Abstract
4-Coumarate:CoA ligase (4CL, EC6.2.1.12), located at the end of the phenylpropanoid metabolic pathway, regulates the metabolic direction of phenylpropanoid derivatives and plays a pivotal role in the biosynthesis of flavonoids, lignin, and other secondary metabolites. In order to understand the molecular characteristics and potential biological functions of the 4CL gene family in the pomegranate, a bioinformatics analysis was carried out on the identified 4CLs. In this study, 12 Pg4CLs were identified in the pomegranate genome, which contained two conserved amino acid domains: AMP-binding domain Box I (SSGTTGLPKGV) and Box II (GEICIRG). During the identification, it was found that Pg4CL2 was missing Box II. The gene cloning and sequencing verified that this partial amino acid deletion was caused by genome sequencing and splicing errors, and the gene cloning results corrected the Pg4CL2 sequence information in the ‘Taishanhong’ genome. According to the phylogenetic tree, Pg4CLs were divided into three subfamilies, and each subfamily had 1, 1, and 10 members, respectively. Analysis of cis-acting elements found that all the upstream sequences of Pg4CLs contained at least one phytohormone response element. An RNA-seq and protein interaction network analysis suggested that Pg4CL5 was highly expressed in different tissues and may participate in lignin synthesis of pomegranate. The expression of Pg4CL in developing pomegranate fruits was analyzed by quantitative real-time PCR (qRT-PCR), and the expression level of Pg4CL2 demonstrated a decreasing trend, similar to the trend of flavonoid content, indicating Pg4CL2 may involve in flavonoid synthesis and pigment accumulation. Pg4CL3, Pg4CL7, Pg4CL8, and Pg4CL10 were almost not expressed or lowly expressed, the expression level of Pg4CL4 was higher in the later stage of fruit development, suggesting that Pg4CL4 played a crucial role in fruit ripening. The expression levels of 4CL genes were significantly different in various fruit development stages. The results laid the foundation for an in-depth analysis of pomegranate 4CL gene functions.
1. Introduction
The emergence of lignin enables terrestrial plants to stand upright and transport water for a long distance. Flavonoids are a crucial class of secondary metabolites of plants [1]. These two metabolites participate in regulating various physiological functions and improving the adaptability to environmental stress [2,3]. The 4CL gene can encode multiple enzymes and exhibit distinct substrate affinities that appear to coincide with different metabolic functions [4]. Different 4CL isozymes can selectively catalyze cinnamic acid, p-coumaric acid and other substances to produce corresponding CoA thioesters. Some CoA thioesters can be synthesized by cinnamoyl-CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) catalyze the reaction into the specific pathway of lignin biosynthesis [5], or through chalcone synthase (CHS) and chalcone isomerase (CHI) to guide the reaction into the specific pathway of flavonoid biosynthesis [6]. Members of the 4CL gene family have now been identified in several species, such as Arabidopsis [7], rice [8], cotton [9], Eucalyptus grandis [10], and pear [11]. The 4CL proteins all contain two amino acid conserved structural domains: Box I (SSGTTGLPKGV) and Box II (GEICIRG) [12]; Box I is the AMP-binding functional domain, and this region is not only conserved in 4CL but is also a conserved sequence typical to all acyl-activated enzymes, and can be directly involved in the catalytic reaction, whereas Box II is only highly conserved in 4CL and is not directly involved in the catalysis; it may be related to the spatial conformation of the enzyme [13].
Lavhale et al. [13] found that the 4CL family may evolve separately after monocots and eudicots were isolated by phylogenetic analysis of 4CL protein sequences in different species. The 4CL family genes in eudicots are divided into two groups, type I and type II [8]. Type I 4CL members of eudicots, such as Arabidopsis At4CL1, At4CL2, At4CL4, and E. grandis Egr4CL1, play a crucial role in regulating lignin biosynthesis; Type II 4CL members are closely related to flavonoid biosynthesis, At4CL3 and Egr4CL2 participate in flavonoid biosynthesis [3,10,14]. In addition, some genes contain the same highly conserved AMP structural domain as 4CL, and are classified as 4CL-like [7,15]. Although these genes are similar to 4CL in structure and can be successfully clustered with 4CL in phylogeny, their evolutionary relationships and functions are still unknown [16]. Recently, it has been reported that 4CL and 4CL-like genes are able to regulate various physiological functions and improve the ability to adapt to the environment [17].
The pomegranate (Punica granatum) is not only of considerable ornamental value but it is also a medicinal and food fruit [18,19]. Pomegranates are rich in antioxidants, such as flavonoids, which are extremely effective against tumors, diabetes, and other diseases [20,21,22,23]. According to the differences in seed hardness, pomegranates are classified into soft-seeded, semi-soft-seeded, and hard-seeded varieties, and it was found that the lignin content of the pericarp of pomegranate was positively correlated with seed hardness [18]. Soft-seeded pomegranates are highly edible and more popular with the market and consumers. The characters of soft-seeded and high flavonoid content will be a new target of pomegranate quality breeding in the future. The identification and evolutionary analysis of the 4CL gene family has been reported on in recent years, but research on the 4CL gene family in the pomegranate has not been reported on. The completion of pomegranate genome sequencing and the publication of data [24,25,26] provides data support for pomegranate gene function research. This study aimed to identify Pg4CLs gene members in the pomegranate ‘Taishanhong’ genome by bioinformatics methods, analyze their phylogeny, gene structure, conserved motifs, cis-acting elements, protein interaction network and tissue expression specificity, and explore their potential roles in pomegranate growth and development with different tissues. The gene Pg4CL2 (Pg001327.1) was cloned, the reason for Box II deletion was analyzed, and its function was further studied.
2. Results
2.1. Identification of Pg4CL Family Genes and Gene Cloning of Pg001327.1
In this study, 16 candidate members were selected from the pomegranate genome of ‘Taishanhong’ by using the hidden Markov file of AMP binding domain (PF00501), and their AMP binding domain was verified by Pfam, NCBI, and SMART, respectively; the sequences of Box I and Box II without the 4CL amino acid conserved domain were eliminated. During the identification, it was found that although Box II of the Pg001327.1 amino acid sequence was missing, At4CL3 had the highest homology with Pg001327.1 (61.79%) in the genome of ‘Taishanhong’. To explore whether Pg001327.1 belongs to the 4CL gene family, multiple alignments were performed between Pg001327.1, At4CL3, and homologous proteins of other species (FASTA S1, Figure 1), and only Box II of Pg001327.1 was deleted; Box I and Box II of amino acid sequences of other species were highly conserved. In order to explore the reasons for the deletion of the Pg001327.1 amino acid sequence of Box II, a gene cloning experiment was carried out.
