Cleavage of Early Mouse Embryo with Damaged DNA
Abstract
1. Introduction
2. Results
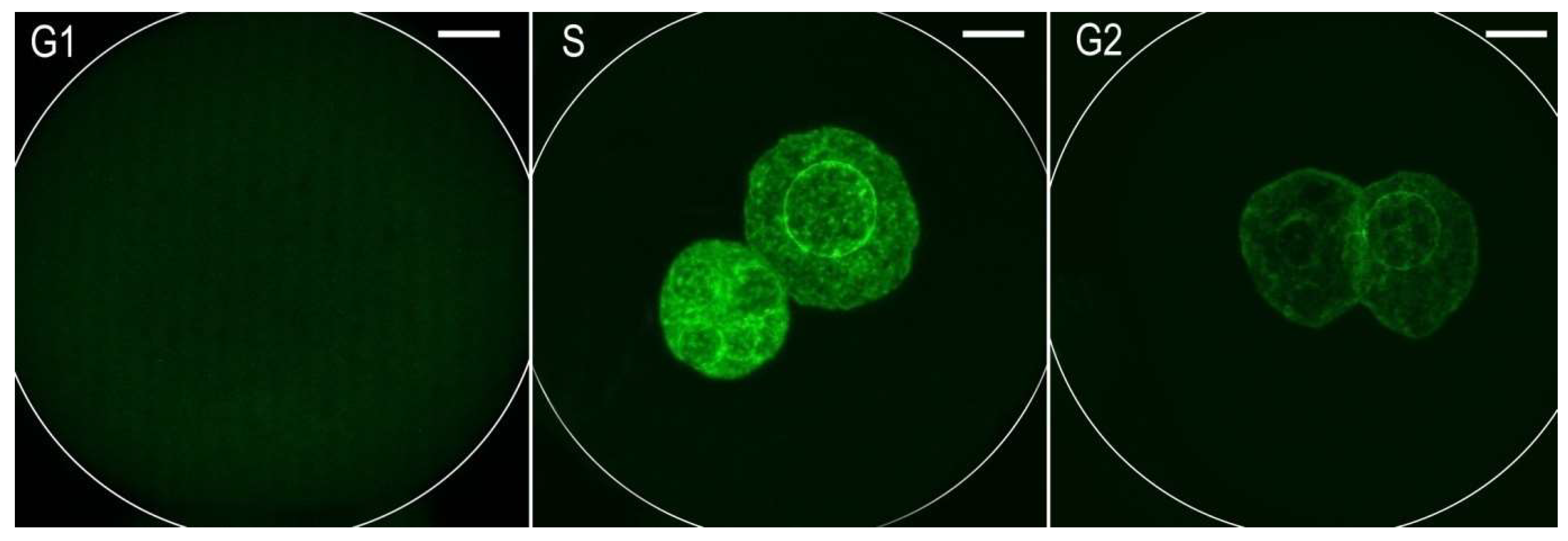
2.1. Incorporation of EdU into DNA in Individual Cell Cycle Phases of One-Cell Embryos
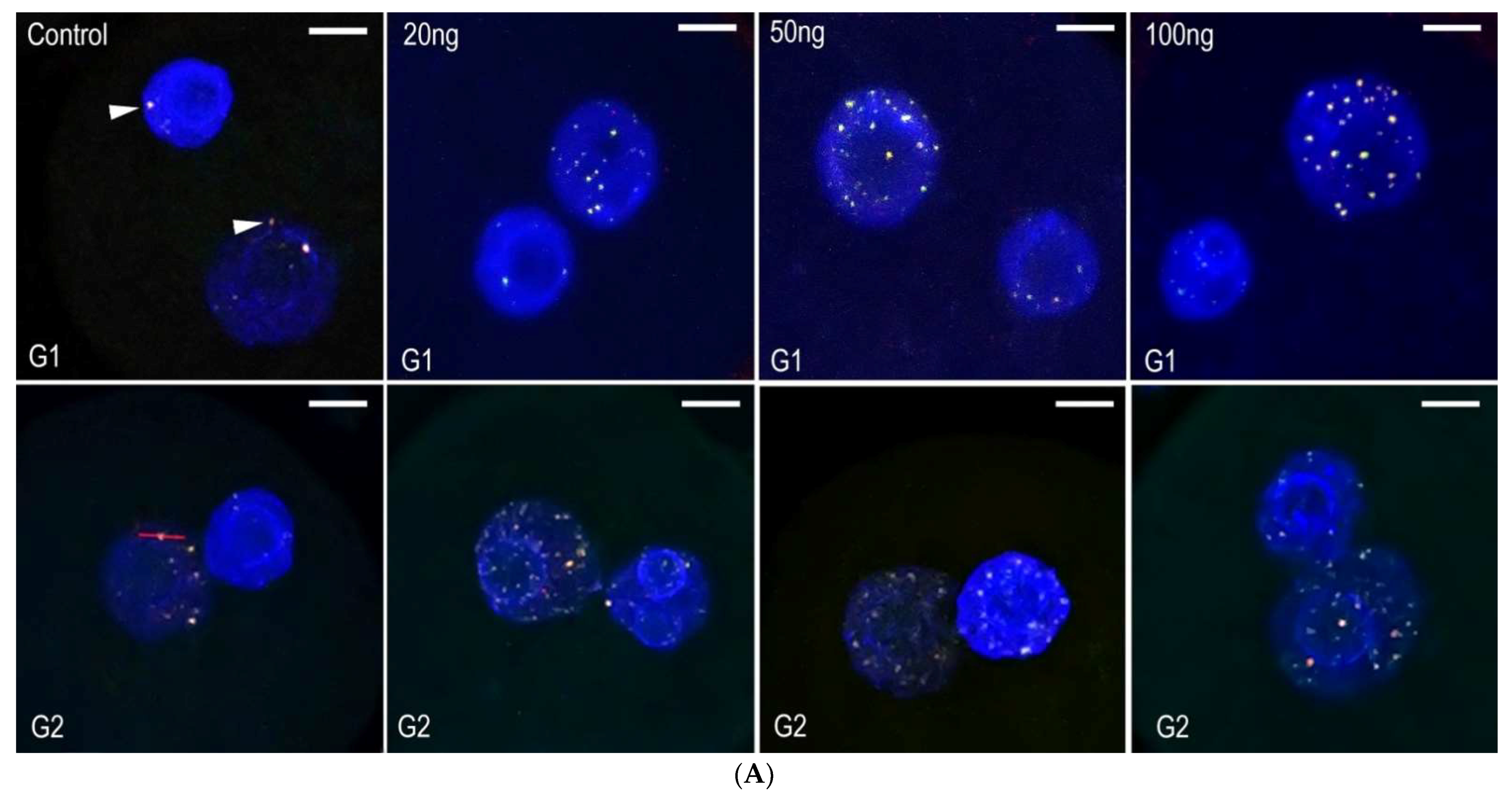
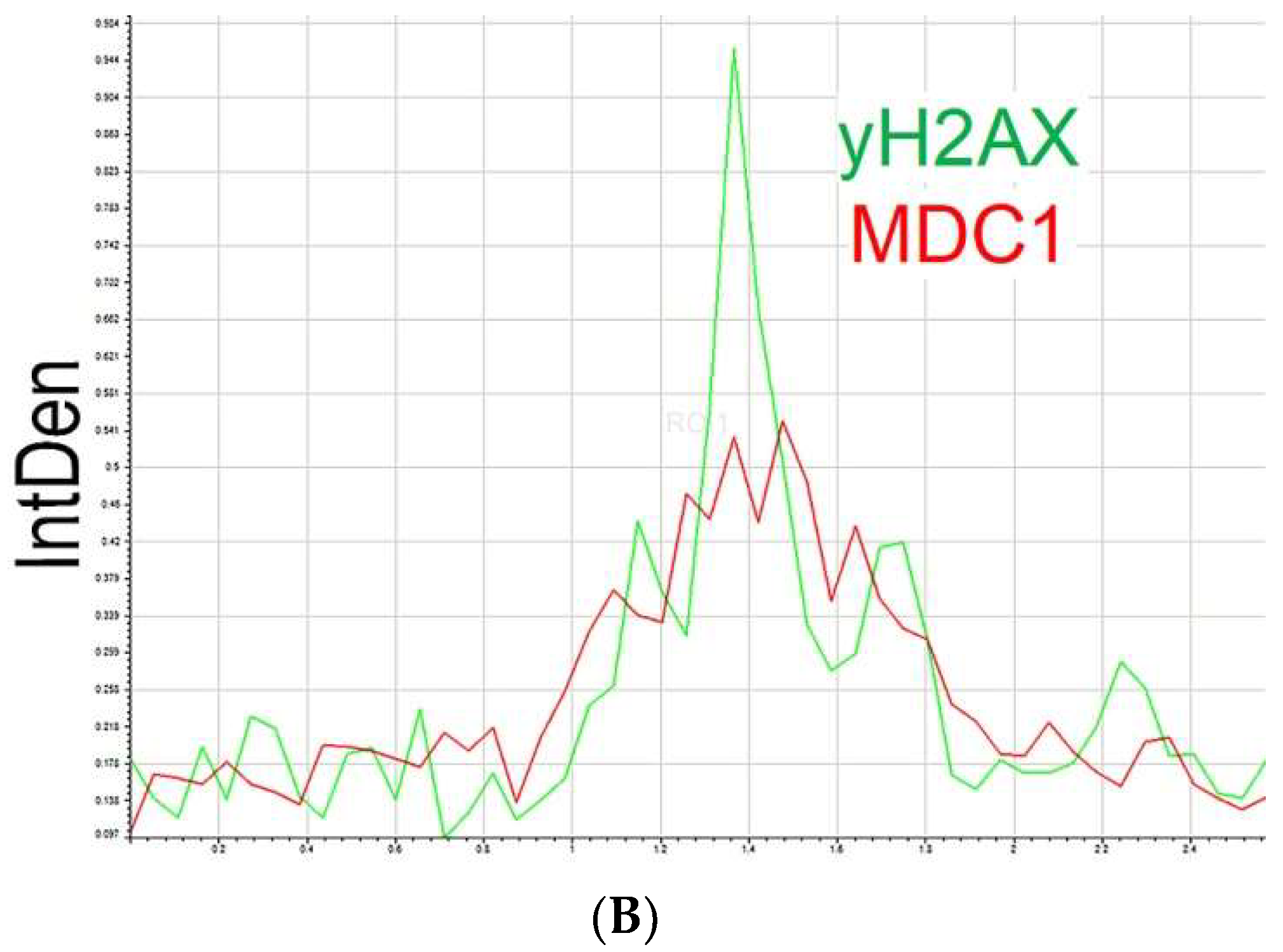
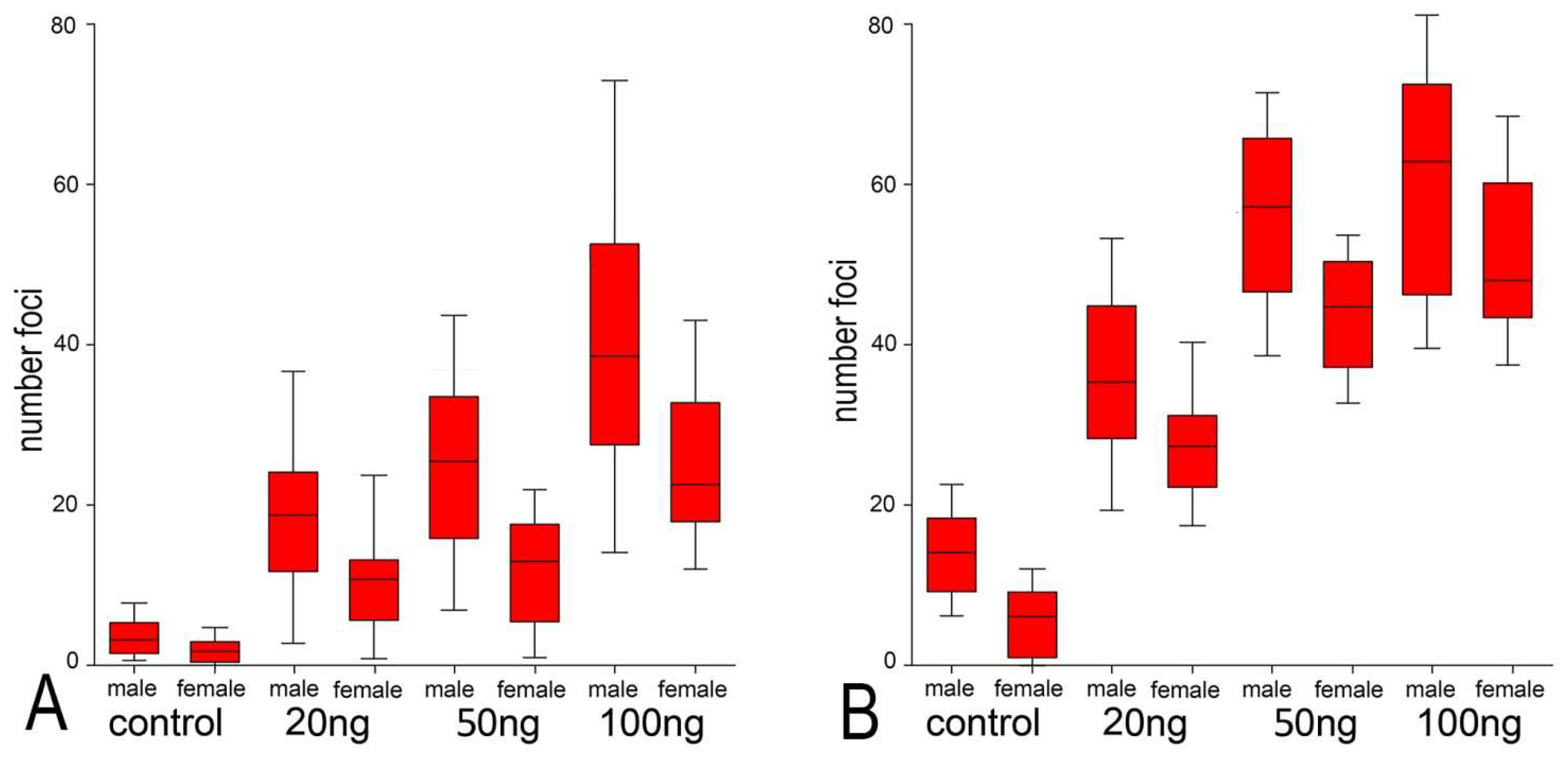
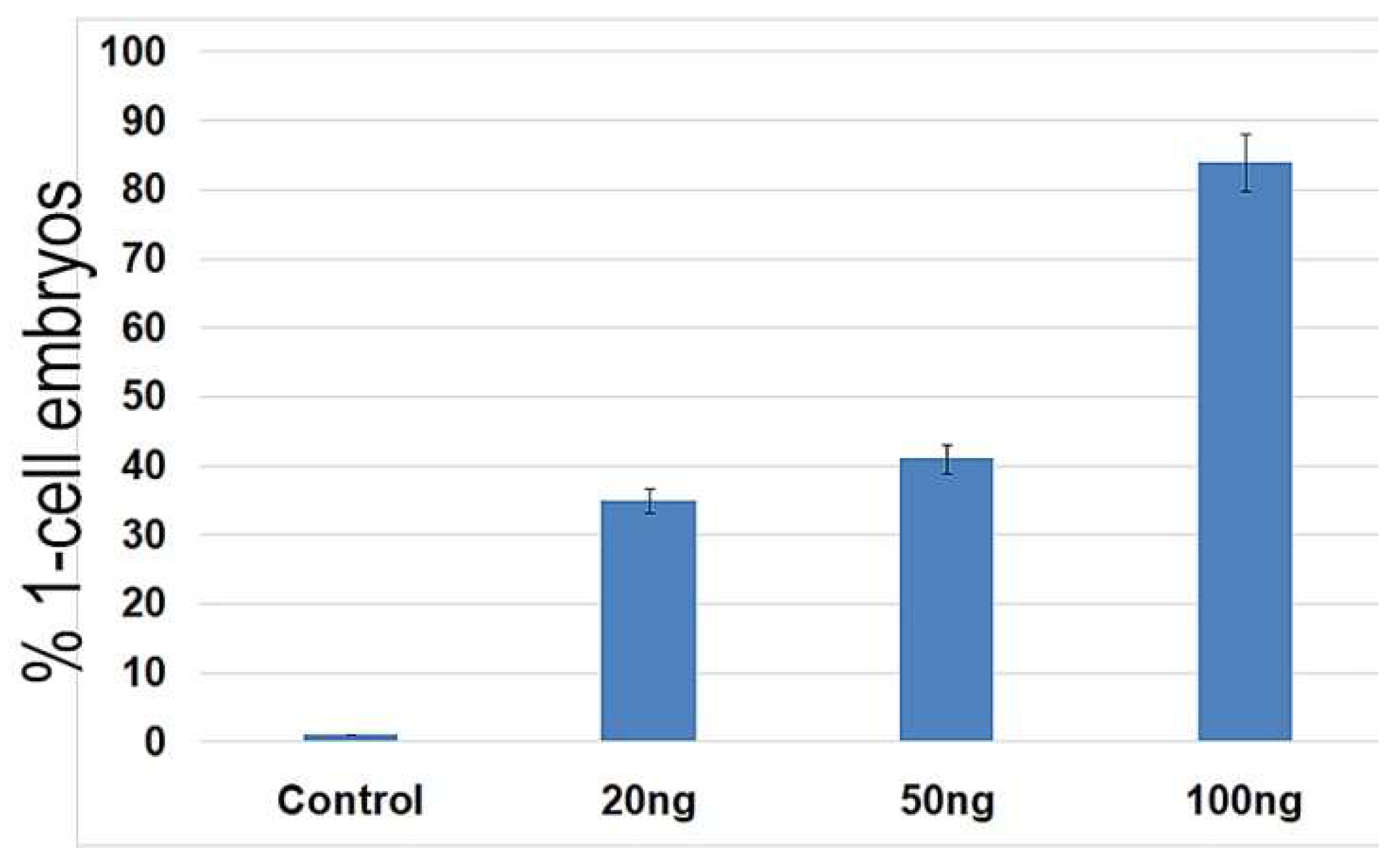
2.2. Neocarzinostatin Increasesthe Incidence of yH2AXSer139-Positive Foci during the First Embryonic Cell Cycle
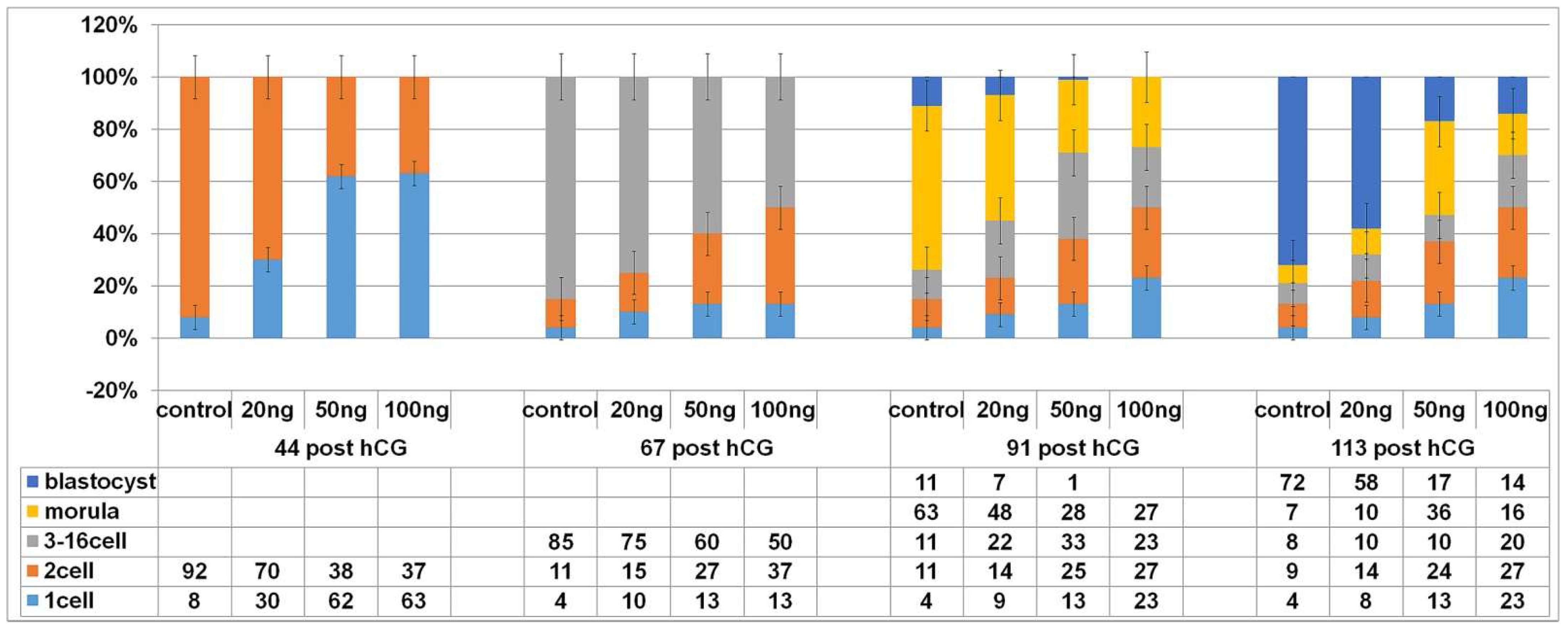
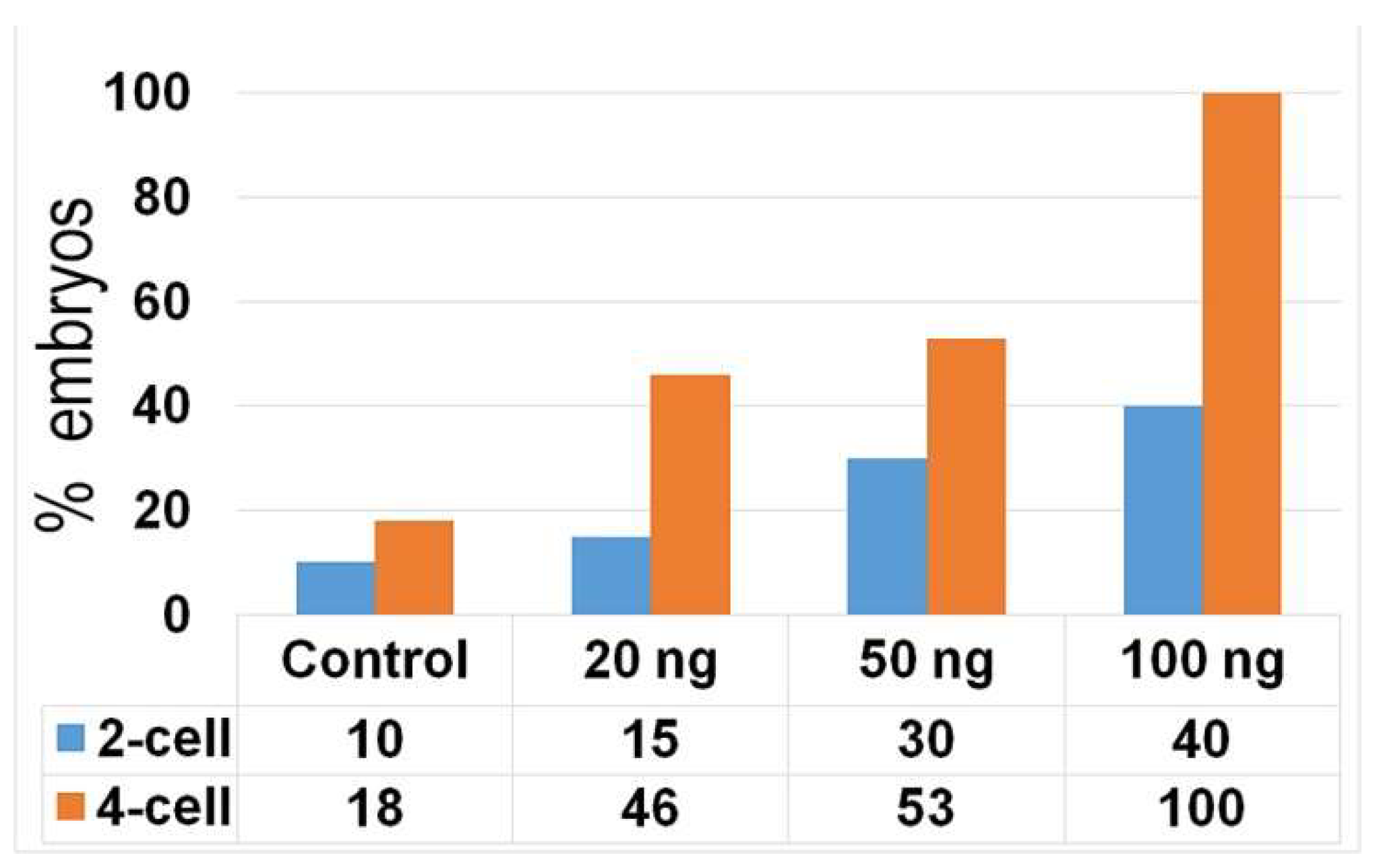
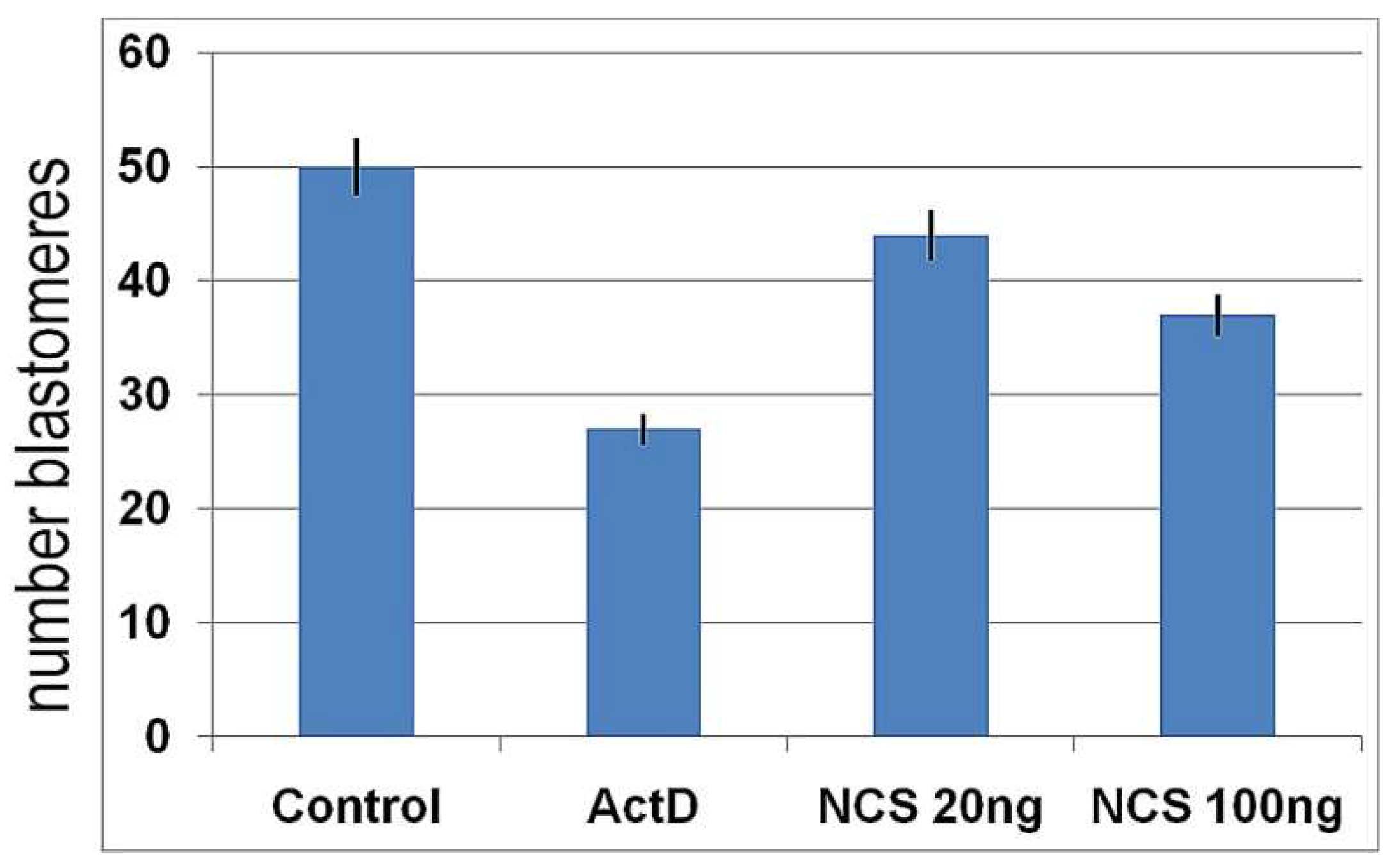
2.3. DNA Damage Reduce the Cleavage Ability of Early Embryos
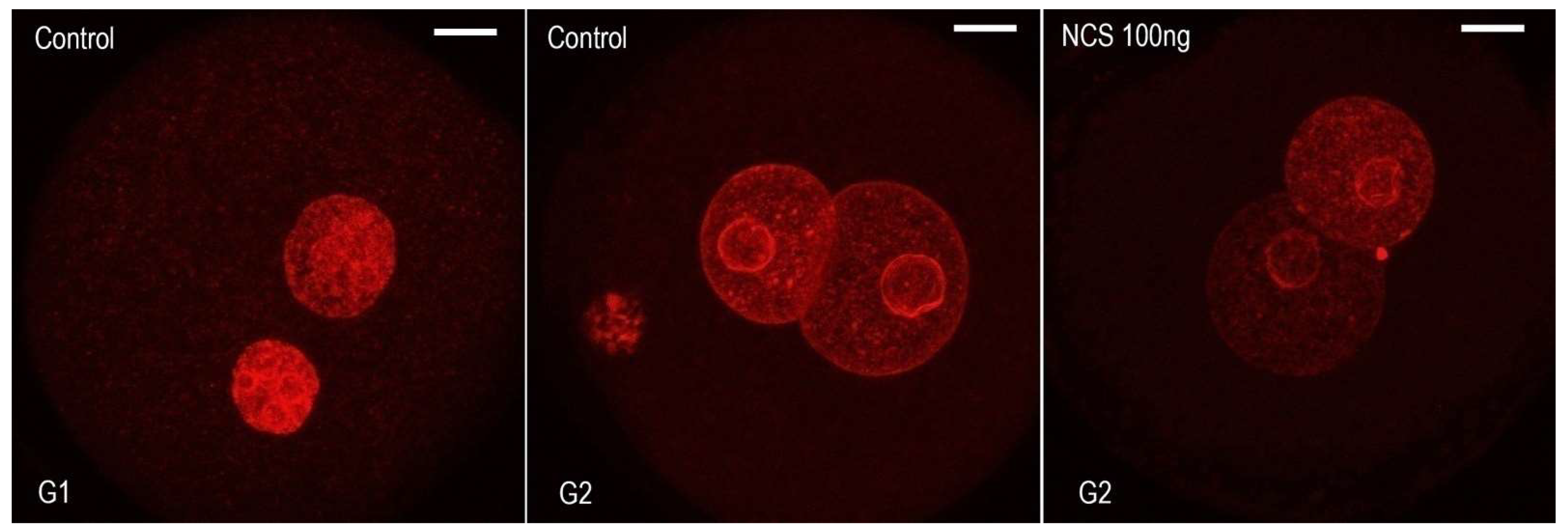
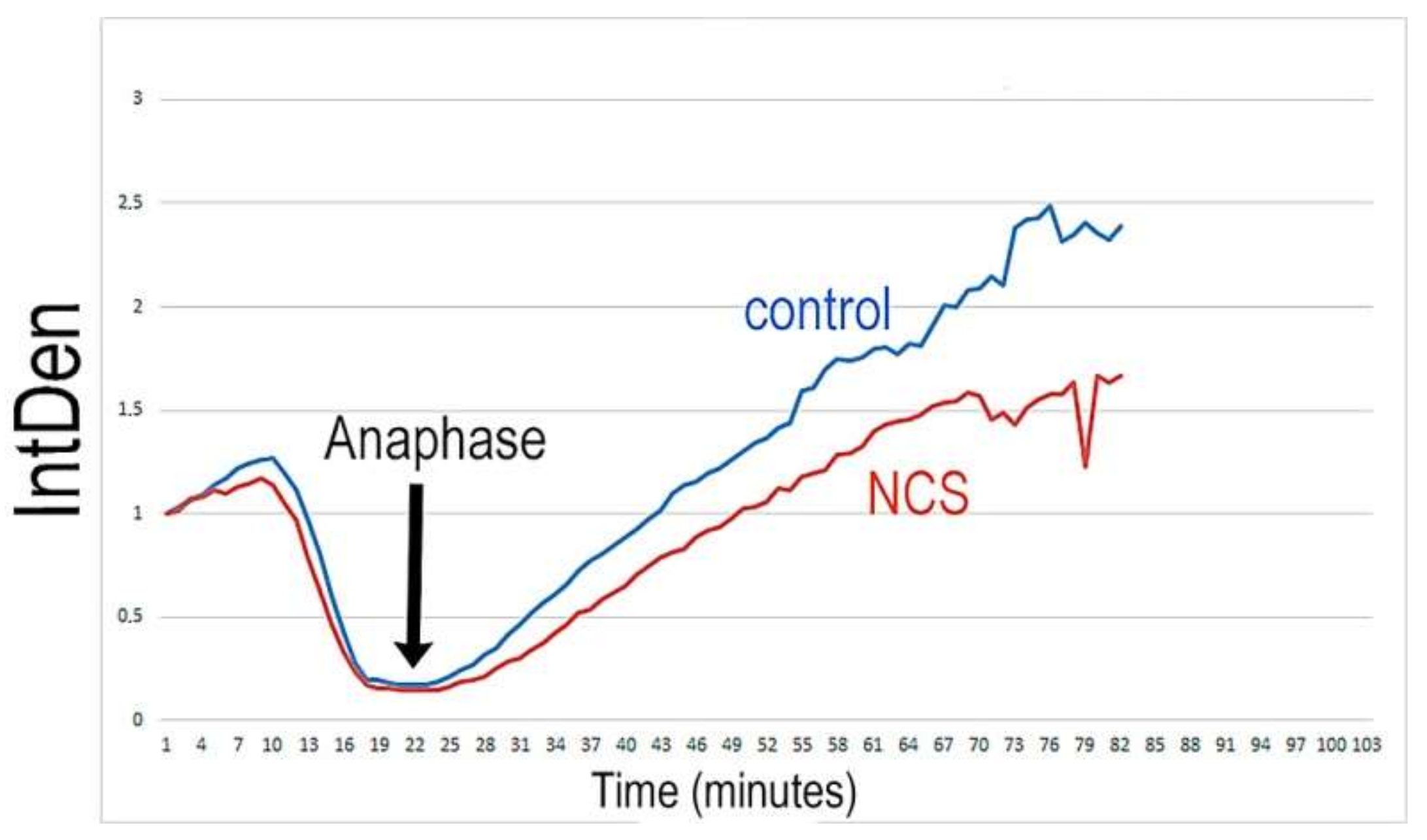
2.4. Higher DNA Damage Level Cause Decreased Volume of DNA after the End of the S-Phase
2.5. DNA Damage CanInduce Chromosomal Missegregation
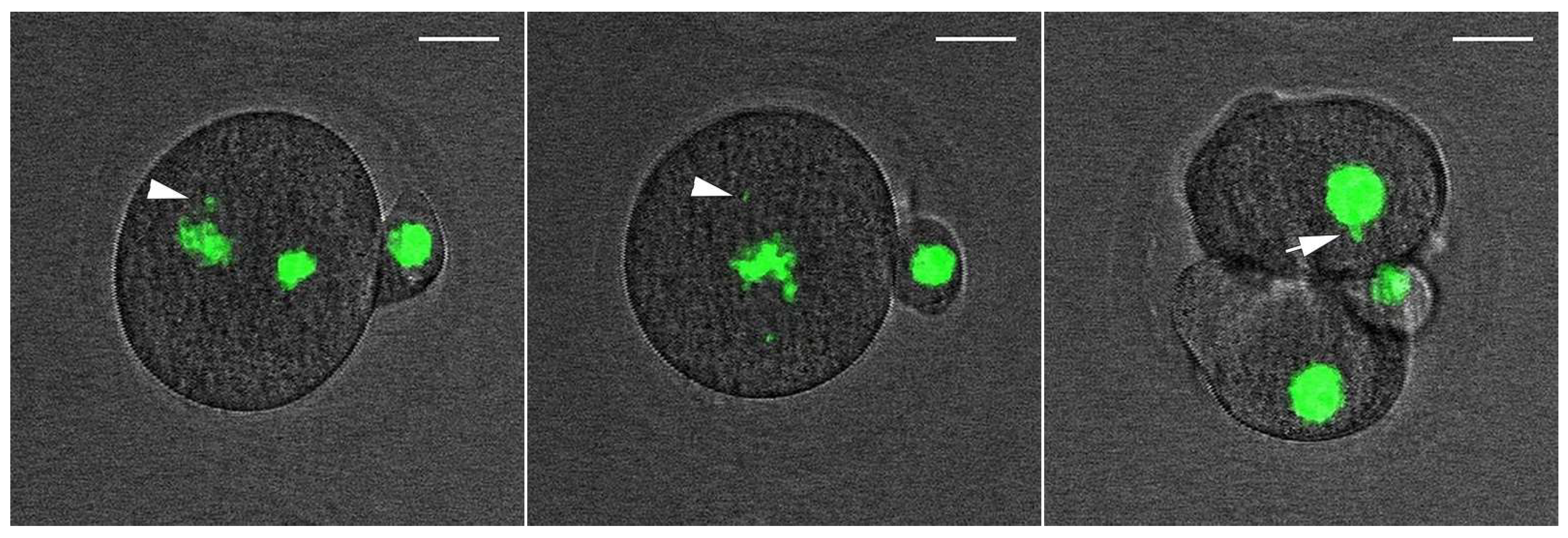
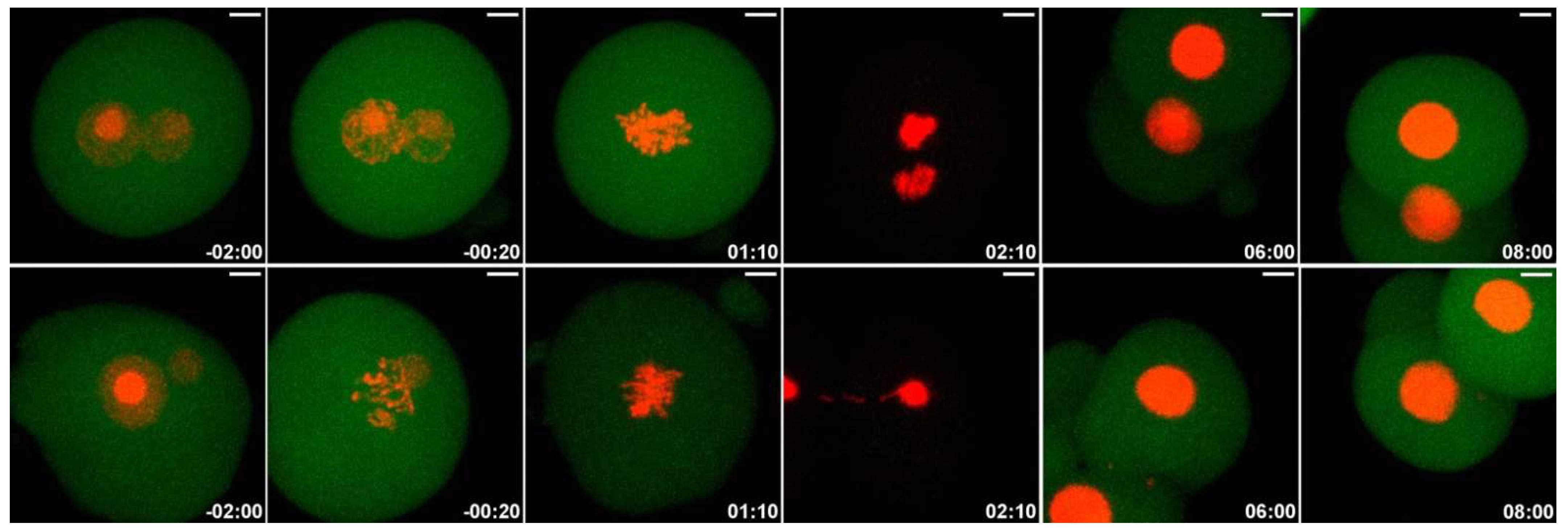
2.6. Low Dose of NCS Does Not Affect Entry into Anaphase but Induces Micronuclei Formation
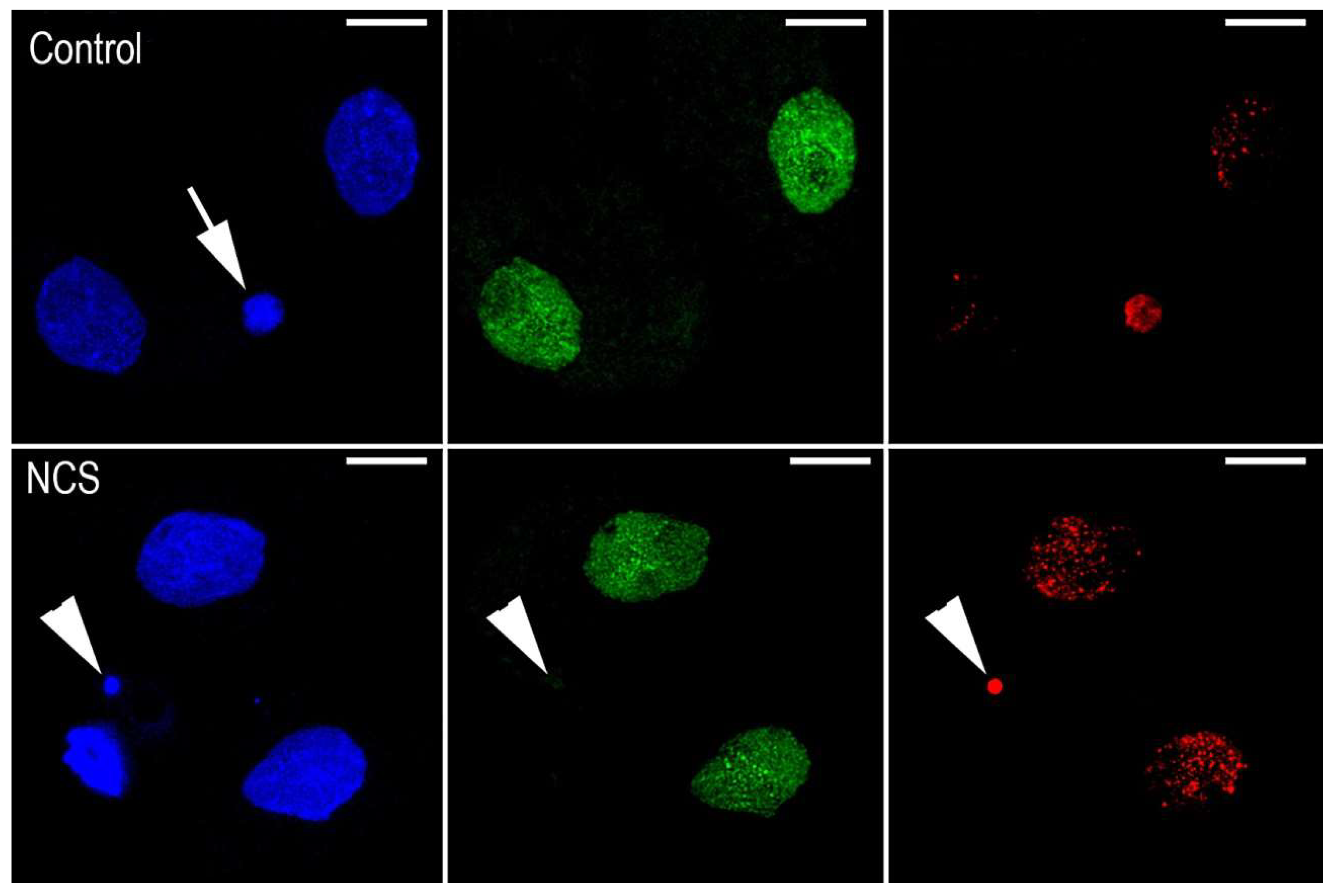
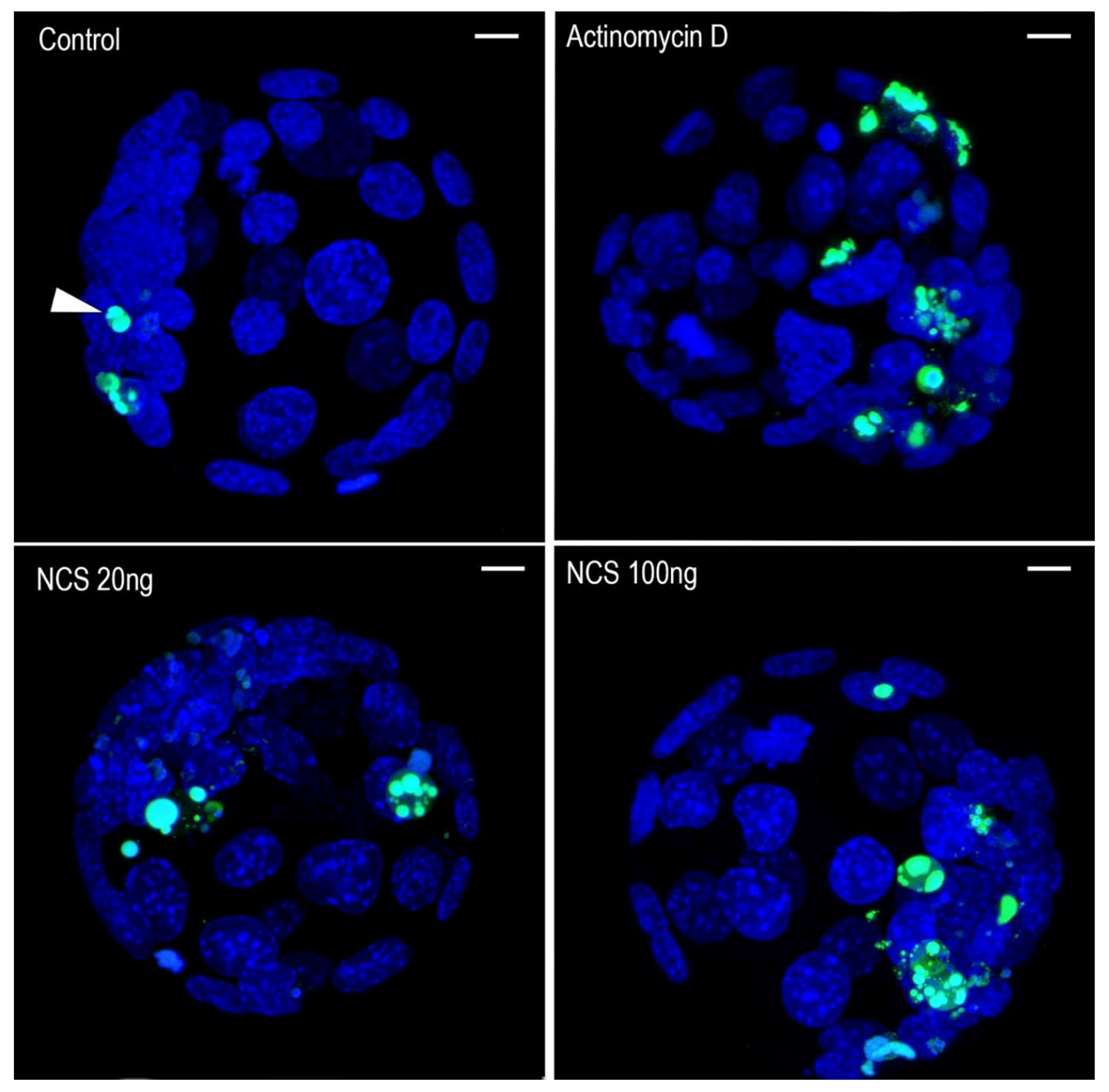
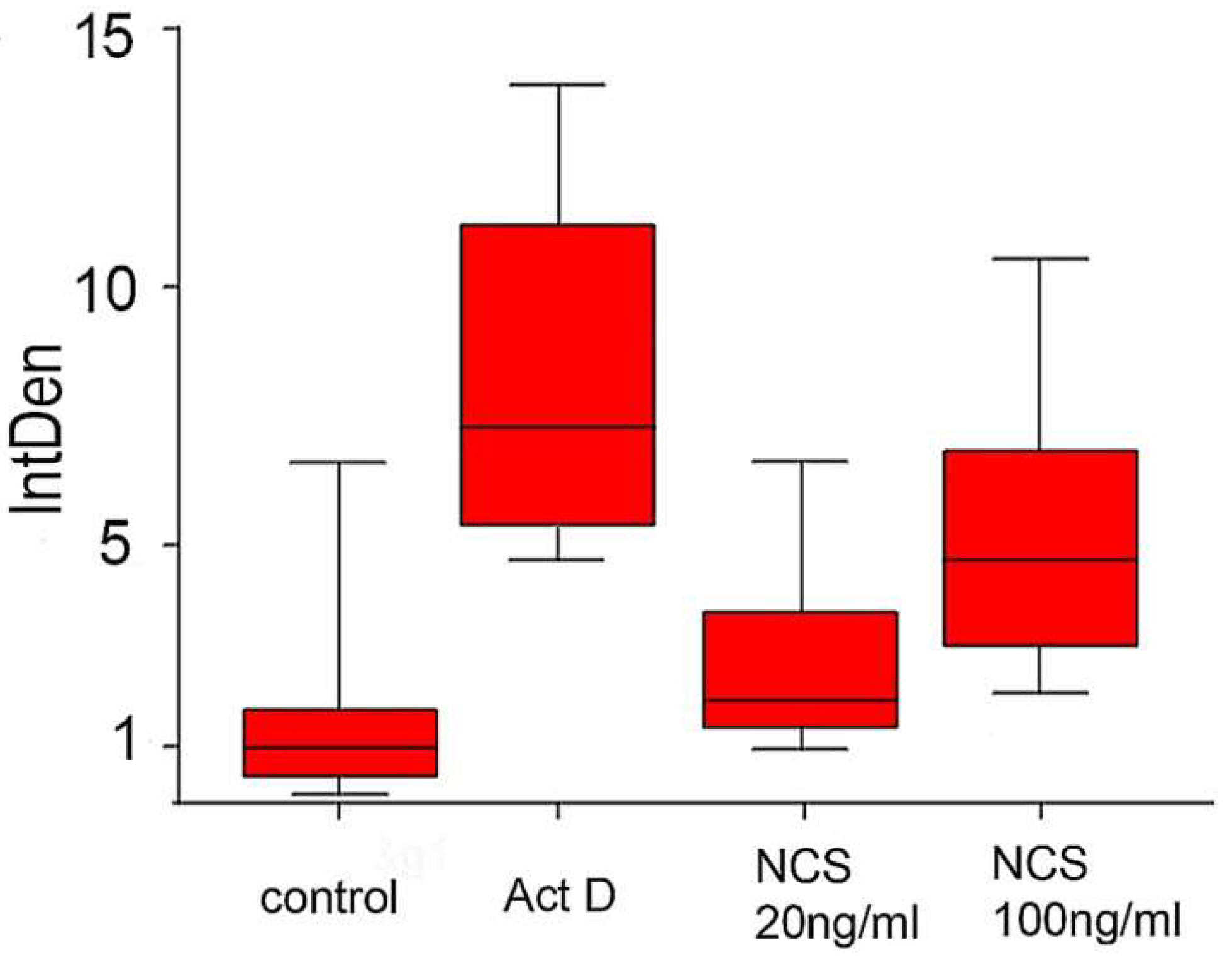
2.7. DNA Damage Increases Chromatin Fragmentation and Reduces the Number of Blastomeres
3. Discussion
4. Materials and Methods
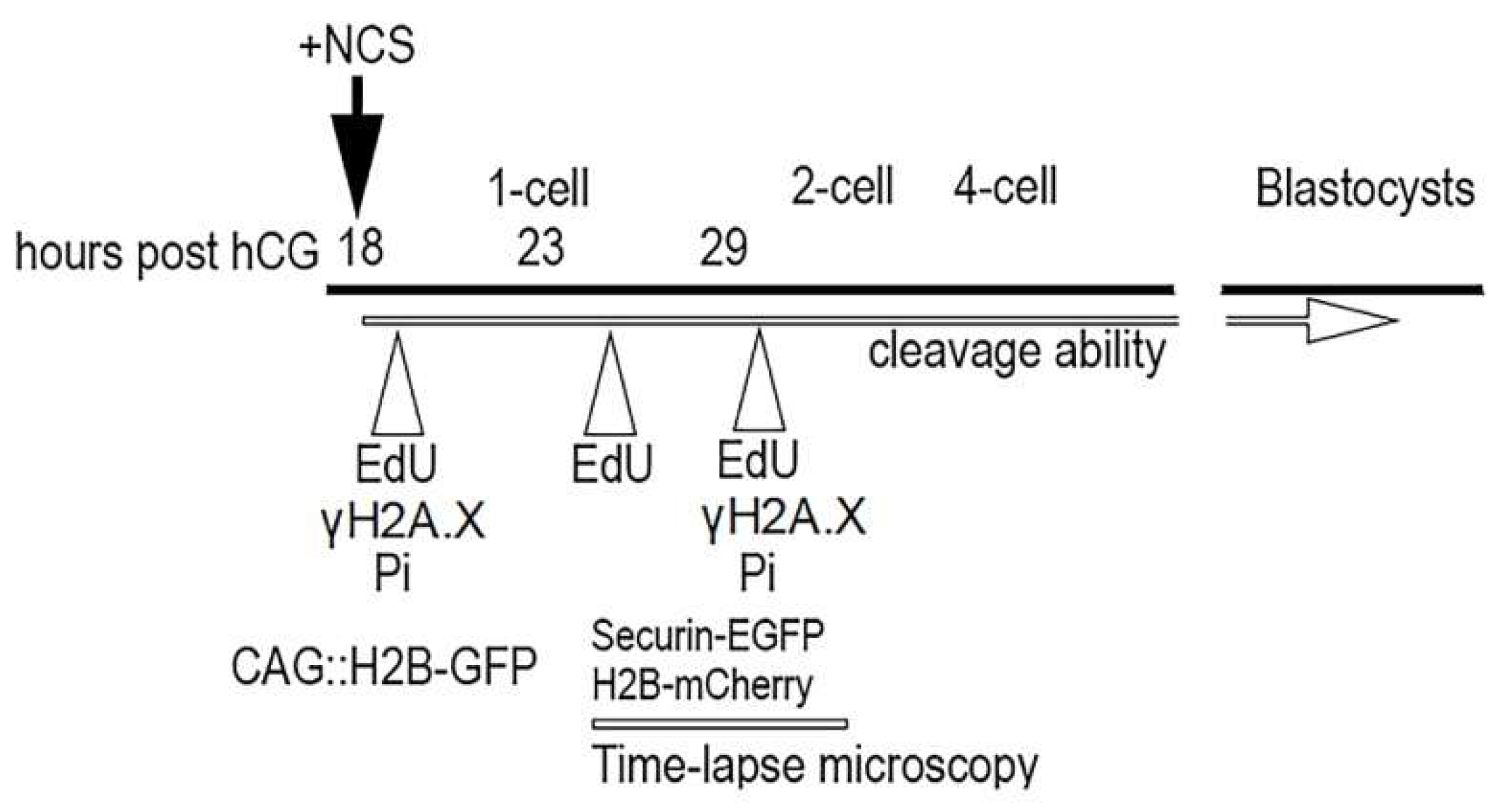
4.1. Embryo Collection and In Vitro Culture
4.2. Evaluation of the Ability to Cleavage
4.3. Immunocytochemistry
4.4. Propidium Staining
4.5. Time-Lapse Recording
4.6. TUNEL Assay
4.7. Image Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| APC/C | Anaphase-promoting complex/cyclosome |
| DAPI | 4’,6-diamidino-2-phenylindole |
| DSBs | Double-stranded breaks |
| eCG | Pregnant mare’s serum gonadotropin |
| EdU | 5-ethynyl-2’-deoxyuridine |
| EGFP | Enhanced green fluorescent protein |
| FITC | Fluorescein.5-isothiocyanate |
| hCG | Human chorionic gonadotropin |
| MN/MNs | Micronuleus/Micronuclei |
| NCS | Neocarzinostatin |
| NEBD | Nuclear envelope breakdown |
