. [Google Scholar] [CrossRef] [Green Version]
- Smýkalová, I.; Vrbová, M.; Cvečková, M.; Plačková, L.; Žukauskaitė, A.; Zatloukal, M.; Hrdlička, J.; Plíhalová, L.; Doležal, K.; Griga, M. The effects of novel synthetic cytokinin derivatives and endogenous cytokinins on the in vitro growth responses of hemp (Cannabis sativa L.) explants. Plant Cell Tiss. Organ. Cult. 2019, 139, 381–394. [Google Scholar] [CrossRef]
- Bairu, M.W.; Novák, O.; Doležal, K.; Van Staden, J. Changes in endogenous cytokinin profiles in micropropagated Harpagophytum procumbens in relation to shoot-tip necrosis and cytokinin treatments. Plant Growth Regul. 2011, 63, 105–114. [Google Scholar] [CrossRef]
- Bajguz, A.; Piotrowska, A. Conjugates of auxin and cytokinin. Phytochemistry 2009, 70, 957–969. [Google Scholar] [CrossRef]
- Letham, D.S.; Palni, L.M.S. The biosynthesis and metabolism of cytokinins. Annu. Rev. Plant Physiol. 1983, 34, 163–197. [Google Scholar] [CrossRef]
- Frebort, I.; Kowalska, M.; Hluska, T.; Frebortova, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis lonely guy (LOG) multiplemutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012, 69, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Sakakibara, H.; Sugiyama, T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 26405–26410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmüling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [Green Version]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Vankova, R.; Tanaka, M.; Seki, M.; Ham, L.H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.P. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS ONE 2012, 7, e42411. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, S.; Kikuchi, K.; Fukuda, M.; Honda, I.; Imanishi, S. Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 2012, 63, 5569–5579. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.H.; Schaller, G.E.; Kieber, J.J. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.N.; Lv, Y.X.; Zhang, M.; Liu, Y.; Kong, L.J.; Zou, M.H.; Lu, G.; Cao, J.S.; Yu, X.L. Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 2013, 14, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, Y.T.; Chen, W.C.; Chen, C.Y.; Ho, H.Y.; Yeh, C.H.; Kuo, Y.Z.; Su, C.L.; Yen, S.H.; Hsueh, H.Y.; Yeh, J.H.; et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome providesnew insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018, 16, 2027–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, M.C.; Mor, D.W.S.; Armstrong, D.J.; Shudo, K.; Okamoto, T. Cytokinin activity of N-phenil-N′-1,2,3-thidiazol-5-ylurea (thidiazuron). Phytochemistry 1982, 21, 1509–1511. [Google Scholar] [CrossRef]
- Chang, C.; Chang, W.C. Plant regeneration from callus of Cymbidium ensifolium var ‘misericors’. Plant Cell Rep. 1998, 17, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Chen, J.T.; Chang, W.C. Multiple shoot formation and plant regeneration from stem nodal explants of Paphiopedilum orchids. In Vitro Cell. Dev. Biol. Plant 2002, 38, 595–597. [Google Scholar] [CrossRef]
- Park, S.Y.; Yeung, E.C.; Chakrabarty, D.; Paek, K.Y. An efficient direct induction of protocorm-like bodies from leaf subepidermal cells of Doritaenopsis hybrid using thin-section culture. Plant Cell Rep. 2002, 21, 46–51. [Google Scholar]
- Lee, Y.I.; Lee, N. Plant regeneration from protocorm-derived callus of Cypripedium formosanum. In Vitro Cell. Dev. Biol. Plant 2003, 39, 475–479. [Google Scholar] [CrossRef]
- Capelle, S.C.; Mok, D.W.S.; Kirchner, S.C.; Mok, M.C. Effects of TDZ on cytokinin autonomy and the metabolism of N6-(Δ2-isopentenyl)[8-14C]adenosine in callus tissues of Phaseolus lunatus L. Plant Physiol. 1983, 73, 796–802. [Google Scholar] [CrossRef] [Green Version]
- Mok, M.C.; Mok, D.W.S. The metabolism of [14C]-thidiazuron in callus tissues of Phaseolus lunatus. Physiol. Plant 1985, 65, 427–432. [Google Scholar] [CrossRef]
- Hare, P.D.; van Staden, J. Inhibitory effect of thidiazuron on theactivity of cytokinin oxidase isolated from soybean callus. Plant Cell Physiol. 1994, 35, 1121–1125. [Google Scholar] [CrossRef]
- Murthy, B.N.S.; Murch, S.J.; Saxena, P.K. TDZ-induced somatic embryogenesis in intact seedlings of peanut (Arachis hypogaea): Endogenous growth regulator levels and significance of cotyledons. Physiol. Plant. 1995, 94, 268–276. [Google Scholar] [CrossRef]
- Hutchinson, M.J.; Saxena, P.K. Role of purine metabolism in TDZ-induced somatic embryogenesis of geranium (Pelargonium xhorturum) hypocotyl cultures. Physiol. Plant. 1996, 98, 517–522. [Google Scholar] [CrossRef]