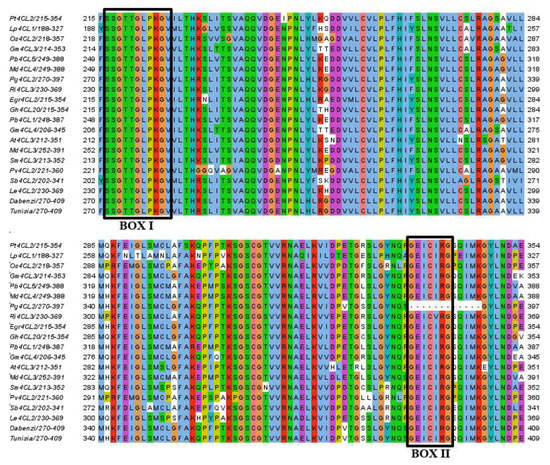
Figure 1.
Multiple sequences alignment of At4CL3 with other plant homologous proteins. Note: the prefixes Pt, Lp, Os, Gm, Pb, Md, Ri, Egr, Gh, At, Sm, Pv, Sb, and Le stand for Populus tremuloides, Loblolly pine, Oryza Sativa, Glycine max, Pyrus bretschneideri, Malus domestica, Rubus idaeus, Eucalyptus grandis, Gossypium hirsutum, Arabidopsis thaliana, Salvia miltiorrhiza, Panicum virgatum, Sorghum bicolor, and Lithospermum erythrorhizon, respectively. ‘Dabenzi’ and ‘Tunisia’ belong to pomegranate.
The Pg001327.1 gene was amplified from the cDNA of ‘Taishanhong’ by PCR (Figure 2). The DNA was collected and sequenced after cloning. The total length of the PCR products was 1738 bp. The sequencing results were compared with pomegranate ‘Taishanhong’ genome sequence Pg001327.1 and ‘Tunisian’ genome sequence XP_ 031382283.1 (Figure 3), reaching 90.14% and 92.05%, respectively, with high homology. The Pg001327.1 gene was missing 36 nucleotides within positions 1404–1439 bp. Following the nucleotide positions of Pg001327.1 and XP_031382283.1, the Pg001327.1 nucleotide deletion sequence ‘GGTGAAATTTGTATTCGAGGCCAACAGATTATGAAAGG’ was translated, and the amino acid sequence was ‘GEICIRGQQIMK’, which contained Box II (GEICIRG), indicating that the deletion of amino acid sequence Box II of Pg001327.1 was caused by genome sequencing and assembly errors. According to the sequencing results, the sequence information of the genome Pg001327.1 was supplemented with missing nucleotides, and the corrected sequence was used for subsequent bioinformatics analysis.
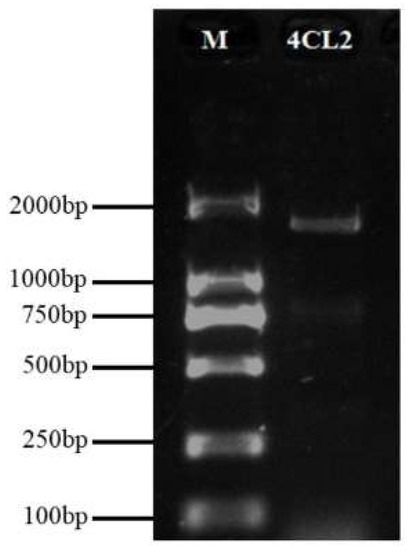
Figure 2.
Detection of Pg001327.1 PCR products by electrophoresis. Note: M: DL2000 DNA Marker; 4CL2: Target gene.

Figure 3.
Multiple alignments of Pg001327.1, XP_031382283.1 and sequencing results. This square represented that Pg001327.1 was missing 36 nucleotides within positions 1404–1439 bp.
2.2. Analysis of Physical and Chemical Properties
This study finally identified 12 candidate Pg4CLs genes in the pomegranate genome and named the Pg4CLs gene sequence as Pg4CL1–Pg4CL12 in the order of gene ID (Table 1). An analysis of the physical and chemical properties of Pg4CLs showed that the coding sequence of Pg4CLs was between 1629 and 2343 bp. In addition, the number of exons of Pg4CLs members was between 4 (Pg4CL8) and 14 (Pg4CL1), most of which were 6 exons. The Pg4CLs protein contains 542 (Pg4CL6) to 780 (Pg4CL1) amino acids, and the isoelectric point was between 5.49 (Pg4CL8) and 8.91 (Pg4CL12). A total of 58.3% of the Pg4CLs protein had a pI value greater than 7, which was slightly alkaline. The instability index of Pg4CLs protein ranged from 26.44 (Pg4CL6) to 49.09 (Pg4CL4), and 42% of Pg4CLs protein had good structural stability (less than 40 was stable). The total grand average of hydropathicity (GRAVY) of Pg4CL proteins ranged from −0.057 (Pg4CL4) to 0.159 (Pg4CL3). A total of 66.7% of Pg4CLs proteins were hydrophobic proteins (positive values were expressed as hydrophobic proteins). The analysis found that none of the Pg4CL proteins contained signal peptides and belonged to intracellular proteins. The prediction results of subcellular localization showed that most of the Pg4CL proteins (Pg4CL1, Pg4CL3, Pg4CL6, Pg4CL7, Pg4CL8, Pg4CL11, Pg4CL12) were localized in the plasma membrane, while the others were localized in the cytosol, extracellular, and peroxisome, respectively.

Table 1.
The basic information of the Pg4CLs gene family in the pomegranate.
The prediction of the tertiary structure of proteins contributes to the study of their functions. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. The Pg4CL protein tertiary structure model had a very high confidence and great similarity (Figure 4).
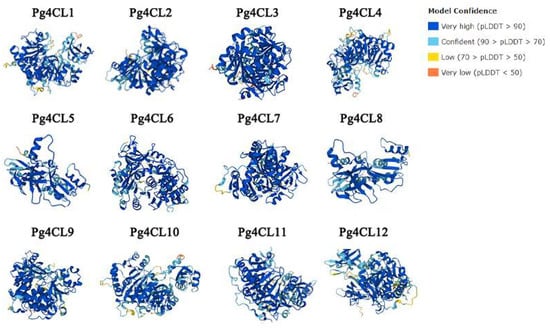
Figure 4.
