Machine-Learning Analysis of Serum Proteomics in Neuropathic Pain after Nerve Injury in Breast Cancer Surgery Points at Chemokine Signaling via SIRT2 Regulation
Abstract
:1. Introduction
2. Results
2.1. Participants and Descriptive Data
| Protein | Baseline | Post-OP | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-PPSNP | PPSNP | Non-PPSNP | PPSNP | ||||||||||
| Mean and SD | Range | Mean and SD | Range | Wilcoxon P | Mean and SD | Range | Mean and SD | Range | Wilcoxon P | ||||
| Demographics | |||||||||||||
| Age | 57.43 ± 7.84 | 33–68 | 53.85 ± 6.06 | 42–65 | 0.01941 | 64.03 ± 7.49 | 41–74 | 60.33 ± 5.84 | 48–71 | 0.01461 | |||
| BMI | 23.82 ± 3.52 | 17.8–30.8 | 25.22 ± 4.34 | 18.6–34.9 | 0.24 | 23.58 ± 3.72 | 16.8–30.12 | 25.97 ± 4.2 | 19.72–37.34 | 0.05718 | |||
| Proteins | |||||||||||||
| Variable name | Standard names | NCBI | UNIPROT | ||||||||||
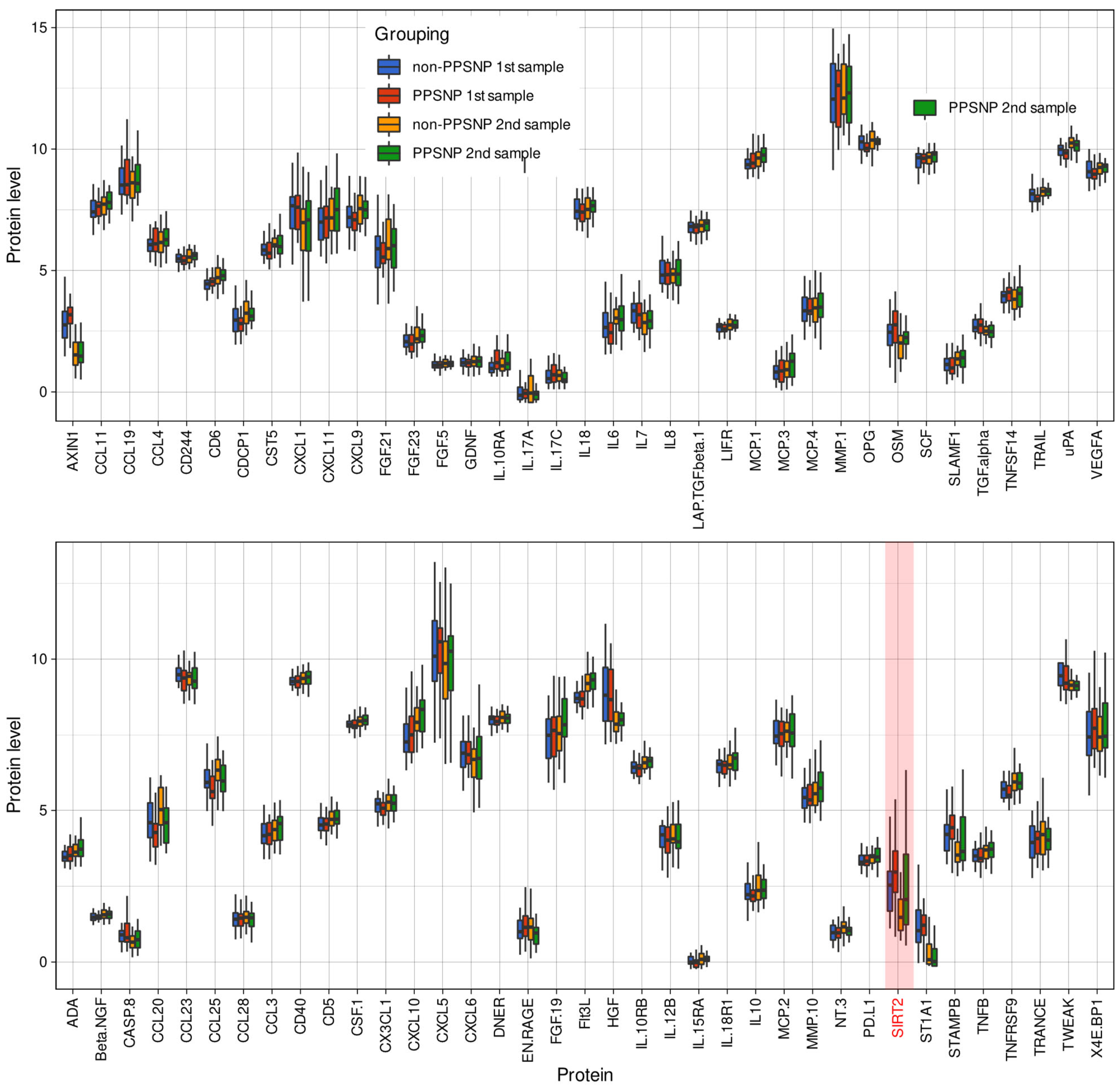
| ADA | ADA | 100 | A0A0S2Z381 | 3.55 ± 0.41 | 3.09–5.1 | 3.64 ± 0.5 | 3.05–5.39 | 0.5307 | 3.71 ± 0.48 | 2.84–5.66 | 3.88 ± 0.65 | 3.14–6.03 | 0.5412 |
| AXIN1 | AXIN1 | 8312 | A0A0S2Z4R0 | 2.86 ± 0.86 | 1.47–4.75 | 3.08 ± 0.57 | 1.62–4.05 | 0.1122 | 1.75 ± 0.99 | 0.55–4.29 | 1.84 ± 1.09 | 0.51–4.51 | 0.7936 |
| Beta.NGF | NGF | 841 | A0A024R3Z8 | 1.5 ± 0.19 | 1.22–2.12 | 1.58 ± 0.39 | 1.3–3.06 | 0.8803 | 1.62 ± 0.27 | 1.23–2.22 | 1.6 ± 0.3 | 1.24–2.88 | 0.7331 |
| CASP.8 | CASP8 | 6356 | P51671 | 0.93 ± 0.41 | 0.32–2.24 | 1 ± 0.58 | 0.34–2.55 | 0.8553 | 0.8 ± 0.55 | 0.16–2.99 | 0.98 ± 0.81 | 0.2–3.57 | 0.5951 |