| PBS | Phosphate buffer saline |
| RT | Room temperature |
| SAC | Spindle assembly checkpoint |
| TRITC | Tetramethylrhodamine |
References
- Musson, R.; Gąsior, Ł.; Bisogno, S.; Ptak, G.E. DNA damage in preomplantation embryos and gametes: Specification, clinical relevance and repair strategies. Hum. Reprod. Update 2022, dmab046. [Google Scholar] [CrossRef]
- Munisha, M.; Schimenti, J.C. Genome maintenance during embryogenesis. DNA Repair 2021, 106, 103195. [Google Scholar] [CrossRef]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic reprogamming of DNA methylation in the early mouse embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Wossidlo, M.; Arand, J.; Sebastiano, V.; Leikhov, K.; Boiani, M.; Reihardt, R.; Scholler, H.; Walter, J. Dynamic link od DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010, 29, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Menezo, Y.; Dale, B.; Cohen, M. DNA damage and repair in human oocyte and embryos: A review. Zygote 2010, 18, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Bazrgar, M.; Gourabi, H.; Yazdi, P.E.; Vazirinasab, H.; Fakhri, M.; Hassani, F.; Valojerdi, M.R. DNA repair signalling pathway genes are overexpressed in poor-quality pre-implantation human embryos with complex aneuploidy. Eur. J. Obs. Gynecol. Reprod. Biol. 2014, 175, 152–156. [Google Scholar] [CrossRef]
- Ladstatter, S.; Tachibana-Konwalski, K. A Surveillance mechanism ensure repair of DNA lesions during zygotic reprogramming. Cell 2016, 167, 1774–1787. [Google Scholar] [CrossRef]
- Shukla, V.; Høffding, M.K.; Hoffmann, E.R. Genome diversity and instability in human germ cells and preimplantation embryos. Semin. Cell Dev. Biol. 2021, 113, 132–147. [Google Scholar] [CrossRef]
- Girardi, L.; Serdarogullari, M.; Patassini, C.; Poli, M.; Fabiani, M.; Caroselli, S.; Coban, O.; Findikli, N.; Boynukalin, F.K.; Bahceci, M.; et al. Incidence, origin, and predictive model for the detection and clinical management of segmental aneuploidies in human embryos. Am. J. Hum. Genet. 2020, 106, 525–534. [Google Scholar] [CrossRef]