Tertiary structure of Pg4CL proteins. The tertiary structure of the protein was predicted through the AlphaFold. Model confidences are indicated by different colors. Blue, light blue, yellow, and red represent very high, confident, low, and very low confidence, respectively.
2.3. Phylogenetic Analysis and Classification of the Pg4CL Gene Family
In order to investigate the phylogenetic relationships of 4CL proteins, a phylogenetic tree was constructed using the neighbor-joining (NJ) method for a total of 38 amino acid sequences of 4CLs from A. thaliana, E. grandis, and P. granatum (Figure 5). The results showed that Pg4CL5 successfully clustered with At4CL1, At4CL2, At4CL4, and Egr4CL1, indicating that Pg4CL5 belonged to Group 1; Pg4CL2 clustered with At4CL3/Egr4CL2 and belonged to Group 2; the remaining 10 Pg4CLs clustered with At4CL-like/Egr4CL-like gene sequences and belonged to Group 3. Moreover, the functional studies of Group 1 and Group 2 genes in A. thaliana and E. grandis showed that they were involved in lignin synthesis and flavonoid synthesis. Therefore, it speculated that Pg4CL5 and Pg4CL2 play a pivotal role in lignin and flavonoid biosynthesis in the pomegranate.
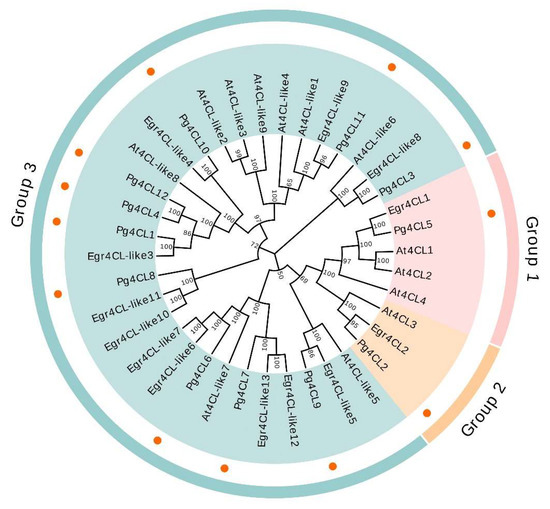
Figure 5.
The phylogenetic tree of the 4CL gene family in A. thaliana, E. grandis, and P. granatum. The phylogenetic tree was constructed by using the neighbor-joining method with 1000 bootstrap replications. The prefixes At, Egr, and Pg stand for A. thaliana, E. grandis, and P. granatum.
2.4. Analysis of Conserved Motifs and Gene Structures of Pg4CL Gene Family
A total of 10 conserved motifs were identified, numbered motifs 1–10 (Figure S1, Table 2). Pfam analysis showed that among these 10 motifs, five motifs (motif 1, motif 2, motif 4, motif 5 and motif 6) encoded AMP structural domains, of which motif 5 and motif 2 contained the conserved structural domains Box I (SSGTTGLPKGV) and Box II (GEICIRG), respectively. The conserved motif distribution of Pg4CLs protein was constructed based on the results of motif analysis (Figure 6B). The results showed that all Pg4CLs amino acid sequences contained all motifs, and the same subgroup contained similar motif distributions. The gene structure was visualized by TBtools (Figure 6C). The analysis showed that the length of the Pg4CLs gene was 2.1–12 kb. Among the 12 Pg4CLs, 9 of them consisted of 6 exons and 5 introns. In addition, Pg4CL1 contained the most exons (14), resulting in a significantly longer gene length than other members. Group 1 and group 2 4CL members (Pg4CL5, Pg4CL2) had 6 exons. Because of their longer introns, their gene lengths were longer than that of members of group 3 (except for Pg4CL1). The number of introns ranged from 4 (Pg4CL8) to 14 (Pg4CL1).

Table 2.
Information of the 10 motifs of Pg4CLs proteins.
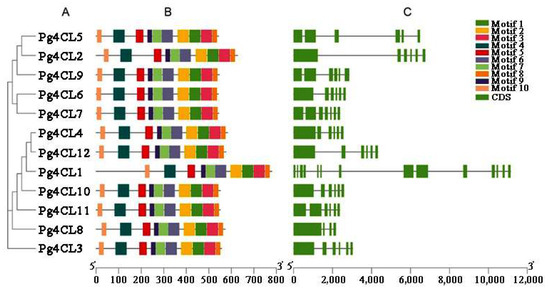
Figure 6.
Phylogenetic tree, conservative protein motifs and gene structure of the Pg4CLs in pomegranate. (A) The phylogenetic tree of 12 Pg4CLs. (B) Conserved motifs in the 12 Pg4CLs. The motifs were identified by the MEME Suite. Different conserved motifs, numbers 1–10, are displayed in different colored boxes. (C) Gene structures of the 12 Pg4CLs.
2.5. Cis-Acting Elements in the Promoter Region of Pg4CLs
The cis-acting elements are the binding sites of transcription factors and play a crucial role in regulating gene expression function. Visualizing the identified cis-acting elements of Pg4CLs, all Pg4CLs contained at least one phytohormone response element (Figure 7A), including abscisic acid response element (ABRE), growth hormone response elements (TGA-element, AuxRR-core, TGA-box), MeJA response elements (CGTCA-motif, TGACG-motif), gibberellin response elements (TATC-box, GARE-motif, P-box), and salicylic acid response elements (TCA-elements). TGA-element was an auxin-responsive element and TGA-box was part of an auxin-responsive element. Among the 12 Pg4CLs, 8 contained MeJA response elements, 6 contained gibberellin response elements, 10 contained abscisic acid response elements, 9 contained salicylic acid response elements, and 7 contained auxin response elements. There were also low-temperature response elements (LTR) and drought response elements (MBS) in the promoter regions of some Pg4CLs, 6 contained LTR and 6 contained MBS. These Pg4CLs may respond to plant biotic and abiotic stresses. In addition, there was an MYB binding site (MBSI) involved in gene regulation of flavonoid biosynthesis in the promoter region of the Pg4CL6. Some Pg4CLs promoter regions also contained regulatory elements, such as endosperm expression (GCN4_motif) and meristem expression-related (CAT-box) (Figure 7A). These Pg4CLs may be closely related to plant growth and development. In addition, G-box and G-Box were cis-acting regulatory elements involved in light responsiveness. The binding sites of G-box and G-box transcription factors were different, the former was CACGTG and the latter was CACGTT. A total of 24 light-responsive elements appeared in this study (Figure 7B), of which the promoter regions of the Pg4CLs all contained a large number (5–17) of light-responsive elements, indicating that the Pg4CLs may be regulated by light.
