Loss of Interleukin-13-Receptor-Alpha-1 Induces Apoptosis and Promotes EMT in Pancreatic Cancer
Abstract
1. Introduction
2. Results
2.1. Expression of IL-13Rα1, IL-4Rα and γc Chains in Cultured Human Pancreatic Cancer Cells
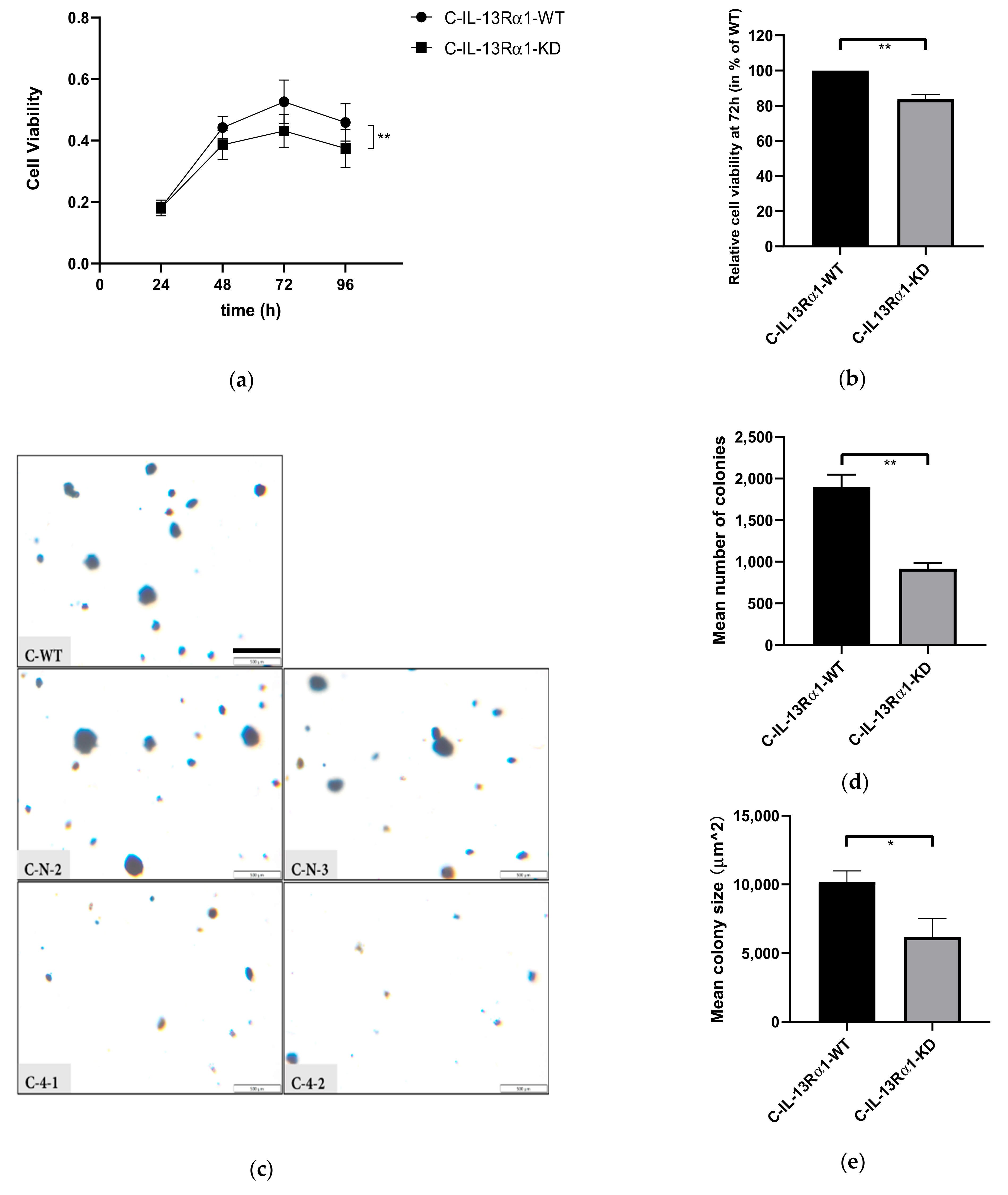
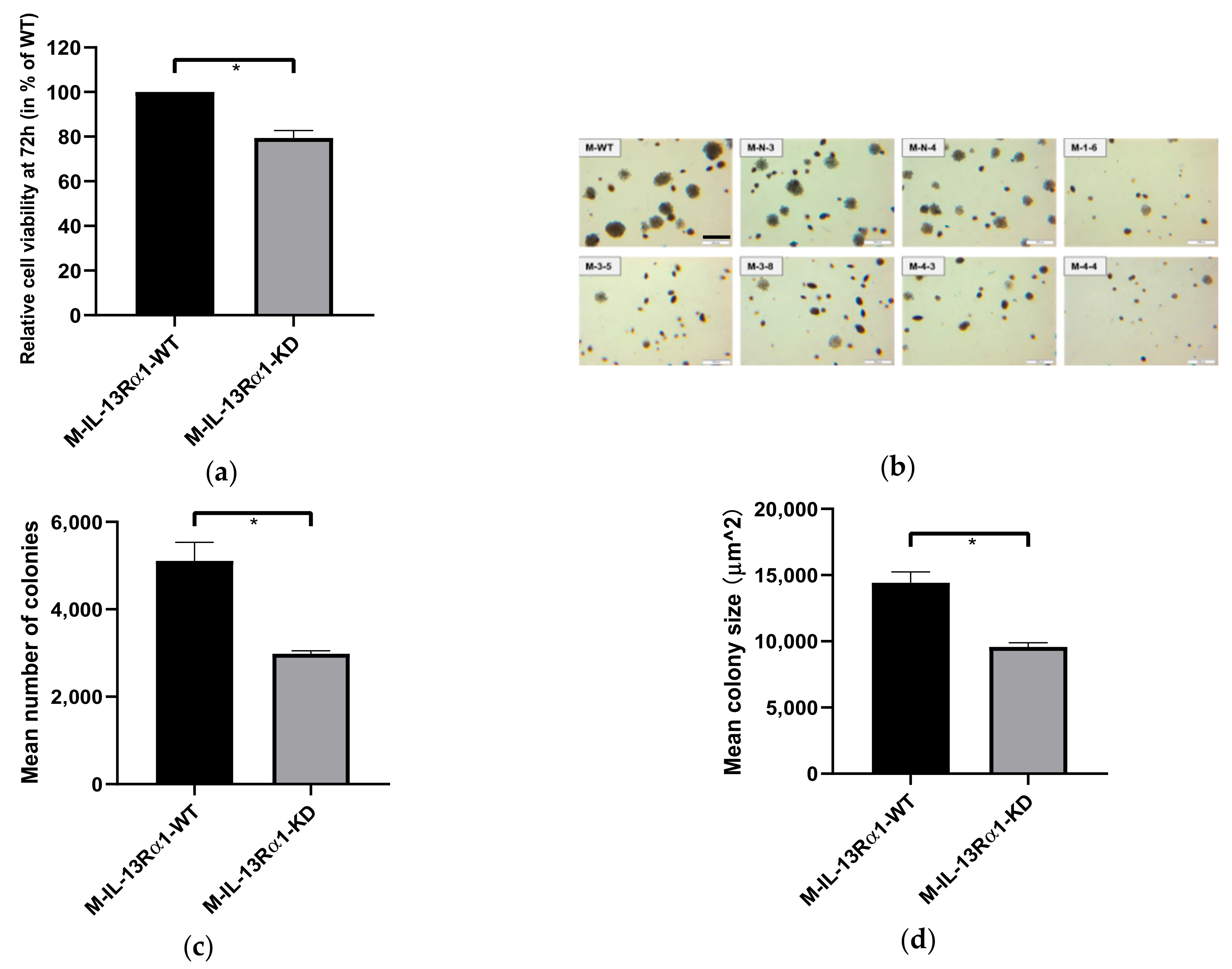
2.2. Effects of IL-13Rα1-Downregulation on the Malignant Phenotype in Pancreatic Cancer Cells
2.2.1. Effect of IL-13Rα1-Downregulation on Cell Growth
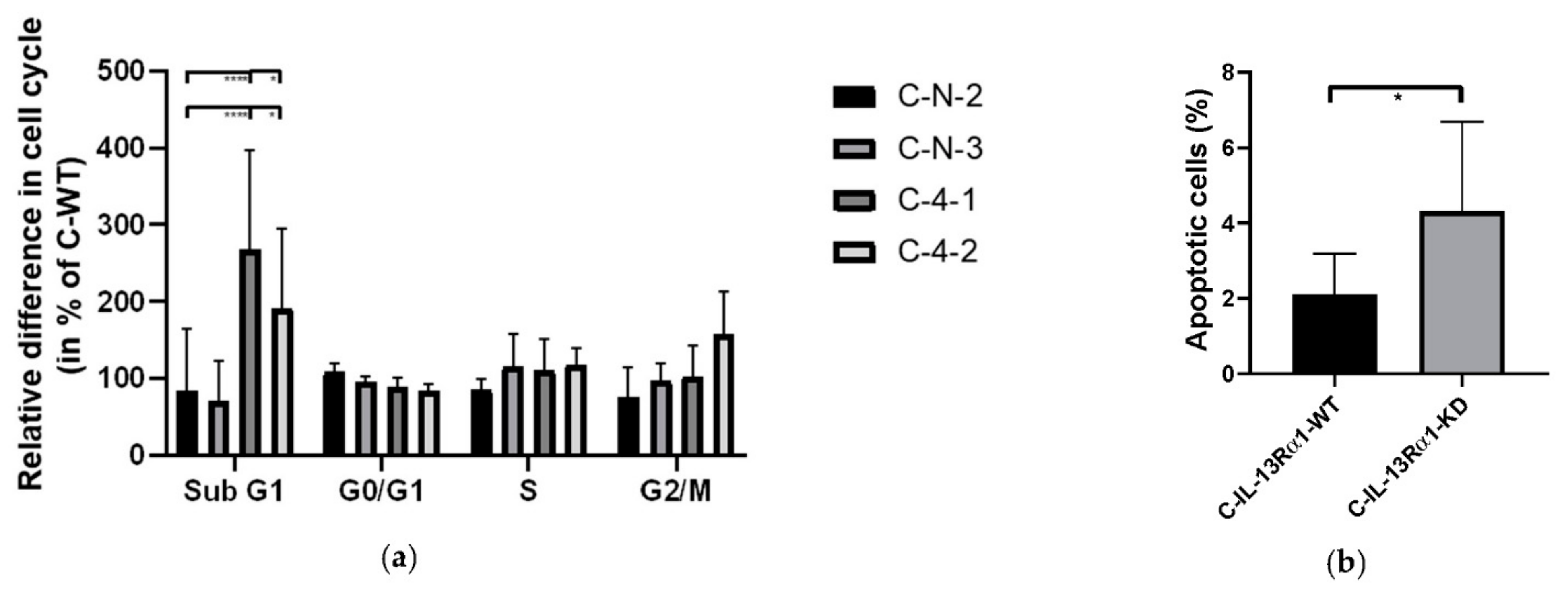
2.2.2. Effect of IL-13Rα1-Downregulation on the Cell Cycle
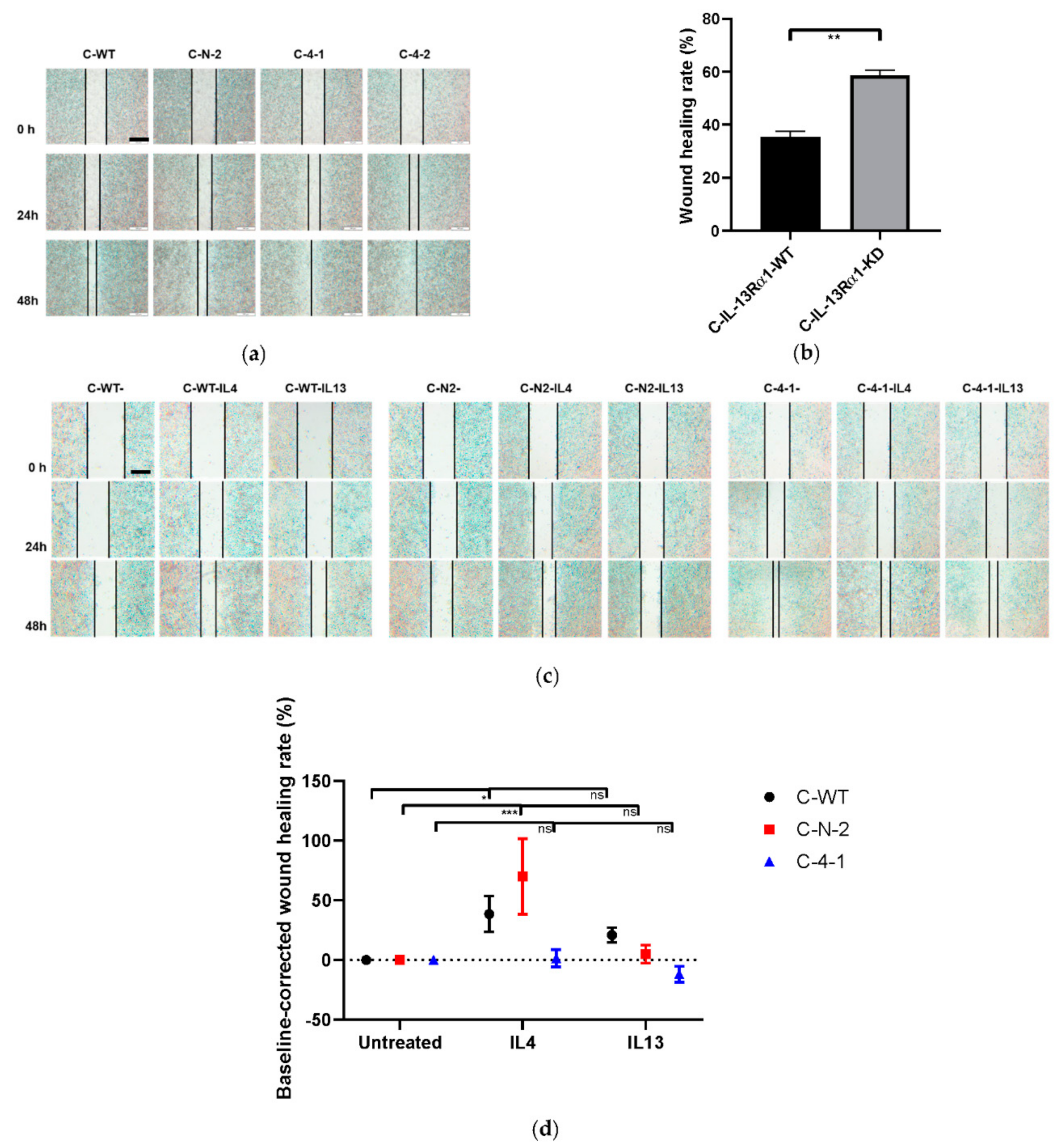
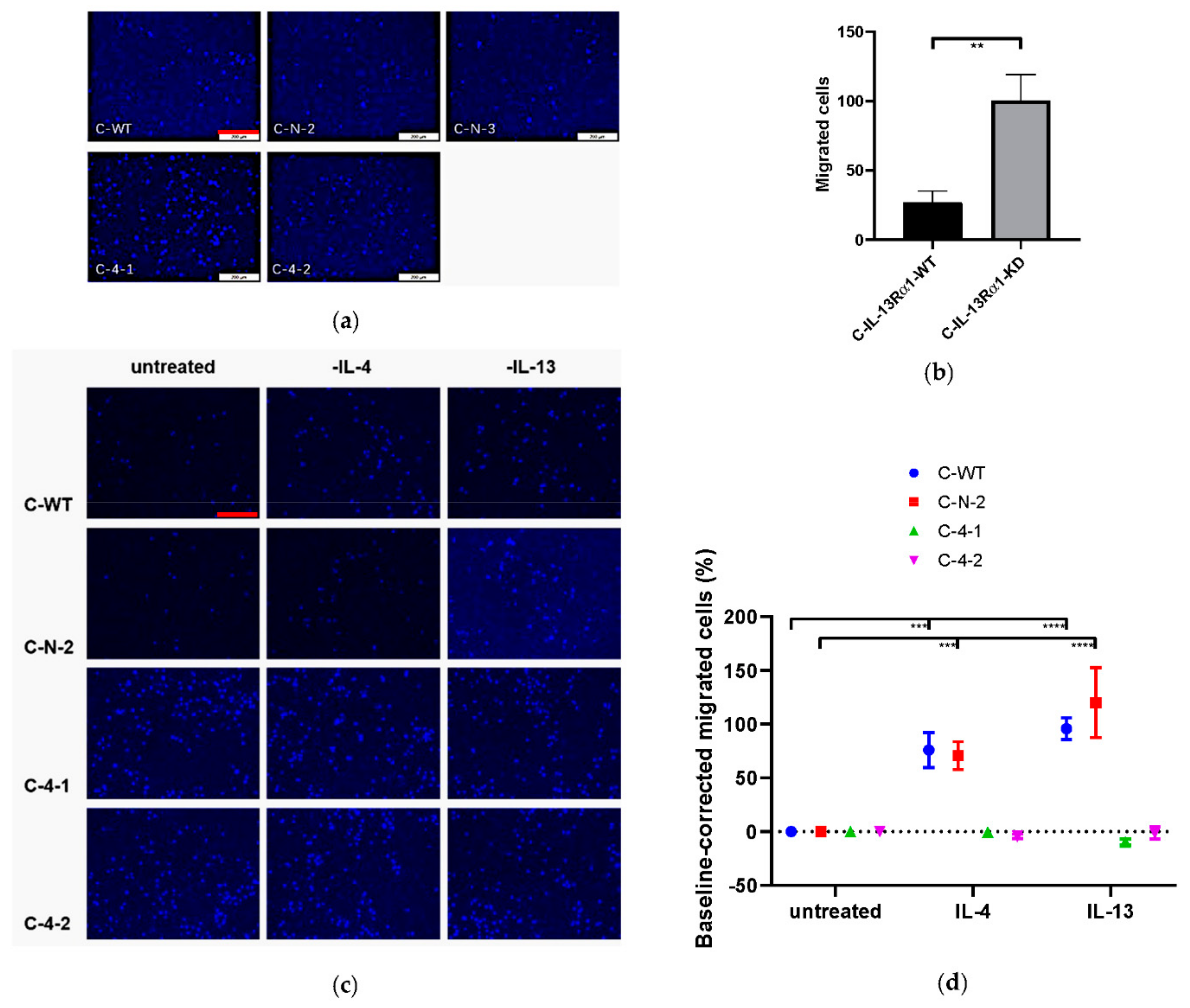
2.2.3. Effect of IL-13Rα1-Downregulation on Cell Mobility and Migration
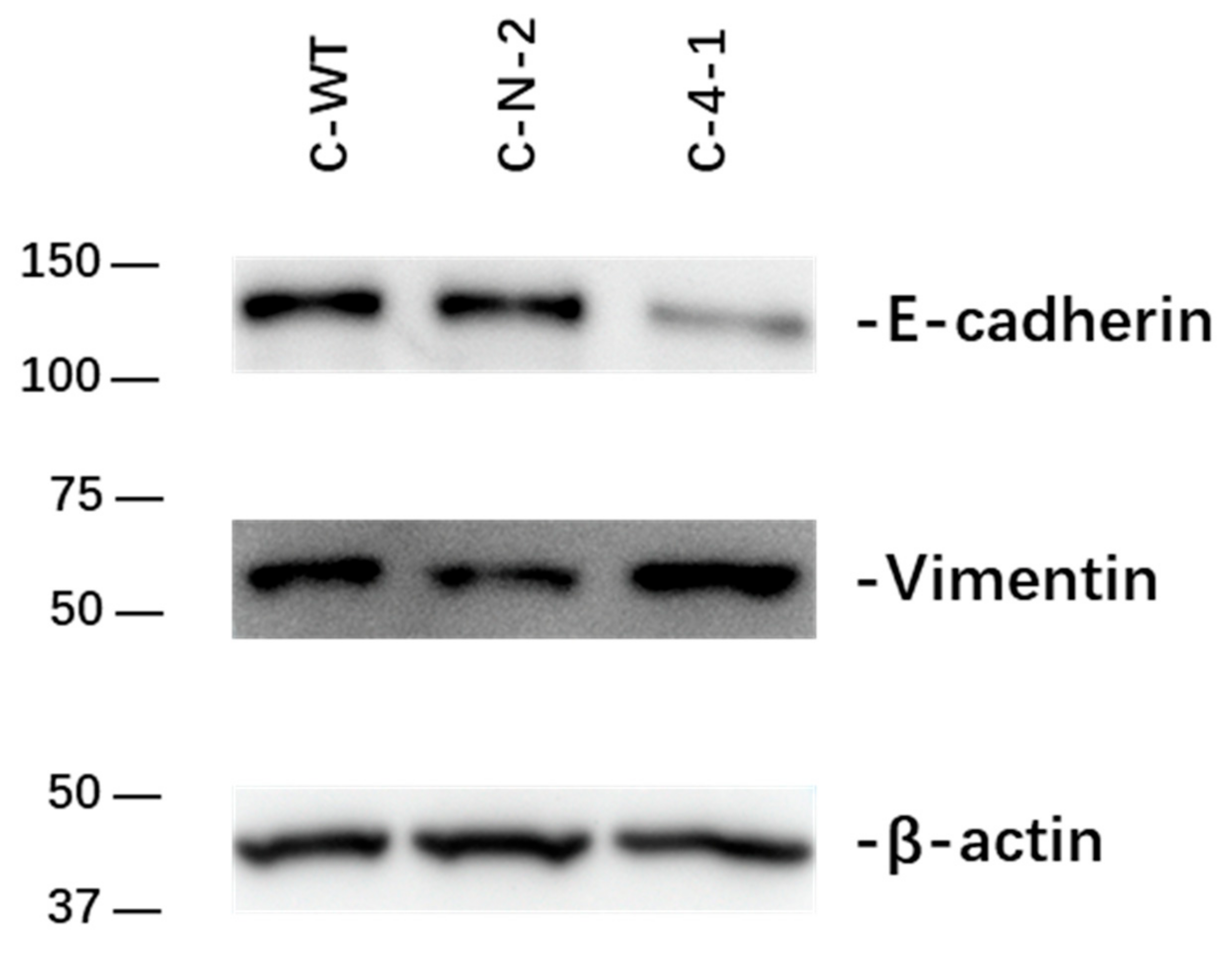
2.2.4. Effect of IL-13Rα1-Downregulation on Epithelial-to-Mesenchymal Transition (EMT)
2.3. Effect of IL-13Rα1-Downregulation on IL-4 and IL-13 Signalling
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Immunoblotting
4.3. Transfection
4.4. Cell Growth Assay
4.5. Cell Migration Assay
4.6. Giemsa Staining Assay
4.7. Cell Cycle and Apoptosis Analysis
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Akt | Protein kinase B |
| C-KD | Capan-1-IL-13Rα1-knockdown clone |
| C-N | Capan-1 sham-transfected cells |
| C-WT | Capan-1 wild type |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DPBS | Dulbecco’s Phosphate Buffered Saline |
| EMT | Epithelial–mesenchymal transition |
| ERK | Extracellular signal-regulated kinase |
| FCS | Fetal calf serum |
| γc | common gamma chain |
| GSDMD | Gasdermin D |
| HPF | High power field |
| IL | Interleukin |
| IL-4Rα | IL-4-receptor alpha |
| IL-13Rα | IL-13-receptor alpha |
| IRS | Insulin receptor substrate |
| KD | knockdown |
| kDa | kilodalton |
| MAPK | Mitogen-activated protein kinase |
| MMPs | Matrix metalloproteinases |