- Palmer, N.; Kaldis, P. Regulation of the embryonic cell cycle during mammalian preimplantation development. Curr. Top. Dev. Biol. 2016, 120, 2–53. [Google Scholar] [CrossRef]
- Mu, X.F.; Jin, X.L.; Farnham, M.M.J.; Li, Y.; O’Neil, C. DNA damage-sensing kinases mediate the mouse 2-cell embryo’s response to genotoxic stress. Biol. Reprod. 2011, 85, 524–535. [Google Scholar] [CrossRef] [PubMed]
- House, N.C.M.; Koch, M.R.; Freudenreich, C.H. Chromatin modification and DNA repair beyond double-strand breaks. Front. Genet. 2014, 5, 296. [Google Scholar] [CrossRef] [PubMed]
- Derijck, A.H.A.; van der Heijden, G.W.; Giele, M.; Philippens, M.E.P.; van Bavel, C.A.W.; de Boer, P. γH2AX signalling during sperm chromatin remodelling in the mouse zygote. DNA Repair 2006, 5, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, M.; Oda, S.; Mitani, H.; Nagata, M.; Aoki, F. Deficiency in response to DNA double-strand breaks in mouse early preimplantation embryos. Biochem. Biophys. Res. Commun. 2007, 358, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Ma, X.S.; Ma, J.Y.; Luo, Y.B.; Lin, F.; Wang, Z.B.; Fan, H.Y.; Schatten, H.; Sun, Q.Y. Laser microbeam-induced DNA damage inhibits cell division in early fertilized eggs and early embryos. Cell Cycle 2013, 12, 3336–3344. [Google Scholar] [CrossRef]
- Ma, J.Y.; Ou-Yang, Y.C.; Wang, Z.W.; Wang, Z.B.; Jing, Z.Z.; Luo, S.H.; Hou, Y.; Liu, Y.H.; Schatten, H.; Sun, Q.Y. The effect of DNA double-strand breaks on mouse oocyte meiotic maturation. Cell Cycle 2013, 12, 1233–1241. [Google Scholar] [CrossRef][Green Version]
- Marchetti, F.; Bishop, J.B.; Lowe, X.; Generoso, W.M.; Hozier, J.; Wyrobek, A.J. Etoposide induces heritable chromosomal aberration and aneuploidy during male meiosis in mouse. Proc. Natl. Acad. Sci. USA 2001, 98, 3953–3957. [Google Scholar] [CrossRef]
- Mayer, A.; Baran, V.; Sakakibara, Y.; Brzakova, A.; Ferencova, I.; Motlik, J.; Kitajima, T.S.; Schultz, R.M.; Solc, P. DNA damage response during mouse oocyte maturation. Cell Cycle 2016, 15, 546–558. [Google Scholar] [CrossRef]