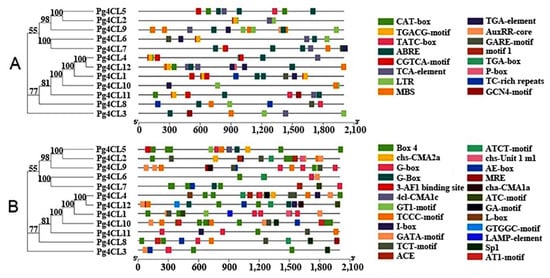
Figure 7.
Cis-acting elments of Pg4CLs. (A) Hormone, stress, growth and development response elements of Pg4CLs; (B) Light response element of Pg4CLs. Note: abscisic acid response element (ABRE), growth hormone response elements (TGA-element, AuxRR-core, TGA-box), MeJA response elements (CGTCA-motif, TGACG-motif), gibberellin response elements (TATC-box, GARE-motif, P-box), and salicylic acid response elements (TCA-elements).
2.6. Protein Interaction Networks of Pg4CL Gene Family
The protein interaction network prediction of 12 Pg4CLs showed no direct interaction between the proteins, which indicated that these proteins had no direct regulatory relationship and might play their functions. The potential functions of 4CL proteins Pg4CL5 and Pg4CL2 were predicted by protein interaction network analysis (Figure 8). The results are shown in Figure 8A,B, Pg4CL5 had high similarity with A. thaliana 4CL1 (E-value was 5.51 × 10−234), and 4CL1 was co-expressed with lignin synthesis proteins (C4H, IRX4, CYB84A1). Pg4CL2 was similar to A. thaliana 4CL3 (E-value was 3.9 × 10−212); 4CL3 was co-expressed with flavonoid synthesis proteins (TT4, TT5, OMT1, F3H, and FLS1). In addition, 4CL1 and 4CL3 were co-expressed with proteins in other metabolite synthesis pathways (At1g80820, HCT, LysoPL2).
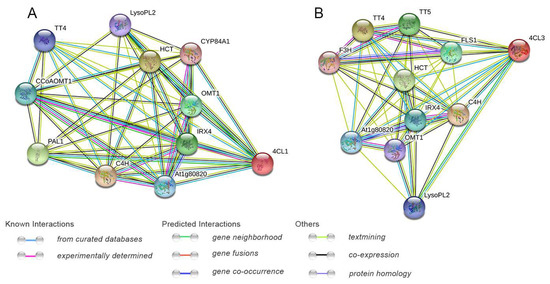
Figure 8.
Function interaction network of Pg4CL5 (A)/Pg4CL2 (B) proteins. Note: TT4: chalcone synthase; CCoAOMT1: caffeoyl coenzyme A-O-methyltransferase; At1g80820: cinnamoyl-CoA reductase 2; C4H: cinnamate 4-hydroxylase; IRX4: cinnamoyl-CoA reductase 1; PAL1: phenylalanine ammonia-lyase 1; OMT1: flavone 3′-O-methyltransferase 1; HCT: shikimate O-hydroxycinnamoyltransferase; CYP84A1: ferulate 5-hydroxylase; LysoPL2: caffeoyl shikimate esterase; TT5: chalcone-flavanone isomerase; F3H: flavanone-3-hydroxylase; FLS1: flavonol synthase.
2.7. Expression Analysis of Pg4CL Genes with RNA-Seq
To investigate the expression pattern of Pg4CL genes in different tissues of pomegranate, the expression of Pg4CLs family genes was analyzed based on published transcriptome data of different tissues of the pomegranate. The results are shown in Figure 9, Pg4CL genes were expressed in leaves, roots, flowers, seed coats, and pericarps, but there were significant differences in the expression levels of genes in different subfamilies (Figure 9A). The class I 4CL gene (Pg4CL5) was highly expressed in different tissues of pomegranate, with the highest expression in the roots and pericarps. Class II 4CL gene (Pg4CL2) was highly expressed in the leaves, flowers, and exocarps of ‘Dabenzi’ and the pericarps of ‘Tunisia’ and ‘Baiyushizi’, but the lower expression in the roots and pericarps of ‘Dabenzi’. In addition, some of the 4CL-like genes (Pg4CL1, Pg4CL4, Pg4CL6, and Pg4CL11) were highly expressed in various tissues of pomegranate, but Pg4CL8, Pg4CL9, and Pg4CL10 were lowly expressed or not expressed in various tissues, and the expression levels of 4CL-like genes were significantly different in various tissues. These results suggest that 4CL-like genes may be functionally differentiated.
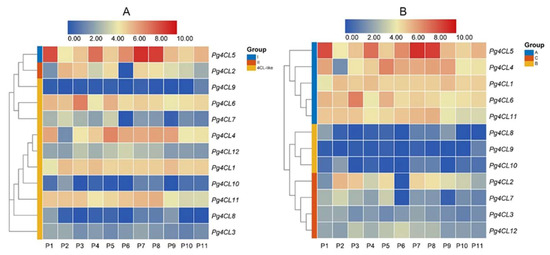
Figure 9.
Heatmap of Pg4CLs gene expression in different tissues. Note: (A) Gene expression patterns of different subfamilies genes; (B) hierarchical clustering of gene expression patterns. P1: root; P2: leaf; P3: flower; P4, P7, P8: pericarp; P5: exocarp; P6, P9: pericarp; P10–P11: a mixture of flowers, leaves, fruits, and roots (cultivars P1–P6 are ‘Dabenzi’, cultivars P7, P8, P9, and P10 are ‘Tunisia’, ‘Baiyushizi’, ‘Wonderful’, ‘nana’ and ‘Black127’, respectively.