| CCL11 | CCL11 | 6357 | Q99616 | 7.49 ± 0.48 | 6.46–8.56 | 7.58 ± 0.44 | 6.9–8.41 | 0.5951 | 7.7 ± 0.52 | 6.66–8.73 | 7.85 ± 0.42 | 6.93–8.52 | 0.3051 |
| CCL19 | CCL19 | 6363 | Q6IBD6 | 8.72 ± 0.85 | 7.3–11.21 | 8.91 ± 1.01 | 7.72–11.23 | 0.7936 | 8.54 ± 0.71 | 7.02–9.7 | 8.93 ± 1.06 | 7.67–12.26 | 0.2621 |
| CCL20 | CCL20 | 6364 | P78556 | 4.71 ± 0.91 | 3.31–7.57 | 4.44 ± 0.92 | 3.2–6.76 | 0.1584 | 5.13 ± 0.89 | 3.83–7.97 | 4.84 ± 1.3 | 3.58–9.29 | 0.04785 |
| CCL23 | CCL23 | 6368 | P55773 | 9.52 ± 0.33 | 9.05–10.46 | 9.34 ± 0.41 | 8.53–10.29 | 0.1017 | 9.39 ± 0.38 | 8.64–10.41 | 9.36 ± 0.44 | 8.52–10.24 | 0.6859 |
| CCL25 | CCL25 | 6370 | O15444 | 6.02 ± 0.54 | 4.98–7.22 | 5.74 ± 0.51 | 4.49–6.67 | 0.0646 | 6.33 ± 0.61 | 5.01–7.79 | 6.02 ± 0.57 | 4.97–6.99 | 0.0773 |
| CCL28 | CCL28 | 56,477 | A0N0Q3 | 1.49 ± 0.48 | 0.75–2.92 | 1.44 ± 0.34 | 0.78–2.34 | 0.9179 | 1.55 ± 0.52 | 0.91–3.63 | 1.4 ± 0.31 | 0.64–1.99 | 0.6512 |
| CCL3 | CCL3 | 6348 | A0N0R1 | 4.23 ± 0.44 | 3.39–5.19 | 4.24 ± 0.44 | 3.39–4.87 | 0.9053 | 4.35 ± 0.47 | 3.58–5.26 | 4.52 ± 0.58 | 3.55–6.11 | 0.3051 |
| CCL4 | CCL4 | 6351 | P13236 | 6.11 ± 0.46 | 5.33–7.21 | 6.24 ± 0.68 | 5.18–8.47 | 0.6512 | 6.15 ± 0.55 | 5.13–7.31 | 6.38 ± 0.69 | 5.29–8.45 | 0.2369 |
| CD244 | CD244 | 6354 | P80098 | 5.49 ± 0.24 | 4.94–5.9 | 5.4 ± 0.28 | 4.72–5.99 | 0.2056 | 5.58 ± 0.3 | 5.05–6.08 | 5.58 ± 0.3 | 4.92–6.19 | 0.7451 |
| CD40 | CD40 | 6355 | P80075 | 9.33 ± 0.31 | 8.69–10.44 | 9.3 ± 0.35 | 8.79–10.14 | 0.6061 | 9.37 ± 0.33 | 8.86–10.4 | 9.42 ± 0.4 | 8.76–10.66 | 0.6285 |