- Gawecka, J.E.; Marh, J.; Ortega, M.; Yamauchi, Y.; Ward, M.A.; Ward, W.S. Mouse zygotes respond sperm DNA damage by delaying paternal DNA replication and embryonic development. PLoS ONE 2013, 8, e56385. [Google Scholar] [CrossRef]
- Maragos, P.; Carroll, J. Oocytes progress beyond prophase in the presence DNA damage. Curr. Biol. 2012, 22, 989–994. [Google Scholar] [CrossRef]
- Jacquet, P.; Buset, J.; Vankerkom, J.; Baatout, S.; deSaint-Georges, L.; Schoonjansm, W.; Desaintes, C. Mouse one-cell embryo undergoing aradiation-induced G2 arrest may re-enter S-phase in absence of cytokinesis. Can. J. Physiol. Pharm. 2002, 80, 618–624. [Google Scholar] [CrossRef]
- Barton, T.S.; Robaire, B.; Hales, B.F. DNA damage recognition in rat zygote following chronic paternal cyclophospamide exposure. Toxicol. Sci. 2007, 100, 492–503. [Google Scholar] [CrossRef]
- Kort, D.H.; Chia, G.; Treff, N.R.; Tanaka, A.J.; King, T.; Vensand, L.B. Human embryo commonly form abnormal nuclei during development: A mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum. Reprod. 2016, 31, 312–313. [Google Scholar] [CrossRef] [PubMed]
- Golberg, I.H. Free radical mechanisms in neocarzinostatin-induced DNA damage. Free Radic. Biol. Med. 1987, 3, 41–54. [Google Scholar] [CrossRef]
- Dedon, P.C.; Goldberg, I.H. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem. Res. Toxicol. 1992, 5, 311–332. [Google Scholar] [CrossRef]
- Yuen, W.S.; Merriman, J.A.; O’Bryan, M.K.; Jones, K.T. DNA double strand breaks but not interstrand crosslinks prevent progress through meiosis in fully grown mouse oocytes. PLoS ONE 2012, 7, e43875. [Google Scholar] [CrossRef]
- Wrann, S.; Kaufmann, M.R.; Wirthner, R.; Stiel, D.P.; Werner, R.H. HIF mediated and DNA damage independent histone H2A.X phosphorylation in chronic hypoxia. Biol. Chem. 2013, 394, 519–528. [Google Scholar] [CrossRef]
- Tu, W.Z.; Li, B.; Huang, B.; Wang, Y.; Liu, X.D.; Guan, H.; Zhang, S.M.; Tang, Y.; Rang, W.Q.; Zhou, P.K. Gamma H2A.X foci formation in the absence of DNA damage mitotic H2A.X phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 2013, 587, 3437–3443. [Google Scholar] [CrossRef]