| M-KD | MIA PaCa-2-IL-13Rα1-knockdown clone |
| M-N | MIA PaCa-2 sham-transfected cells |
| M-WT | MIA PaCa-2 wild type |
| mTOR | The mechanistic target of rapamycin |
| p-MLKL | phospho-mixed lineage kinase domain-like protein |
| PCR | Polymerase chain reaction |
| PC | Pancreatic cancer |
| PI3K | phosphoinositide 3-kinase |
| qPCR | real-time quantitative polymerase chain reaction |
| RNA | Ribonucleic acid |
| RPMI | Roswell Park Memorial Institute medium |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| ShRNA | Short haipin ribonucleic acid |
| STAT | Signal transducer and activator of transcription |
| TAMs | Tumor-associated macrophages |
| TME | Tumor microenvironment |
| WB | Western blot |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Traub, B.; Link, K.-H.; Kornmann, M. (Eds.) Curing Pancreatic Cancer. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Lei, X.; Lei, Y.; Li, J.-K.; Du, W.-X.; Li, R.-G.; Yang, J.; Li, J.; Li, F.; Tan, H.-B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Parayath, N.; Padmakumar, S.; Nair, S.V.; Menon, D.; Amiji, M.M. Strategies for targeting cancer immunotherapy through modulation of the tumor microenvironment. Regen. Eng. Transl. Med. 2020, 6, 29–49. [Google Scholar] [CrossRef]
- Thakkar, S.; Sharma, D.; Kalia, K.; Tekade, R.K. Tumor microenvironment targeted nanotherapeutics for cancer therapy and diagnosis: A review. Acta Biomater. 2020, 101, 43–68. [Google Scholar] [CrossRef]
- Huber, M.; Brehm, C.U.; Gress, T.M.; Buchholz, M.; Alashkar Alhamwe, B.; von Strandmann, E.P.; Slater, E.P.; Bartsch, J.W.; Bauer, C.; Lauth, M. The immune microenvironment in pancreatic cancer. Int. J. Mol. Sci. 2020, 21, 7307. [Google Scholar] [CrossRef]