| CD5 | CD5 | 51,744 | Q9BZW8 | 4.54 ± 0.3 | 4.05–5.25 | 4.53 ± 0.32 | 3.84–5.12 | 0.981 | 4.73 ± 0.32 | 4.2–5.46 | 4.74 ± 0.31 | 4.07–5.29 | 0.8429 |
| CD6 | CD6 | 29,126 | Q0GN75 | 4.4 ± 0.31 | 3.75–5.1 | 4.5 ± 0.41 | 3.34–5.13 | 0.269 | 4.73 ± 0.45 | 3.83–5.64 | 4.8 ± 0.4 | 4.03–5.7 | 0.8305 |
| CDCP1 | CDCP1 | 958 | A0A0S2Z3C7 | 2.99 ± 0.6 | 1.95–4.39 | 2.79 ± 0.4 | 1.96–3.54 | 0.2422 | 3.33 ± 0.62 | 2.33–4.61 | 3.31 ± 0.63 | 2.58–5.6 | 0.6398 |
| CSF.1 | CSF1 | 921 | P06127 | 7.85 ± 0.17 | 7.56–8.18 | 7.82 ± 0.23 | 7.39–8.35 | 0.4899 | 7.93 ± 0.23 | 7.43–8.44 | 8.01 ± 0.2 | 7.66–8.41 | 0.2173 |
| CST5 | CST5 | 923 | P30203 | 5.93 ± 0.43 | 5.27–7.23 | 5.87 ± 0.53 | 5.04–7.37 | 0.4225 | 6.18 ± 0.38 | 5.5–7.22 | 6.1 ± 0.56 | 5.11–7.34 | 0.3693 |
| CX3CL1 | CX3CL1 | 64,866 | Q9H5V8 | 5.16 ± 0.3 | 4.46–5.66 | 5.04 ± 0.29 | 4.52–5.61 | 0.1446 | 5.26 ± 0.43 | 4.4–6.05 | 5.25 ± 0.35 | 4.6–5.83 | 0.9431 |
| CXCL1 | CXCL1 | 1435 | A0A024R0A1 | 7.4 ± 1.04 | 5.24–9.44 | 7.43 ± 1.1 | 4.52–9.86 | 0.9684 | 6.71 ± 1.27 | 3.72–9.28 | 6.74 ± 1.57 | 3.75–9.05 | 0.7212 |
| CXCL10 | CXCL10 | 1473 | P28325 | 7.43 ± 0.72 | 6.33–9.07 | 7.66 ± 0.88 | 6.56–9.92 | 0.3954 | 8.05 ± 0.6 | 6.92–9.57 | 8.27 ± 0.84 | 7.05–9.81 | 0.3779 |
| CXCL11 | CXCL11 | 1473 | P28325 | 6.98 ± 0.94 | 5.57–8.73 | 7.1 ± 0.88 | 5.29–9.32 | 0.5732 | 7.22 ± 0.78 | 5.66–8.45 | 7.48 ± 1.09 | 5.69–9.82 | 0.3363 |
| CXCL5 | CXCL5 | 6376 | A0N0N7 | 10.26 ± 1.41 | 7.24–13.22 | 10.32 ± 1.17 | 7.39–12.56 | 0.8678 | 9.49 ± 1.91 | 4.65–13.04 | 9.67 ± 1.61 | 5.9–12.5 | 0.6627 |
| CXCL6 | CXCL6 | 2919 | P09341 | 6.98 ± 0.77 | 5.65–9.22 | 6.97 ± 0.75 | 5.37–8.5 | 0.9305 | 6.64 ± 0.77 | 4.93–8.86 | 6.8 ± 1.05 | 5.08–9.17 | 0.4701 |