Based on the differences in expression patterns, the Pg4CLs family was subjected to cluster analysis (Figure 9B), classified into three groups, A, B, and C. The gene expression level of group A was significantly higher than those of groups B and C and had higher expression in various tissues. The genes in group B were low or not expressed in all tissues, suggesting that these genes may have lost some of their functions during evolution. Genes in group C were expressed in leaves, flowers, and fruits, and the expression of different genes had some tissue specificity. Cluster analysis showed that different subfamily genes might have similar expression patterns, such as class I Pg4CL5 and 4CL-like genes (Pg4CL1, Pg4CL4, Pg4CL6, Pg4CL11), which had high expression in pomegranate roots, leaves, flowers, seed coats, and pericarps.
2.8. qRT-PCR Analysis of Pg4CLs during Fruit Development in Pomegranate
To explore the putative function of Pg4CLs in fruit development, the expression patterns of Pg4CLs in pericarp of pomegranate were analyzed by qRT-PCR (Figure 10). The expression level of Pg4CL1 showed an increasing trend in S1–S4 stages, the highest expression level was reached at the S4 stage. With the ripening of pomegranate fruit, the flavonoid content gradually decreased, and the expression level of Pg4CL2 and flavonoid content demonstrated a similar trend, indicating that Pg4CL2 was involved in the synthesis of flavonoids. The expression level of Pg4CL4 was higher in the later stage of fruit development, suggesting that Pg4CL4 played a crucial role in fruit ripening. The expression of Pg4CL5 was higher in S5 stages. In addition, Pg4CL3, Pg4CL7, Pg4CL8, and Pg4CL10 were almost not expressed or lowly expressed in S2–S7, and the expression levels in the S1 stage were significantly higher than those in S2–S7, and the expression levels of 4CL-like genes were significantly different in various fruit development stages.
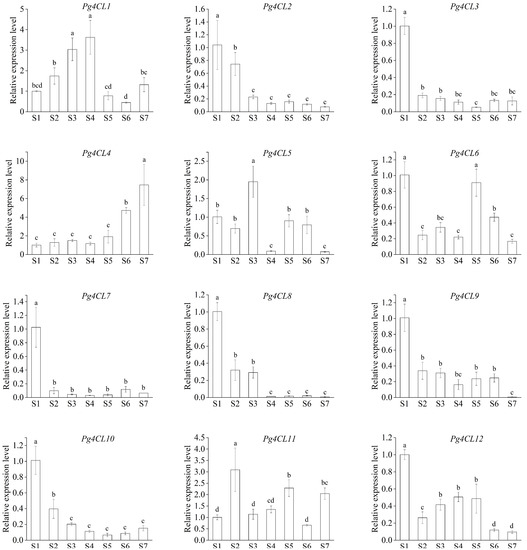
Figure 10.
Relative expression levels of 12 Pg4CL from the pericarp during fruit development. Note: the seven stages of fruit development were S1–S7, and the specific dates of the seven stages were 15 July, 28 July, 10 August, 23 August, 5 September, 18 September, and 1 October, respectively. The vertical bar in the bar graph is standard error. Bars with different letters (a–d) indicate significant differences at p < 0.05 according to Duncan’s test.
3. Discussion
Multiple gene families encode 4CLs in higher plants, and the number of their gene members varies depending on the plant species. In recent years, the identification and functional analysis of 4CL gene families have been reported for many species, such as 13 members in A. thaliana [7], 13 members in E. grandis [10], 12 members in P. bretschneideri [11], 14 members in O. sativa [8], etc. In this study, the identification of the 4CL gene family members in the ‘Taishanhong’ genome revealed that A. thaliana At4CL3 had the highest amino acid sequence similarity to Pg001327.1. However, the At4CL3 in the pomegranate ‘Tunisia’ genome and other homologous protein sequence alignment results of different species showed that the Pg001327.1 was missing Box II, a conserved structural domain to the 4CL gene family. It was speculated that the Pg001327.1 might have the sequencing and splicing errors of the genome, which caused the deletion of Box II in the amino acid sequence. The Pg001327.1 gene cloning verified this conjecture, and the genome was corrected. A total of 12 Pg4CLs were identified in the genome of ‘Taishanhong’. Combined with the phylogenetic tree of 4CL amino acid sequences of A. thaliana and E. grandis, it was found that there was 2–4 4CL genes in class I, which was different from dicots plants [13]. In addition, only one gene was contained in the 4CL class I gene of the pomegranate and E. grandis [10]. By clustering with the model plant A. thaliana, the expression of its homologous gene Pg4CL5 may be related to lignin synthesis based on the functions of At4CL1, At4CL2, and At4CL5 [3,4]. Pomegranate, E. grandis, and A. thaliana contained only one 4CL class II gene, and its homologue Pg4CL2 was presumed to be involved in flavonoid synthesis based on the function of At4CL3 [3,4]. In addition, pomegranate contained 10 4CL-like genes, and although the functions of these 4CL-like genes were unknown, results from studies in other species suggested that both 4CL and 4CL-like genes can regulate various physiological functions of plants and improve their ability to adapt to their environment [17].
Based on the conserved motifs of Pg4CLs, it was found that all amino acid sequences of Pg4CLs contained 10 motifs, and similar motif distributions were found in the same subgroup. A gene structure analysis showed that Pg4CL genes contained 4–14 exons, including 6 exons in class I and class II 4CL genes, which was consistent with the number of exons in A. thaliana (3–6) and Gossypium hirsutum (4–6) [9,27], indicating that the structure of 4CL class I and class II genes were relatively conservative. However, the number of exons of 4CL-like varied greatly among different species, such as 1–5 in A. thaliana and 1–11 in Populus pruinosa [27]. This study showed that the members of Pg4CLs were conserved in both gene structure and protein conserved motifs.
Cis-acting elements existed in gene promoters and formed specific binding with transcription factors, which played a principal role in regulating the expression of target genes [28,29]. The promoter region of pomegranate Pg4CLs was enriched in a variety of cis-acting elements related to hormone responses and abiotic stresses, and this result was broadly consistent with the pear and G. hirsutum [9,30], indicating that 4CL gene promoter regulatory elements were somewhat conservative among different species. In addition, 66.7% of the promoter regions of Pg4CLs contained MYB transcription factor binding sites, among which, 6 Pg4CLs contained MYB binding sites (MBS) involved in drought induction, 4 Pg4CLs contained MBS involved in photoreaction, and there were also MBS involved in the regulation of flavonoid biosynthesis genes in the promoter region of Pg4CL6; these three MBS were found in the 4CL gene family of longan [31]. All Pg4CLs contained at least one phytohormone response element, such as the TGA-element, TATC-box, TGACG-motif, and TCA-elements, etc., which indicated that Pg4CLs might respond to auxin, gibberellin, jasmonic acid, salicylic acid, and other hormones involved in plant growth, development, and stress response regulation. In addition, 4CLs involved in stress resistance had been verified in plants, such as Fraxinus mandshurica, G. hirsutum, and poplar [9,17,27].