- Miyabayashi, K.; Baker, L.A.; Deschênes, A.; Traub, B.; Caligiuri, G.; Plenker, D.; Alagesan, B.; Belleau, P.; Li, S.; Kendall, J. Intraductal transplantation models of human pancreatic ductal adenocarcinoma reveal progressive transition of molecular subtypes. Cancer Discov. 2020, 10, 1566–1589. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761. [Google Scholar] [CrossRef]
- Whiteside, T. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef]
- Biffi, G.; Oni, T.E.; Spielman, B.; Hao, Y.; Elyada, E.; Park, Y.; Preall, J.; Tuveson, D.A. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019, 9, 282–301. [Google Scholar] [CrossRef]
- Moraga, I.; Richter, D.; Wilmes, S.; Winkelmann, H.; Jude, K.; Thomas, C.; Suhoski, M.M.; Engleman, E.G.; Piehler, J.; Garcia, K.C. Instructive roles for agonist binding parameters in determining the functional bandwidth of cytokine receptor signaling. Science signaling 2015, 8, ra114. [Google Scholar] [CrossRef] [PubMed]
- Leonard, W.J.; Lin, J.-X.; O’Shea, J.J. The γc family of cytokines: Basic biology to therapeutic ramifications. Immunity 2019, 50, 832–850. [Google Scholar] [CrossRef] [PubMed]
- Autenshlyus, A.; Davletova, K.; Studenikina, A.; Mikhaylova, E.; Varaksin, N.; Zhurakovsky, I.; Proskura, A.; Sidorov, S.; Lyakhovich, V. Cytokine production by blood immune cells, tumor and its microenvironment, characteristic of extracellular matrix in patients with invasive ductal carcinoma of no special type. Biomeditsinskaya Khimiya 2019, 65, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Ben-Baruch, A. Host microenvironment in breast cancer development: Inflammatory cells, cytokines and chemokines in breast cancer progression: Reciprocal tumor–microenvironment interactions. Breast Cancer Res. 2002, 5, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Sakurai, M.; Yamamoto, Y.; Suzuki, E.; Tsuda, M.; Kataoka, T.R.; Hirata, M.; Nishie, M.; Nojiri, T.; Kumazoe, M. Alteration of specific cytokine expression patterns in patients with breast cancer. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Braddock, M.; Hanania, N.A.; Sharafkhaneh, A.; Colice, G.; Carlsson, M. Potential risks related to modulating interleukin-13 and interleukin-4 signalling: A systematic review. Drug Saf. 2018, 41, 489–509. [Google Scholar] [CrossRef]
- Ito, S.-E.; Shirota, H.; Kasahara, Y.; Saijo, K.; Ishioka, C. IL-4 blockade alters the tumor microenvironment and augments the response to cancer immunotherapy in a mouse model. Cancer Immunol. Immunother. 2017, 66, 1485–1496. [Google Scholar] [CrossRef]
- Hallett, M.A.; Venmar, K.T.; Fingleton, B. Cytokine stimulation of epithelial cancer cells: The similar and divergent functions of IL-4 and IL-13. Cancer Res. 2012, 72, 6338–6343. [Google Scholar] [CrossRef]
- Kwaśniak, K.; Czarnik-Kwaśniak, J.; Maziarz, A.; Aebisher, D.; Zielińska, K.; Karczmarek-Borowska, B.; Tabarkiewicz, J. Scientific reports concerning the impact of interleukin 4, interleukin 10 and transforming growth factor β on cancer cells. Cent. -Eur. J. Immunol. 2019, 44, 190. [Google Scholar] [CrossRef]
- Seyfizadeh, N.; Seyfizadeh, N.; Gharibi, T.; Babaloo, Z. Interleukin-13 as an important cytokine: A review on its roles in some human diseases. Acta Microbiol. Immunol. Hung. 2015, 62, 341–378. [Google Scholar] [CrossRef][Green Version]
- Lin, X.; Wang, S.; Sun, M.; Zhang, C.; Wei, C.; Yang, C.; Dou, R.; Liu, Q.; Xiong, B. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Song, X.; Traub, B.; Luxenhofer, M.; Kornmann, M. Involvement of IL-4, IL-13 and Their Receptors in Pancreatic Cancer. Int. J. Mol. Sci. 2021, 22, 2998. [Google Scholar] [CrossRef] [PubMed]
- Kornmann, M.; Kleeff, J.; Debinski, W.; Korc, M. Pancreatic cancer cells express interleukin-13 and-4 receptors, and their growth is inhibited by Pseudomonas exotoxin coupled to interleukin-13 and-4. Anticancer. Res. 1999, 19, 125–131. [Google Scholar] [PubMed]