| CXCL9 | CXCL9 | 3627 | A0A024RDA4 | 7.2 ± 0.69 | 5.86–8.89 | 7.04 ± 0.63 | 5.8–8.65 | 0.3693 | 7.59 ± 0.7 | 6.57–8.89 | 7.52 ± 0.6 | 6.41–8.86 | 0.7571 |
| DNER | DNER | 6373 | O14625 | 8.02 ± 0.22 | 7.46–8.44 | 7.98 ± 0.23 | 7.56–8.54 | 0.3779 | 8.1 ± 0.24 | 7.67–8.52 | 8.05 ± 0.23 | 7.58–8.46 | 0.4412 |
| EN.RAGE | S100A12 | 6374 | P42830 | 1.13 ± 0.51 | 0.24–2.83 | 1.22 ± 0.52 | 0.34–2.47 | 0.4899 | 1.24 ± 0.71 | 0.12–3.08 | 1.03 ± 0.57 | 0.3–2.74 | 0.1632 |
| FGF.19 | FGF19 | 6372 | P80162 | 7.5 ± 0.96 | 5.92–10.58 | 7.53 ± 1 | 5.68–9.46 | 0.8059 | 7.73 ± 0.97 | 6.36–10.34 | 7.94 ± 0.81 | 5.91–9.43 | 0.1359 |
| FGF.21 | FGF21 | 3576 | A0A024RDA5 | 5.93 ± 1.26 | 3.6–9.72 | 5.78 ± 0.93 | 3.87–7.72 | 0.6742 | 6.16 ± 1.12 | 3.64–8.14 | 5.87 ± 1.06 | 4.13–7.74 | 0.3363 |
| FGF.23 | FGF23 | 4283 | Q07325 | 2.17 ± 0.48 | 1.51–3.63 | 2.06 ± 0.48 | 1.37–3.47 | 0.3363 | 2.38 ± 0.57 | 1.42–4.21 | 2.39 ± 0.44 | 1.77–3.48 | 0.8305 |
| FGF.5 | FGF5 | 92,737 | Q8NFT8 | 1.13 ± 0.22 | 0.86–1.79 | 1.12 ± 0.19 | 0.67–1.47 | 0.8803 | 1.19 ± 0.21 | 0.87–1.81 | 1.16 ± 0.15 | 0.92–1.5 | 0.6976 |
| Flt3L | FLT3LG | 1978 | Q13541 | 8.73 ± 0.3 | 8–9.29 | 8.71 ± 0.38 | 8.01–9.47 | 0.8553 | 9.25 ± 0.44 | 8.36–10.25 | 9.3 ± 0.48 | 8.41–10.67 | 0.6398 |
| GDNF | GDNF | 9965 | O95750 | 1.23 ± 0.34 | 0.47–2.15 | 1.21 ± 0.3 | 0.64–2.24 | 0.7212 | 1.31 ± 0.36 | 0.64–2.14 | 1.26 ± 0.32 | 0.7–1.88 | 0.8305 |
| HGF | HGF | 26,291 | Q9NSA1 | 8.89 ± 1.05 | 7.19–11.17 | 8.77 ± 1.02 | 7.26–10.52 | 0.7212 | 7.94 ± 0.46 | 7.2–9.01 | 7.98 ± 0.33 | 7.28–8.58 | 0.3779 |