The protein interaction network prediction of 12 Pg4CLs revealed no interaction between their proteins, indicating no direct regulatory relationship between these proteins [13]. The interaction network analysis of Pg4CL5 and Pg4CL2 showed that Pg4CL5 was co-expressed with lignin biosynthesis, combined with a high expression of Pg4CL5 in different tissues and the highest expression in roots and pericarps, indicating that Pg4CL5 may be associated with lignin biosynthesis in various tissues. Pg4CL5 had been cloned and validated in pomegranate ‘Baiyushizi’, and the conclusion is consistent with the present study [32]. In combination with the 4CL class II gene Pg4CL2 co-expressed with flavonoid synthesis, the high expression of Pg4CL2 in flowers, seed coats, and fruit may be associated with flavonoid synthesis and pigment accumulation [33]. In addition, it was also found that Pg4CL2 had high expression in the pericarps of ‘Tunisia’ and ‘Baiyushizi’, but a lower expression in ‘Dabenzi’, which may be due to different genotypes and differences in gene expression patterns and may also be related to fruit characteristics [34]. We found that the expression of Pg4CL2 decreased gradually, which was similar to the change trend of flavonoid content, indicating that Pg4CL2 was involved in the synthesis of flavonoids. This is consistent with the results in Pyrus bretschneideri [30] and Morus alba [35]. To a large extent, the function of the 4CL-like gene is still unknown, and it has been reported that both 4CL and 4CL-like genes can regulate various physiological functions of plants. The expression of Pg4CLs family genes was analyzed based on published transcriptome data of different tissues of pomegranate, some 4CL-like genes (Pg4CL1, Pg4CL6, Pg4CL11) were highly expressed in pomegranate tissues, but some 4CL-like genes (Pg4CL8, Pg4CL9, Pg4CL10) were low or not expressed in various tissues. In addition, the expression of Pg4CL in developing pomegranate fruits was analyzed by quantitative real-time PCR (qRT-PCR), Pg4CL1 showed an increasing trend in S1–S4 stages, the highest expression level was reached at the S4 stage; this is similar to the results in the Chinese pear [30]. Pg4CL3, Pg4CL7, Pg4CL8, and Pg4CL10 were significantly increased, suggesting that these genes might play important roles in the early stage of fruit development [30]. The expression level of the 4CL-like gene in different fruit developments is different, which indicates that the function of the 4CL-like gene may be differentiated.
4. Materials and Methods
4.1. Genome and Transcriptome Data Sources
Pomegranate genome (‘Taishanhong’ ASM286412v1; ‘Tunisia’ ASM765513v2) [24,25], transcriptome data, and homologous amino acid sequences of At4CL3 in different species were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/ (accessed on 4 November 2021)). The 4CL amino acid sequences of E. grandis and Arabidopsis were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 4 November 2021)) and the TAIR database (https://www.arabidopsis.org (accessed on 4 November 2021)).
4.2. Identification of Pg4CL Family Genes
The Hidden Markov model (HMM) files corresponding to the AMP binding domain (PF00501) [9] were downloaded from the Pfam protein family database (http://pfam.xfam.org/ (accessed on 15 November 2021)) [36] and used as probes to screen the amino acid sequences of Pg4CLs (E-value ≤ 1.0 × 10−10). The protein structural domains were verified using Pfam, NCBI CDD, and SMART (http://smart.embl-heidelberg.de/ (accessed on 15 November 2021 )) [37,38], respectively, by removing repetitive sequences and sequences without AMP structural domains. Multiple sequence alignments were performed using MAFFT (Suita, Japan) [39] to identify whether they contained the conserved structural domains Box I (SSGTTGLPKGV) and Box II (GEICIRG).
4.3. Gene Cloning of Pg001327.1
Total RNA was extracted using the BioTeke plant total RNA extraction kit (centrifugal column type) (BioTeke Corporation Co., Ltd., Beijing, China), and cDNA was obtained by the reverse transcription kit (PrimeScriptTM RT reagent Kit with gDNA Eraser, TaKaRa Biomedical Technology Co., Ltd., Tokyo, Japan). Oligo 7 software (Cascade, CA, USA) was used to design cloning primers. Used the cDNA as a template, the upstream and downstream primers (F: 5′-ATGATATCTGTTGCCCCTTCT-3′, R: 5′TTATAAGGGAGTGGAGGAGG-3′) were used for PCR amplification. The PCR reaction system was 50 μL, including 25 μL 2 × Rapid Taq Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China), 2 μL upstream and downstream primers, 2 μL cDNA template, and 21 μL ddH2O. The PCRs were performed on the Applied Biosystems 7500 (Thermo Fisher Scientific Inc., Waltham, MA, USA). The amplification procedures were 95 °C for 3 min; 95 °C for 15 s, 58 °C for 45 s, 72 °C for 1 min, a total of 35 cycles, and 72 °C for 5 min. The similarity between sequencing results and sequences Pg001327.1 and XP_031382283.1 was analyzed using BioXM 2.6 software, and the conservativeness of sequences was analyzed using Jalview software (Dundee, UK) after multiple sequence alignments using MAFFT [39].
4.4. Virtual Prediction of Physical and Chemical Properties
The ExPASy (https://web.expasy.org/protparam/ (accessed on 15 January 2022)) [40] was used to predict the physical and chemical properties of Pg4CL proteins, mainly including amino acid numbers, isoelectric point, instability index, grand average of hydropathicity (GRAVY). In addition, the SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP (accessed on 15 January 2022)) was used to predict the signal peptide of the Pg4CL proteins [41]. Subcellular localization of the Pg4CL proteins was predicted using the Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ (accessed on 19 January 2022)) [42]. Tertiary structures of Pg4CL proteins were predicted using the AlphaFold (https://www.alphafold.ebi.ac.uk/ (accessed on 19 January 2022)), which is based on a massive database and innovative algorithm [43].