- Prokopchuk, O.; Liu, Y.; Henne-Bruns, D.; Kornmann, M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: Evidence for autocrine and paracrine actions. Br. J. Cancer 2005, 92, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Formentini, A.; Prokopchuk, O.; Sträter, J.; Kleeff, J.; Grochola, L.F.; Leder, G.; Henne-Bruns, D.; Korc, M.; Kornmann, M. Interleukin-13 exerts autocrine growth-promoting effects on human pancreatic cancer, and its expression correlates with a propensity for lymph node metastases. Int. J. Colorectal Dis. 2009, 24, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Traub, B.; Sun, L.; Ma, Y.; Xu, P.; Lemke, J.; Paschke, S.; Henne-Bruns, D.; Knippschild, U.; Kornmann, M. Endogenously expressed IL-4Rα promotes the malignant phenotype of human pancreatic cancer in vitro and in vivo. Int. J. Mol. Sci. 2017, 18, 716. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, H.; Gao, W.; Wang, S.; Wu, K.; Lu, C.; Luo, X.; Li, L.; Yu, C. Girdin interaction with vimentin induces EMT and promotes the growth and metastasis of pancreatic ductal adenocarcinoma. Oncol. Rep. 2020, 44, 637–649. [Google Scholar] [CrossRef]
- Chomarat, P.; Banchereau, J. Interleukin-4 and interleukin-13: Their similarities and discrepancies. Int. Rev. Immunol. 1998, 17, 1–52. [Google Scholar] [CrossRef]
- Gandhi, N.A.; Pirozzi, G.; Graham, N.M. Commonality of the IL-4/IL-13 pathway in atopic diseases. Expert Rev. Clin. Immunol. 2017, 13, 425–437. [Google Scholar] [CrossRef]
- Gour, N.; Wills-Karp, M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine 2015, 75, 68–78. [Google Scholar] [CrossRef]
- Kawakami, K.; Kawakami, M.; Husain, S.R.; Puri, R.K. Targeting interleukin-4 receptors for effective pancreatic cancer therapy. Cancer Res. 2002, 62, 3575–3580. [Google Scholar] [PubMed]