| IL.10RA | IL10RA | 8074 | Q9GZV9 | 1.35 ± 1.37 | 0.63–6.65 | 1.48 ± 0.8 | 0.63–3.99 | 0.03419 | 1.44 ± 1.39 | 0.63–6.51 | 1.47 ± 0.8 | 0.63–3.57 | 0.2843 |
| IL.10RB | IL10RB | 2250 | Q8NBG6 | 6.43 ± 0.23 | 6.04–7 | 6.36 ± 0.26 | 5.87–6.91 | 0.3444 | 6.58 ± 0.29 | 5.77–7.31 | 6.62 ± 0.27 | 6.02–7.09 | 0.6061 |
| IL.12B | IL12B | 2323 | B7ZLY4 | 4.08 ± 0.53 | 2.54–4.69 | 4.01 ± 0.63 | 2.78–5.14 | 0.4603 | 4.18 ± 0.51 | 3.12–5.28 | 4.11 ± 0.64 | 3.08–5.34 | 0.4799 |
| IL.15RA | IL15RA | 2668 | A0A0S2Z3V2 | 0.03 ± 0.15 | −0.23–0.31 | −0.03 ± 0.17 | −0.23–0.41 | 0.08132 | 0.1 ± 0.21 | −0.23–0.56 | 0.1 ± 0.17 | −0.23–0.59 | 0.8793 |
| IL.17A | IL17A | 3082 | P14210 | 0.06 ± 0.54 | −0.45–1.76 | −0.01 ± 0.36 | −0.45–0.98 | 0.6834 | 0.14 ± 0.6 | −0.45–1.37 | 0.01 ± 0.48 | −0.45–1.29 | 0.7789 |
| IL.17C | IL17C | 3586 | P22301 | 0.65 ± 0.38 | 0.11–1.5 | 0.81 ± 0.57 | 0.11–2.6 | 0.3416 | 0.72 ± 0.41 | 0.11–1.54 | 0.65 ± 0.49 | 0.11–1.91 | 0.2878 |
| IL.18R1 | IL18R1 | 3587 | Q13651 | 6.47 ± 0.32 | 5.77–7.04 | 6.44 ± 0.32 | 5.55–7.08 | 0.6512 | 6.55 ± 0.37 | 5.79–7.32 | 6.69 ± 0.41 | 6.01–7.74 | 0.3205 |
| IL10 | IL10 | 3588 | Q08334 | 2.32 ± 0.44 | 1.35–3.54 | 2.16 ± 0.4 | 1.13–2.94 | 0.3205 | 2.53 ± 0.66 | 1.64–4.76 | 2.48 ± 0.5 | 1.75–4.08 | 1 |
| IL18 | IL18 | 3593 | P29460 | 7.64 ± 1.01 | 6.64–12.33 | 7.37 ± 0.48 | 6.61–8.37 | 0.3526 | 7.53 ± 0.55 | 6.34–8.45 | 7.65 ± 0.47 | 6.78–8.62 | 0.4412 |
| IL6 | IL6 | 3601 | Q13261 | 2.98 ± 1.14 | 1.54–6.45 | 2.67 ± 0.95 | 1.57–5.25 | 0.2295 | 3.09 ± 0.67 | 1.94–4.99 | 3.17 ± 1.04 | 1.72–6.94 | 0.7692 |