- Borhani, N.; Rajaei, F.; Salehi, Z.; Javadi, A. Analysis od DNA fragmentation in mouse embryos exposed to an low-frequency electromagnetic field. Electromagn. Biol. Med. 2011, 30, 246–252. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, Y.; Li, Z.; Zhou, Y.; Lin, H.; Wu, X.; Chen, M.; Xioa, W. Effects of the insemination of hydrogen peroxide-treated epididymal mouse spermatozoa on γH2AX repair and embryo development. PLoS ONE 2012, 7, e38742. [Google Scholar] [CrossRef][Green Version]
- Grenier, L.; Robaire, B.; Hales, B.F. The activation of DNA damage detection and repair responses in cleavage-stage rat embryos by adameged paternal genome. Toxicol. Sci. 2012, 127, 555–566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Q.L.; Sun, S.C.; Han, J.; Kwak, Y.C.; Kim, N.H.; Cui, X.S. Doxorubicine induces early embryo apoptosis by inhibiting poly(ADP ribose) polymerase. In Vitro 2012, 26, 827–834. [Google Scholar] [PubMed]
- Kuo, L.J.; Yang, L.X. Gamma-H2A.X–a novel biomarker for DNA double-strand breaks. In Vivo 2008, 22, 305–309. [Google Scholar] [PubMed]
- Stucki, M.; Clapperton, J.A.; Mohhamad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 directly binds phosphorylated histone H2A.X to regulate cellular responses to DNA double-strand breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef]
- Strauss, C.; Goldberg, M. Recruitment of protein DNA double-strand breaks. Cell Cycle 2011, 10, 2850–2857. [Google Scholar] [CrossRef] [PubMed]
- Ziegler-Birling, C.; Helmrich, A.; Tora, L.; Torres-Padilla, M.E. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int. J. Dev. Biol. 2009, 53, 1003–1011. [Google Scholar] [CrossRef]
- Chen, Z.X.; Riggs, A.D. DNA methylation and demethylation in mammals. J. Biol. Chem. 2011, 286, 18347–18353. [Google Scholar] [CrossRef]
- Schuermann, D.; Weber, A.R.; Schär, P. Active DNA demethylation by DNA repair: Fact and uncertainties. DNA Repair 2016, 44, 92–102. [Google Scholar] [CrossRef]