- Bilyeu, K.D.; Cole, J.L.; Laskey, J.G.; Riekhof, W.R.; Esparza, T.J.; Kramer, M.D.; Morris, R.O. Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 2001, 125, 378–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Ding, Y.; Wang, Q.; Wang, S. Auxin inhibits the outgrowth of tiller buds in rice (Oryza sativa L.) by downregulating OsIPT expression and cytokinin biosynthesis in nodes. Aust. J. Crop Sci. 2011, 5, 169–174. [Google Scholar]
- Gajdosova, S.; Spíchal, L.; Kamínek, M.; Hoyerová, K.; Novák, O.; Dobrev, P.I.; Galuszka, P.; Klíma, P.; Gaudinová, A.; Žižková, E. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 2011, 62, 2827–2840. [Google Scholar] [CrossRef] [Green Version]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Böttcher, C.; Burbidge, C.A.; Boss, P.K.; Davies, C. Changes in transcription of cytokinin metabolism and signalling genes in grape (Vitis vinifera L.) berries are associated with the ripening-related increase in isopentenyladenine. BMC Plant Biol. 2015, 15, 223. [Google Scholar] [CrossRef] [Green Version]
- Galuszka, P.; Popelková, H.; Werner, T.; Frébortová, J.; Pospíšilová, H.; Mik, V.; Köllmer, I.; Schmülling, T.; Frébort, I.; Galuszka, P.; et al. Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J. Plant Growth Regul. 2007, 26, 255–267. [Google Scholar] [CrossRef]
- Chatfield, J.M.; Armstrong, D.J. Regulation of cytokinin oxidase activity in callus tissues of Phaseolus vulgaris L. cv Great Northern. Plant Physiol. 1986, 80, 493–499. [Google Scholar] [CrossRef] [Green Version]
- Motyka, V.; Faiss, M.; Strnad, M.; Kamínek, M.; Schülling, T. Changes in cytokinin content and cytokinin oxidaseactivity in response to derepression of IPT gene transcription in transgenic tobacco calli and plants. Plant Physiol. 1996, 112, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.H.; Yu, H.; Goh, C.J. Functional characterization of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid. Plant Mol. Biol. 2003, 51, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Piluek, C. Effects of thidiazuron and N6-benzylaminopurine on shoot regeneration of Phalaenopsis. Plant Growth Regul. 1995, 16, 99–101. [Google Scholar] [CrossRef]
- Wang, J.; Tian, C.; Cui, Z.; Shi, B.; Jiao, Y. Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 2017, 29, 1373–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, T.; Higuchi, M.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Kato, T.; Tabata, S.; Shinozaki, K.; Kakimoto, T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 2001, 409, 1060–1063. [Google Scholar] [CrossRef]
- Spíchal, L.; Yu, R.N.; Michael, R.; Takeshi, M.; Romanov, G.A.; Miroslav, S.; Thomas, S. Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 2004, 45, 1299–1305. [Google Scholar] [CrossRef] [Green Version]
- Murthy, B.N.S.; Murch, S.J.; Saxena, P.K. Thidiazuron: A potent regulator of in vitro morphogenesis. In Vitro Cell. Dev. Biol. Plant 1998, 34, 267–275. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Cheng, S.F.; Huang, Y.P.; Wu, Z.R.; Hu, C.C.; Hsu, Y.H.; Tsai, C.H. Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biol. 2010, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bollmark, M.; Kubat, B.; Eliasson, L. Variations in endogenous cytokinin content during adventitious root formation in pea cuttings. J. Plant Physiol. 1988, 132, 262–265. [Google Scholar] [CrossRef]
- Eberle, J.; Wang, T.L.; Cook, S.; Wells, B.; Weiler, E.W. Immunoassay and ultrastructural localization of isopentenyladenine and related cytokinins using monoclonal antibodies. Planta 1987, 172, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Zhang, J.H.; Wang, Z.Q.; Zhu, Q.S.; Wang, W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-Y.; Hao, Z.-G.; Miao, S.; Zhang, X.; Li, J.-Q.; Guo, S.-X.; Lee, Y.-I. Profiles of Cytokinins Metabolic Genes and Endogenous Cytokinins Dynamics during Shoot Multiplication In Vitro of Phalaenopsis. Int. J. Mol. Sci. 2022, 23, 3755. https://doi.org/10.3390/ijms23073755
Li Y-Y, Hao Z-G, Miao S, Zhang X, Li J-Q, Guo S-X, Lee Y-I. Profiles of Cytokinins Metabolic Genes and Endogenous Cytokinins Dynamics during Shoot Multiplication In Vitro of Phalaenopsis. International Journal of Molecular Sciences. 2022; 23(7):3755. https://doi.org/10.3390/ijms23073755
Chicago/Turabian StyleLi, Yuan-Yuan, Zhi-Gang Hao, Shuo Miao, Xiong Zhang, Jian-Qiang Li, Shun-Xing Guo, and Yung-I Lee. 2022. "Profiles of Cytokinins Metabolic Genes and Endogenous Cytokinins Dynamics during Shoot Multiplication In Vitro of Phalaenopsis" International Journal of Molecular Sciences 23, no. 7: 3755. https://doi.org/10.3390/ijms23073755
APA StyleLi, Y.-Y., Hao, Z.-G., Miao, S., Zhang, X., Li, J.-Q., Guo, S.-X., & Lee, Y.-I. (2022). Profiles of Cytokinins Metabolic Genes and Endogenous Cytokinins Dynamics during Shoot Multiplication In Vitro of Phalaenopsis. International Journal of Molecular Sciences, 23(7), 3755. https://doi.org/10.3390/ijms23073755