| IL7 | IL7 | 3605 | Q16552 | 3.44 ± 0.76 | 2.44–5.49 | 3.2 ± 0.67 | 2.13–4.6 | 0.2903 | 2.9 ± 0.84 | 1.64–5.67 | 2.94 ± 0.64 | 1.79–4.02 | 0.6285 |
| IL8 | CXCL8 | 27,189 | Q9P0M4 | 4.95 ± 0.63 | 4.09–6.43 | 4.9 ± 0.85 | 3.84–8.27 | 0.5732 | 4.83 ± 0.59 | 3.6–6.02 | 4.94 ± 0.71 | 3.62–6.22 | 0.6627 |
| LAP.TGF.beta.1 | TGFB1 | 3606 | A0A024R3E0 | 6.82 ± 0.4 | 6.19–7.77 | 6.76 ± 0.35 | 6.06–7.65 | 0.6976 | 6.84 ± 0.43 | 5.79–7.99 | 6.9 ± 0.36 | 6.25–7.86 | 0.7094 |
| LIF.R | LIFR | 8809 | Q13478 | 2.65 ± 0.23 | 2.15–3.04 | 2.62 ± 0.22 | 2.19–3.29 | 0.3363 | 2.74 ± 0.31 | 2.14–3.22 | 2.79 ± 0.2 | 2.47–3.19 | 0.7571 |
| MCP.1 | CST5 | 3569 | B4DVM1 | 9.42 ± 0.35 | 8.76–10.04 | 9.52 ± 0.44 | 8.84–10.63 | 0.5101 | 9.63 ± 0.51 | 8.35–10.57 | 9.75 ± 0.39 | 9.06–10.63 | 0.3609 |
| MCP.2 | CCL8 | 3574 | A8K673 | 7.53 ± 0.63 | 5.57–8.67 | 7.52 ± 0.57 | 6.12–8.41 | 0.8928 | 7.62 ± 0.7 | 5.74–9.13 | 7.59 ± 0.7 | 6.05–8.81 | 0.981 |
| MCP.3 | CCL7 | 4254 | A0A024RBC0 | 0.87 ± 0.51 | 0.17–2.45 | 1.02 ± 1.11 | 0.06–6.02 | 0.9053 | 0.97 ± 0.52 | 0.1–2.06 | 1.16 ± 0.58 | 0.25–2.37 | 0.2234 |
| MCP.4 | CCL13 | 3977 | A8K1Z4 | 3.46 ± 0.83 | 2.14–5.95 | 3.46 ± 0.48 | 2.69–4.61 | 0.7814 | 3.44 ± 0.73 | 2.2–5.01 | 3.56 ± 0.75 | 1.74–4.93 | 0.4318 |
| MMP.1 | MMP1 | 4049 | P01374 | 12.2 ± 1.55 | 9.13–14.97 | 12.18 ± 1.27 | 9.96–13.93 | 0.9053 | 12.32 ± 1.22 | 10.56–14.34 | 12.27 ± 1.38 | 10.16–14.73 | 0.7936 |
| MMP.10 | MMP10 | 4312 | B4DN15 | 5.62 ± 0.82 | 4.59–7.86 | 5.59 ± 0.75 | 4.57–7.55 | 0.8429 | 5.73 ± 0.7 | 4.91–7.71 | 5.8 ± 0.69 | 4.65–7.32 | 0.4701 |