- Kermi, C.; Aze, A.; Maiorano, D. Preserving genome integrity during the early embryonic DNA replication cycle. Genes 2019, 10, 398. [Google Scholar] [CrossRef]
- Jacobs, K.; deVelde, H.; Paepe, C.D.; Serson, K.; Spits, C. Mitotic spindle disruption in human preimplantation embryos activates the spindle assembly checkpoint but not apoptosis until day 5 of development. Molec. Human Reprod. 2017, 23, 321–329. [Google Scholar] [CrossRef]
- Lewis, C.W.; Golsteyn, R.M. Cancer cells that survive checkpoint contain micronuclei that harbor damaged DNA. Cell Cycle 2016, 15, 3131–3145. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, D.; Goldteyn, R.M. G2/M-phase checkpoint adaptation and micronuclei formation as mechanisms that contribute to genomic instability in human cells. Int. J. Mol. Sci. 2017, 18, 2344. [Google Scholar] [CrossRef]
- Vázquez-Diez, C.; Yamagata, K.; Trivedi, S.; Haverfield, J.; FitzHarris, G. Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embyos. Proc. Natl. Acad. Sci. USA 2016, 113, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lentermenn, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Cui, K.; Yi, Q.; Yu, L.; Xu, Y. DNA repair mechanisms in embryonic stem cells. Cell Mol. Life Sci. 2017, 74, 487–493. [Google Scholar] [CrossRef]
- Tian, Y.; Yamauchi, T. Micronucleus formation in 3-day mouse embryos associated with maternal exposure to chlorpyrifos during the early preimplantation period. Reprod. Toxicol. 2003, 17, 401–405. [Google Scholar] [CrossRef]
- Khokhlova, V.; Fesenko, Z.S.; Sopova, J.V.; Leonova, E.I. Features of DNA repair in thre early stages of mammalian embryonic development. Genes 2020, 11, 1138. [Google Scholar] [CrossRef]
- Uehara, R.; Cerritelli, S.M.; Hasin, N.; Sakhuja, K.; London, M.; Iranzo, J.; Chon, H.; Grinberg, A.; Crouch, R.J. Two RNase H2 mutants with differential RNMP processing activity reveal a threshold of ribonucleotide tolerance for embryonic development. Cell Rep. 2018, 25, 1135–1145.e5. [Google Scholar] [CrossRef]