4.5. Multiple Sequences Alignment and Phylogenetic Analysis
Multiple sequences alignment was performed by MAFFT [39] using a total of 38 full-length protein sequences, including Arabidopsis thaliana (At, 13 sequences), Punica granatum (Pg, 12 sequences), and Eucalyptus grandis (Egr, 13 sequences). To investigate the phylogenetic relationships of the 4CL proteins, MEGA7.0 software [44] was used to construct a phylogenetic tree using the NJ method, Bootstrap 1000 repeats, and the others were set as default parameters. The online software EvolView (https://evolgenius.info/ (accessed on 11 February 2022)) was used to beautify the evolutionary tree [45]. According to the 4CL classification of Arabidopsis and E. grandis, Pg4CLs were classified and predicted.
4.6. Analysis of Gene Structure and Protein Conserved Motifs
According to the obtained pomegranate 4CL protein sequences and gene sequences (including introns, exons and upstream and downstream sequences), the online software GSDS 2.0 (http://gsds.gao-lab.org/ (accessed on 15 February 2022)) was used to analyze its gene structure [46]. The protein conserved motifs of Pg4CLs were analyzed online by MEME (http://meme-suite.org/tools/meme (accessed on 15 February 2022)) with a maximum number of motifs of 10 [47]. The phylogenetic tree of Pg4CLs, the obtained motifs, and the gene structure, were visualized using TBtools (Guangdong, China) [48].
4.7. Analysis of Cis-Acting Elements and Protein–Protein Interaction Networks
The promoter region contains conserved sequences required for specific binding and transcription initiation of RNA polymerase, which is generally located 1500–2000 bp upstream of the gene. The 2000 bp upstream base sequence of Pg4CL genes were extracted from the pomegranate genome database using TBtools [48]. The promoter characteristics and cis-acting elements of Pg4CLs were analyzed by the online website PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 15 February 2022)) [49], and the predicted results were sorted and simplified. Finally, TBtools was used to visualize cis-acting elements. The co-expressions of Pg4CLs, Pg4CL5, and Pg4CL2 in the pomegranate were analyzed (A. thaliana was selected as the model species) by using String (https://string-db.org (accessed on 15 February 2022)) [50].
4.8. RNA-Seq Analysis of 4CL Gene Family in Pomegranate
RNA-Seq data of six pomegranate varieties (‘Dabenzi’, ‘Tunisia’, ‘Baiyushizi’, ‘Wonderful’, ‘Nana’, and ‘Black127’) were downloaded from the NCBI database, including root, leaf, flower, pericarp, exocarp, peel, and mixed samples of roots, leaf, flower, and fruit, as shown in Table 3. The TPM value was calculated by Kallisto v0.44.0 (CA, USA) [51], and different transcriptome data were homogenized by Log2 (TPM + 1), and then the expression heat map was drawn by TBtools [48].

Table 3.
RNA-Seq data of pomegranate.
4.9. Expression Patterns by Quantitative Real-Time PCR (qRT-PCR)
We collected samples of seven stages S1, S2, S3, S4, S5, S6, and S7 of fruit development in the pomegranate orchard of Baimashi Village, Tai’an City, Shandong Province. The specific dates of the seven stages were 15 July, 28 July, 10 August, 23 August, 5 September, 18 September, and 1 October. RNA was extracted from the exocarp of the samples, and cDNA was further obtained. Specific primers of Pg4CL2 and Pg4CL5 were designed (Table 4). The pomegranate PgActin was used as the internal reference gene. There were three biological replicates for each treatment. The qRT-PCR reaction system was 20 μL, including 10 μL Hieff® qPCR SYBR Green Master Mix (Low Rox Plus) (Yeasen Biotechnology Co.,Ltd., Shanghai, China), 0.4 μL upstream and downstream primers, 2 μL cDNA template, and 7.2 μL ddH2O. The amplification procedures were 95 °C for 5 min; 95 °C for 15 s, 58 °C for 45 s, a total of 40 cycles. The relative expression level was analyzed by 2−ΔΔCT method. The data were analyzed and plotted using SPSS 23.0 (Santa Clara, CA, USA) and Origin 2018 software (Newton, MA, USA), respectively.

Table 4.
Primers used in this study.
5. Conclusions
In this study, 12 Pg4CLs were identified in the pomegranate genome, all of which contained AMP binding domains. According to the established 4CL classification of A. thaliana and E. grandis, Pg4CLs were divided into three subfamilies, I, II, and 4CL-like. Each subfamily had 1, 1, and 10 members, respectively. Box I and box II were highly conserved in class I and II members and relatively conserved in 4CL-like members. Among them, 4CL class I gene Pg4CL5 was involved in lignin synthesis, and class II Pg4CL2 was related to flavonoid synthesis. The results of gene cloning corrected the sequence information of Pg4CL2 in the ‘Taishanhong’ genome and laid a foundation for its functional research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23073509/s1.