| NT.3 | NTF3 | 4319 | P09238 | 1.08 ± 0.64 | 0.46–3.35 | 0.97 ± 0.25 | 0.33–1.5 | 0.9179 | 1.2 ± 0.6 | 0.25–3.77 | 1.11 ± 0.55 | 0.64–3.58 | 0.1236 |
| OPG | TNFRSF11B | 4803 | P01138 | 10.25 ± 0.4 | 9.39–11.01 | 10.12 ± 0.31 | 9.68–10.91 | 0.09834 | 10.37 ± 0.45 | 9.29–11.11 | 10.36 ± 0.3 | 9.93–11.22 | 0.6742 |
| OSM | OSM | 4908 | P20783 | 2.48 ± 0.79 | 1–4.52 | 2.64 ± 0.95 | 0.37–4.14 | 0.3127 | 1.96 ± 0.66 | 0.82–3.24 | 2.22 ± 0.73 | 0.44–3.73 | 0.1359 |
| PD.L1 | CD274 | 5008 | B5MCX1 | 3.39 ± 0.3 | 2.9–4.07 | 3.32 ± 0.27 | 2.79–3.81 | 0.9431 | 3.5 ± 0.52 | 2.65–5.51 | 3.48 ± 0.33 | 2.8–4.13 | 0.8429 |
| SCF | KITLG | 5328 | P00749 | 9.49 ± 0.48 | 8.08–10.08 | 9.53 ± 0.44 | 8.14–9.98 | 0.9557 | 9.64 ± 0.31 | 8.94–10.25 | 9.68 ± 0.32 | 8.99–10.26 | 0.4507 |
| SIRT2 | SIRT2 | 6283 | P80511 | 2.61 ± 1.18 | 1.11–6.12 | 3.12 ± 1.1 | 0.83–5.37 | 0.05166 | 1.89 ± 1.27 | 0.71–5.38 | 2.58 ± 1.9 | 0.54–8.22 | 0.1782 |
| SLAMF1 | SLAMF1 | 22,933 | A0A0A0MRF5 | 1.2 ± 0.56 | 0.32–3.31 | 1.21 ± 0.89 | 0.49–5.24 | 0.3609 | 1.41 ± 0.67 | 0.2–3.97 | 1.48 ± 0.74 | 0.34–4.32 | 0.8305 |
| ST1A1 | SULT1A1 | 6504 | Q13291 | 1.21 ± 0.82 | −0.03–3.21 | 1.21 ± 0.59 | 0–2.1 | 0.6398 | 0.39 ± 0.64 | −0.13–2.72 | 0.38 ± 0.69 | −0.13–2.31 | 0.5136 |
| STAMPB | STAMBP | 10,617 | A0A140VK54 | 4.22 ± 0.79 | 3.22–6.65 | 4.55 ± 0.73 | 2.93–6.17 | 0.1051 | 3.84 ± 0.98 | 2.83–7.12 | 4.2 ± 1.36 | 3–8.56 | 0.3693 |
| TGF.alpha | TGFA | 6817 | P50225 | 2.72 ± 0.37 | 2.17–3.94 | 2.74 ± 0.4 | 1.88–3.66 | 0.7692 | 2.57 ± 0.46 | 1.82–4.45 | 2.47 ± 0.28 | 1.81–2.91 | 0.6742 |