- Bolton, H.; Graham, S.J.L.; Van der Aa, N.; Kumar, P.; Theunis, K.; Fernandez Gallaro, E.; Voet, T.; Zernicka-Goetz, M. Mouse of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat. Commun. 2016, 7, 11165. [Google Scholar] [CrossRef]
- Pisko, J.; Spirkova, A.; Cikos, S.; Olexikova, L.; Kovarikova, V.; Sefcikova, Z.; Fabian, D. Apoptotic cells in mouse blastocysts are eliminated by neighbouringblastomeres. Sci. Rep. 2021, 11, 9228. [Google Scholar] [CrossRef]
- Mashiko, D.; Ikeda, Z.; Yao, T.; Tokoro, M.; Fukunaga, N.; Asada, Y.; Yamagata, K. Chromosome segregation error during early cleavage in mouse pre-implantation embryo does not necessarily cause developmental failure after blastocyst stage. Sci. Rep. 2020, 10, 854. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Ibeas, P.; Gimeno, I.; Cañón-Beltrán, K.; Gutiérrez-Adán, A.; Rizos, D.; Gómez, E. Senescence and apoptosis during in vitro embryo develpmen in abovine model. Front. Cell Dev. Biol. 2020, 8, 619902. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; van Tol, H.T.A.; Stout, T.A.E.; Roelen, B.A.J. Cellular Fragments in the Perivitelline Space Are Not a Predictor of Expanded Blastocyst Quality. Front. Cell Dev. Biol. 2021, 8, 616801. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.L.; Bauer, G.B.; Povirk, L.F. DNA damage induced by bleomycin, neocarzinostatin, and melphalan in a precisely positioned nucleosome. Asymmetry in protection at the periphery of nucleosome-bound DNA. J. Biol. Chem. 1994, 269, 30587–30594. [Google Scholar] [CrossRef]
- Neef, A.B.; Luedtke, N.W. Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. Proc. Natl. Acad. Sci. USA 2011, 108, 20404–20409. [Google Scholar] [CrossRef]
- Herbert, M.; Levasseur, M.; Homer, H.; Yallop, K.; Murdoch, A.; McDougall, A. Homologue disjunction in mouse oocytes requires proteolysis of securin an cyclin B1. Nat. Cell Biol. 2003, 5, 1023–1025. [Google Scholar] [CrossRef]
- Hadjantonakis, A.K.; Pappaioannou, V. Dynamic in vivo imaging and cell tracking sing a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004, 4, 33. [Google Scholar] [CrossRef]