- Wu, Y.; Konaté, M.M.; Lu, J.; Makhlouf, H.; Chuaqui, R.; Antony, S.; Meitzler, J.L.; Difilippantonio, M.J.; Liu, H.; Juhasz, A. IL-4 and IL-17A cooperatively promote hydrogen peroxide production, oxidative DNA damage, and upregulation of dual oxidase 2 in human colon and pancreatic cancer cells. J. Immunol. 2019, 203, 2532–2544. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://dcc.icgc.org/q?q=data%20obtained%20on%20February%2028th%202022 (accessed on 28 February 2022).
- Dey, P.; Li, J.; Zhang, J.; Chaurasiya, S.; Strom, A.; Wang, H.; Liao, W.T.; Cavallaro, F.; Denz, P.; Bernard, V.; et al. Oncogenic KRAS-Driven Metabolic Reprogramming in Pancreatic Cancer Cells Utilizes Cytokines from the Tumor Microenvironment. Cancer Discov. 2020, 10, 608–625. [Google Scholar] [CrossRef] [PubMed]
- Rajamani, D.; Bhasin, M.K. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016, 8, 38. [Google Scholar] [CrossRef]
- Ayars, M.; O’Sullivan, E.; Macgregor-Das, A.; Shindo, K.; Kim, H.; Borges, M.; Yu, J.; Hruban, R.H.; Goggins, M. IL2RG, identified as overexpressed by RNA-seq profiling of pancreatic intraepithelial neoplasia, mediates pancreatic cancer growth. Oncotarget 2017, 8, 83370–83383. [Google Scholar] [CrossRef]
- Fu, C.; Jiang, L.; Hao, S.; Liu, Z.; Ding, S.; Zhang, W.; Yang, X.; Li, S. Activation of the IL-4/STAT6 signaling pathway promotes lung cancer progression by increasing M2 myeloid cells. Front. Immunol. 2019, 10, 2638. [Google Scholar] [CrossRef]
- Kim, S.D.; Baik, J.S.; Lee, J.-H.; Mun, S.-W.; Yi, J.M.; Park, M.-T. The malignancy of liver cancer cells is increased by IL-4/ERK/AKT signaling axis activity triggered by irradiated endothelial cells. J. Radiat. Res. 2020, 61, 376–387. [Google Scholar] [CrossRef]
- Nelms, K.; Keegan, A.D.; Zamorano, J.; Ryan, J.J.; Paul, W.E. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999, 17, 701–738. [Google Scholar] [CrossRef]