Author Contributions
Conceptualization, Y.W., L.G. and Z.Y.; experimental operation—Y.W. and L.G.; writing—original draft preparation, Y.W. and L.G.; writing—review and editing, Y.W., L.G., Y.Z. and X.Z.; project administration, Z.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Initiative Project for Talents of Nanjing Forestry University [GXL2014070, GXL2018032], the Priority Academic Program Development of Jiangsu High Education Institutions (PAPD), the Natural Science Foundation of Jiangsu Province (BK20180768), China Scholarship Council (CSC, 202108320304). These funding bodies took part in the design of the study and collection, analysis, and interpretation of data, and the writing of the manuscript, as well as in the open access payment.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author and the public pomegranate transcriptomes presented in this study are available in the insert article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sun, Y.; Qiao, L.; Shen, Y.; Jiang, P.; Chen, J.; Ye, X. Phytochemical Profile and Antioxidant Activity of Physiological Drop of Citrus Fruits. J. Food Sci. 2013, 78, C37–C42. [Google Scholar] [CrossRef] [PubMed]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four isoforms of Arabidopsis thaliana 4-coumarate: CoA ligase (4CL) have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamberger, B.; Hahlbrock, K. The 4-coumarate: CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Li, M.; Li, D.; Zhang, J.; Jin, Q.; Sheng, L.; Cai, Y.; Lin, Y. Correction: Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open 2020, 9, 9. [Google Scholar] [CrossRef]
- Dixon, R.A.; Steele, C.L. Flavonoids and isoflavonoids—A gold mine for metabolic engineering. Trends Plant Sci. 1999, 4, 394–400. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-Wide Characterization of the Lignification Toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [Green Version]
- Gui, J.; Shen, J.; Li, L. Functional Characterization of Evolutionarily Divergent 4-Coumarate:Coenzyme A Ligases in Rice. Plant Physiol. 2011, 157, 574–586. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.-C.; Xiong, X.-P.; Zhang, X.-L.; Feng, H.-J.; Zhu, Q.-H.; Sun, J.; Li, Y.-J. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. BMC Plant Biol. 2020, 20, 125. [Google Scholar] [CrossRef]
- Carocha, V.; Soler, M.; Hefer, C.; Cassan-Wang, H.; Fevereiro, P.; Myburg, A.A.; Paiva, J.A.; Grima-Pettenati, J. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytol. 2015, 206, 1297–1313. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Li, X.; Jiang, L. Integrative Analysis of the Core Fruit Lignification Toolbox in Pear Reveals Targets for Fruit Quality Bioengineering. Biomology 2019, 9, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, K.; Hövel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.-P. The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavhale, S.G.; Kalunke, R.M.; Giri, A.P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 2018, 248, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Hamberger, B.; Ellis, M.; Friedmann, M.; de Azevedo Souza, C.; Barbazuk, B.; Douglas, C.J. Genome-wide analyses of phe-nylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and con-servation and diversification of angiosperm gene families. Botany 2007, 85, 1182–1201. [Google Scholar]
- Cukovica, D.; Ehlting, J.; Van Ziffle, J.A.; Douglas, C.J. Structure and Evolution of 4-Coumarate:Coenzyme A Ligase (4CL) Gene Families. Biol. Chem. 2001, 382, 76. [Google Scholar] [CrossRef]
- Wang, B.; Sun, W.; Li, Q.; Li, Y.; Luo, H.; Song, J.; Sun, C.; Qian, J.; Zhu, Y.; Hayward, A.; et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 2014, 241, 711–725. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Li, X.; Ma, K.; Zhan, Y.; Zeng, F. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4 (4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biol. 2019, 19, 231. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Liu, C.; Li, J.; Qi, Y.; Gao, Z.; Zhang, X.; Yi, X.; Pan, H.; Ming, R.; Xu, Y. Diversity of metabolite accumulation patterns in inner and outer seed coats of pomegranate: Exploring their relationship with genetic mechanisms of seed coat development. Hortic. Res. 2020, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Saminathan, T.; Bodunrin, A.; Singh, N.V.; Devarajan, R.; Nimmakayala, P.; Jeff, M.; Aradhya, M.; Reddy, U.K. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 2016, 16, 122. [Google Scholar] [CrossRef] [Green Version]
- Pirzadeh, M.; Caporaso, N.; Rauf, A.; Shariati, M.A.; Yessimbekov, Z.; Khan, M.U.; Imran, M.; Mubarak, M.S. Pomegranate as a source of bioactive constituents: A review on their characterization, properties and applications. Crit. Rev. Food Sci. Nutr. 2021, 61, 982–999. [Google Scholar] [CrossRef]
- Patel, C.; Dadhaniya, P.; Hingorani, L.; Soni, M. Safety assessment of pomegranate fruit extract: Acute and subchronic toxicity studies. Food Chem. Toxicol. 2008, 46, 2728–2735. [Google Scholar] [CrossRef] [PubMed]
- Kandylis, P.; Kokkinomagoulos, E. Food Applications and Potential Health Benefits of Pomegranate and its Derivatives. Foods 2020, 9, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, S.; Ismail, T.; Fraternale, D.; Sestili, P. Pomegranate peel and peel extracts: Chemistry and food features. Food Chem. 2015, 174, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2017, 16, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.-H.; Ma, T.; Luo, W.-C.; Xu, J.-M.; Liu, J.-Q.; Wan, D.-S. Identification of 4CL Genes in Desert Poplars and Their Changes in Expression in Response to Salt Stress. Genes 2015, 6, 901–917. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Geng, Z.; Zhang, C.; Wang, K.; Jiang, X. Whole-genome characterization of Rosa chinensis AP2/ERF transcription factors and analysis of negative regulator RcDREB2B in Arabidopsis. BMC Genom. 2021, 22, 90. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear. Genes 2016, 7, 89. [Google Scholar] [CrossRef] [Green Version]
- Hong, P.; Xu, X.; Wang, J.; Chen, X.; Shen, X.; Lin, Y.; Lai, Z. Identification and functional analysis of 4CL gene family during early somatic embryogenesis in Dimocarpus longan Lour. Chin. J. Trop. Crops 2021, 42, 909–919. (In Chinese) [Google Scholar]
- Dong, L.; Abdurazak, I.; Hu, Q.; Li, C.; Zhang, S. Cloning and Expression Analysis of Pomegranate 4-coumarate-CoA Ligase Pg4CL. Acta Bot. Sin. 2016, 36, 2146–2151. (In Chinese) [Google Scholar]
- Luo, X.; Cao, D.; Li, H.; Zhao, D.; Xue, H.; Niu, J.; Chen, L.; Zhang, F.; Cao, S. Complementary iTRAQ-based proteomic and RNA sequencing-based transcriptomic analyses reveal a complex network regulating pomegranate (Punica granatum L.) fruit peel colour. Sci. Rep.-UK 2018, 8, 12362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldi, P.; Moser, M.; Brilli, M.; Pindo, M.; Si-Ammour, A.; Vrhovsek, U. Fine-tuning of the flavonoid and monolignol pathways during apple early fruit development. Planta 2017, 245, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Yu, J.; Cai, Y.-X.; Zhu, P.-P.; Liu, C.-Y.; Zhao, A.-C.; Lü, R.-H.; Li, M.-J.; Xu, F.-X.; Yu, M.-D. Characterization and functional analysis of 4-coumarate: CoA ligase genes in mulberry. PLoS ONE 2016, 11, e0155814. [Google Scholar]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 2016, 32, 1933–1942. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Ono, N.N.; Britton, M.T.; Fass, J.N.; Nicolet, C.M.; Lin, D.; Tian, L. Exploring the Transcriptome Landscape of Pomegranate Fruit Peel for Natural Product Biosynthetic Gene and SSR Marker Discovery F. J. Integr. Plant Biol. 2011, 53, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Ophir, R.; Sherman, A.; Rubinstein, M.; Eshed, R.; Schwager, M.S.; Harel-Beja, R.; Bar-Ya’Akov, I.; Holland, D. Single-Nucleotide Polymorphism Markers from De-Novo Assembly of the Pomegranate Transcriptome Reveal Germplasm Genetic Diversity. PLoS ONE 2014, 9, e88998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).