- Kudo, N.R.; Anger, M.; Peters, A.H.; Stemmann, O.; Theussl, H.C.; Helmhart, W.; Kudo, H.; Heyting, C.; Nasmyth, K. Role of cleavage by separase of the Rec8 kleisin subunit of cohesion during mammalian meiosis I. J. Cell Sci. 2009, 122, 2686–2698. [Google Scholar] [CrossRef]
- Solc, P.; Kitajima, T.S.; Yoshida, S.; Brzakova, A.; Kaido, M.; Baran, V.; Mayer, A.; Samalova, P.; Motlik, J.; Ellenberger, J. Multiple requirements of PLK1 during mouse oocyte maturation. PLoS ONE 2015, 10, e0116783. [Google Scholar] [CrossRef]
- Baran, V.; Brzakova, A.; Rehak, P.; Kovarikova, V.; Solc, P. PLK1 regulates spindle formation kinetics and APC/C activation in mouse zygote. Zygote 2015, 24, 338–345. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Friese, E.; Kayning, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Ruaden, C.; Saafeld, S.; Schmid, B.; et al. FiJi: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Luisier, F.; Vonesch, C.; Blue, T.; Unser, M. Fast interscale wavelet denoising of Poisoon-corrupted images. Signal. Proc. 2010, 90, 415–427. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baran, V.; Ďuríček, T.; Pisko, J.; Drutovič, D.; Šolc, P. Cleavage of Early Mouse Embryo with Damaged DNA. Int. J. Mol. Sci. 2022, 23, 3516. https://doi.org/10.3390/ijms23073516
Baran V, Ďuríček T, Pisko J, Drutovič D, Šolc P. Cleavage of Early Mouse Embryo with Damaged DNA. International Journal of Molecular Sciences. 2022; 23(7):3516. https://doi.org/10.3390/ijms23073516
Chicago/Turabian StyleBaran, Vladimír, Tomáš Ďuríček, Jozef Pisko, Dávid Drutovič, and Petr Šolc. 2022. "Cleavage of Early Mouse Embryo with Damaged DNA" International Journal of Molecular Sciences 23, no. 7: 3516. https://doi.org/10.3390/ijms23073516
APA StyleBaran, V., Ďuríček, T., Pisko, J., Drutovič, D., & Šolc, P. (2022). Cleavage of Early Mouse Embryo with Damaged DNA. International Journal of Molecular Sciences, 23(7), 3516. https://doi.org/10.3390/ijms23073516