- Timme, S.; Ihde, S.; Fichter, C.; Waehle, V.; Bogatyreva, L.; Atanasov, K.; Kohler, I.; Schöpflin, A.; Geddert, H.; Faller, G. STAT3 expression, activity and functional consequences of STAT3 inhibition in esophageal squamous cell carcinomas and Barrett’s adenocarcinomas. Oncogene 2014, 33, 3256–3266. [Google Scholar] [CrossRef]
- Datta, S.R.; Brunet, A.; Greenberg, M.E. Cellular survival: A play in three Akts. Genes Dev. 1999, 13, 2905–2927. [Google Scholar] [CrossRef]
- Nicholson, K.M.; Anderson, N.G. The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal. 2002, 14, 381–395. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhang, J.; Liu, H.; Wan, L.; Zhang, H.; Huang, Q.; Xu, E.; Lai, M. IL-13/STAT6 signaling plays a critical role in the epithelial-mesenchymal transition of colorectal cancer cells. Oncotarget 2016, 7, 61183. [Google Scholar] [CrossRef] [PubMed]
- Gann, P.H.; Deaton, R.J.; McMahon, N.; Collins, M.H.; Dellon, E.S.; Hirano, I.; Hua, S.Y.; Rodriguez, C.; Harris, S. An anti–IL-13 antibody reverses epithelial-mesenchymal transition biomarkers in eosinophilic esophagitis: Phase 2 trial results. J. Allergy Clin. Immunol. 2020, 146, 367–376.e3. [Google Scholar] [CrossRef]
- Kunwar, S.; Chang, S.; Westphal, M.; Vogelbaum, M.; Sampson, J.; Barnett, G.; Shaffrey, M.; Ram, Z.; Piepmeier, J.; Prados, M.; et al. Phase III randomized trial of CED of IL13-PE38QQR vs. Gliadel wafers for recurrent glioblastoma. Neuro Oncol. 2010, 12, 871–881. [Google Scholar] [CrossRef]
- Hurwitz, H.; Uppal, N.; Wagner, S.A.; Bendell, J.; Thaddeus, B.J.; Wade, S.; Nemunaitis, J.; Stella, P.; Pipas, J.M.; Wainberg, Z.; et al. Results from a Phase 2 Study of Ruxolitinib or Placebo with Capecitabine as Second-Line Therapy in Patients with Metastatic Pancreatic Cancer: The Recap Trial. Ann. Oncol. 2014, 25, ii115. [Google Scholar] [CrossRef]


| STAT6 | STAT3 | ERK | Akt | PI3K | |
|---|---|---|---|---|---|
| C-WT | 100 | 100 | 100 | 100 | 100 |
| C-N-2 | 117.6 | 110.6 | 151.8 | 83.3 | 114.5 |
| C-4-1 | 119.6 | 83.3 | 97.7 | 41.2 | 105.1 |
| p-STAT6 | p-STAT3 | p-ERK | p-Akt | PI3K | |
|---|---|---|---|---|---|
| C-WT-IL4 | 1479.3 | 196.9 | 182.8 | 251.2 | 129.4 |
| C-WT-IL13 | 1769.0 | 225.3 | 209.1 | 275.6 | 133.4 |
| C-N-2-IL4 | 778.7 | 166.2 | 202.3 | 215.6 | 104.4 |
| C-N-2-IL13 | 837.4 | 148.9 | 162.7 | 211.7 | 126.5 |
| C-4-1-IL4 | 435.9 | 141.2 | 169.9 | 168.4 | 131.9 |
| C-4-1-IL13 | 348.9 | 114.4 | 180.9 | 140.3 | 145.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Shen, X.; Kang, Q.; Yang, X.; Denzinger, M.; Kornmann, M.; Traub, B. Loss of Interleukin-13-Receptor-Alpha-1 Induces Apoptosis and Promotes EMT in Pancreatic Cancer. Int. J. Mol. Sci. 2022, 23, 3659. https://doi.org/10.3390/ijms23073659
Shi J, Shen X, Kang Q, Yang X, Denzinger M, Kornmann M, Traub B. Loss of Interleukin-13-Receptor-Alpha-1 Induces Apoptosis and Promotes EMT in Pancreatic Cancer. International Journal of Molecular Sciences. 2022; 23(7):3659. https://doi.org/10.3390/ijms23073659
Chicago/Turabian StyleShi, Jingwei, Xiao Shen, Qi Kang, Xing Yang, Maximilian Denzinger, Marko Kornmann, and Benno Traub. 2022. "Loss of Interleukin-13-Receptor-Alpha-1 Induces Apoptosis and Promotes EMT in Pancreatic Cancer" International Journal of Molecular Sciences 23, no. 7: 3659. https://doi.org/10.3390/ijms23073659
APA StyleShi, J., Shen, X., Kang, Q., Yang, X., Denzinger, M., Kornmann, M., & Traub, B. (2022). Loss of Interleukin-13-Receptor-Alpha-1 Induces Apoptosis and Promotes EMT in Pancreatic Cancer. International Journal of Molecular Sciences, 23(7), 3659. https://doi.org/10.3390/ijms23073659