| TNFB | LTA | 7039 | P01135 | 3.65 ± 0.93 | 2.49–7.66 | 3.47 ± 0.41 | 2.64–4.27 | 0.7692 | 3.65 ± 0.44 | 2.59–4.65 | 3.68 ± 0.37 | 2.91–4.35 | 0.6859 |
| TNFRSF9 | TNFRSF9 | 7040 | P01137 | 5.74 ± 0.37 | 5.26–6.74 | 5.62 ± 0.38 | 5–6.49 | 0.2114 | 5.96 ± 0.42 | 5.18–7.07 | 5.95 ± 0.38 | 5.22–6.56 | 0.9557 |
| TNFSF14 | TNFSF14 | 4982 | O00300 | 3.9 ± 0.46 | 2.99–4.99 | 4.02 ± 0.49 | 2.93–4.93 | 0.2621 | 3.86 ± 0.51 | 2.94–4.77 | 3.99 ± 0.56 | 3.06–5.22 | 0.4225 |
| TRAIL | TNFSF10 | 3604 | Q07011 | 8.19 ± 0.54 | 7.39–10.41 | 7.95 ± 0.23 | 7.45–8.42 | 0.02957 | 8.35 ± 0.53 | 7.68–10.64 | 8.25 ± 0.18 | 7.95–8.61 | 0.8429 |
| TRANCE | TNFSF11 | 8743 | P50591 | 3.9 ± 0.62 | 2.77–5 | 3.97 ± 0.68 | 2.28–5.31 | 0.7212 | 4.19 ± 0.74 | 3.02–6.08 | 4.05 ± 0.53 | 3.1–5.46 | 0.4799 |
| TWEAK | TNFSF12 | 8600 | O14788 | 9.48 ± 0.49 | 8.62–10.38 | 9.41 ± 0.59 | 8.52–10.66 | 0.4999 | 9.1 ± 0.34 | 8.05–9.69 | 9.1 ± 0.3 | 8.4–9.71 | 0.9053 |
| uPA | PLAU | 8742 | O43508 | 9.95 ± 0.33 | 8.97–10.46 | 9.79 ± 0.26 | 9.23–10.28 | 0.0168 | 10.26 ± 0.38 | 9.41–10.97 | 10.14 ± 0.31 | 9.43–10.62 | 0.276 |
| VEGFA | VEGFA | 8740 | O43557 | 9.15 ± 0.47 | 8.27–10.01 | 9.01 ± 0.33 | 8.32–9.87 | 0.3205 | 9.24 ± 0.42 | 8.47–10.33 | 9.24 ± 0.41 | 8.5–10.32 | 0.8182 |
| X4E.BP1 | EIF4EBP1 | 7422 | A0A087WUD8 | 7.66 ± 1.14 | 5.5–11.07 | 8.09 ± 1.4 | 6.41–10.94 | 0.3526 | 7.73 ± 1.32 | 6.15–11.62 | 8.05 ± 1.66 | 6.08–11.69 | 0.6285 |
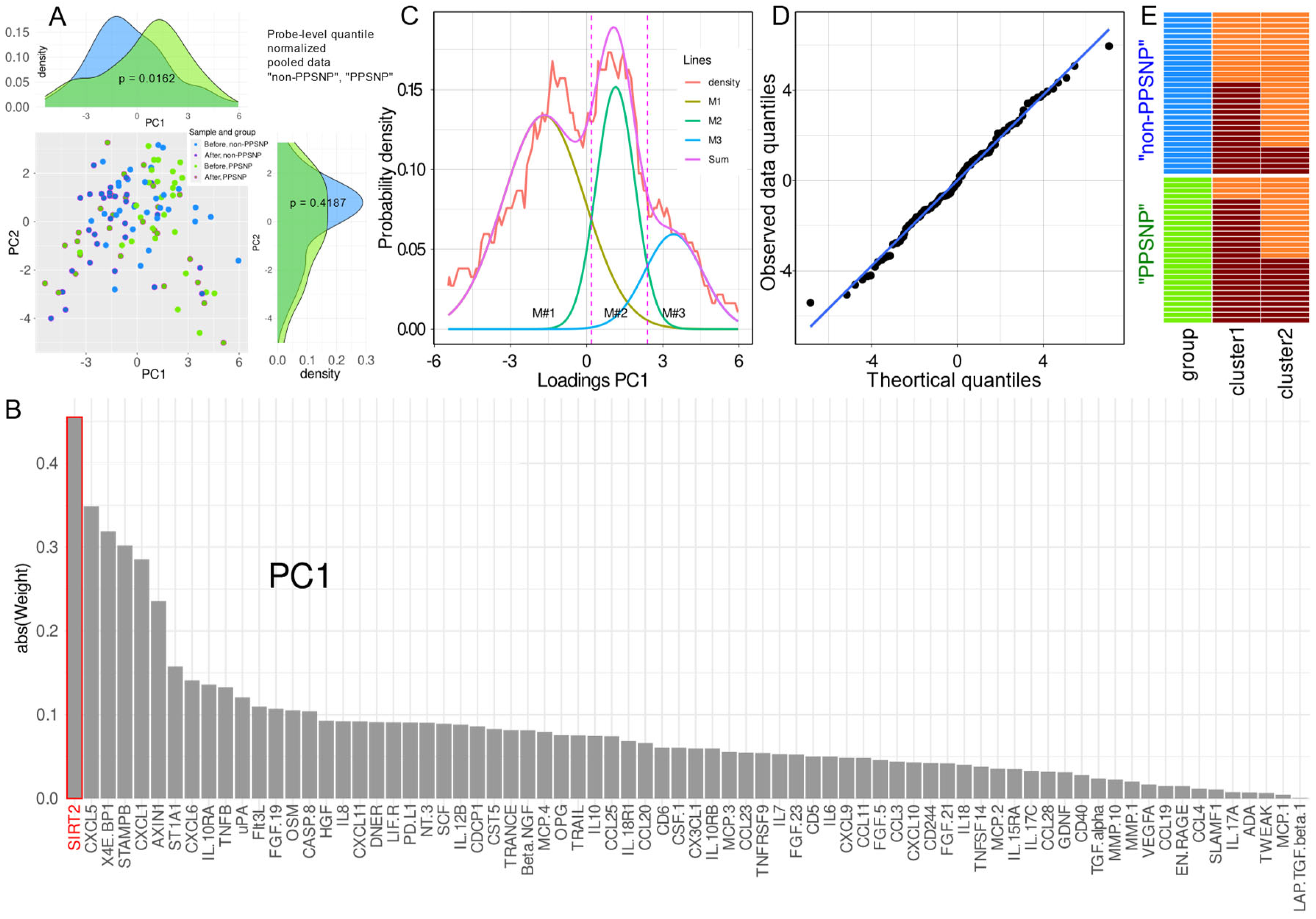
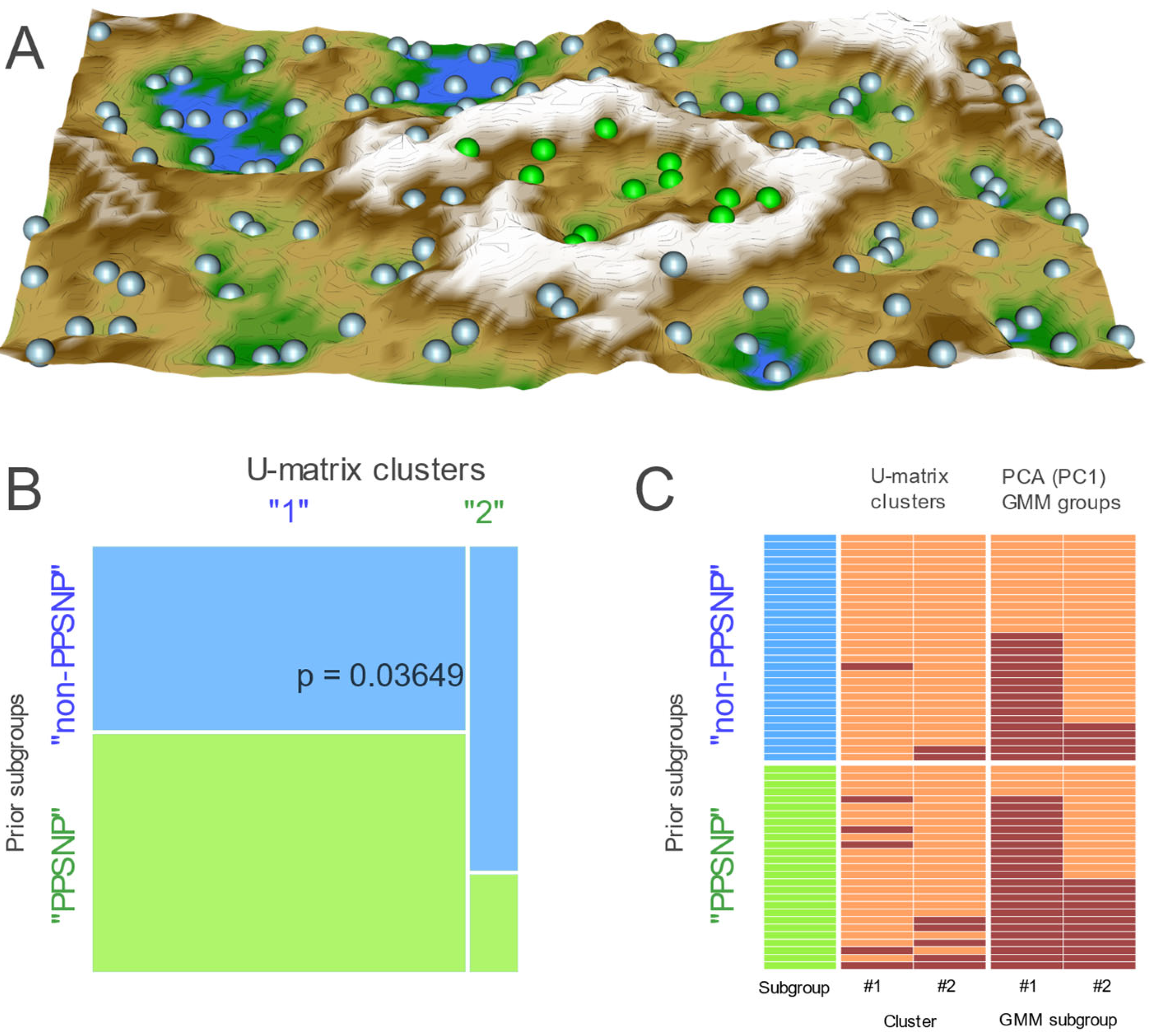
2.2. Data Projection-Based Protein Marker Patterns Relevant to Pain-Related Subgroup Separation
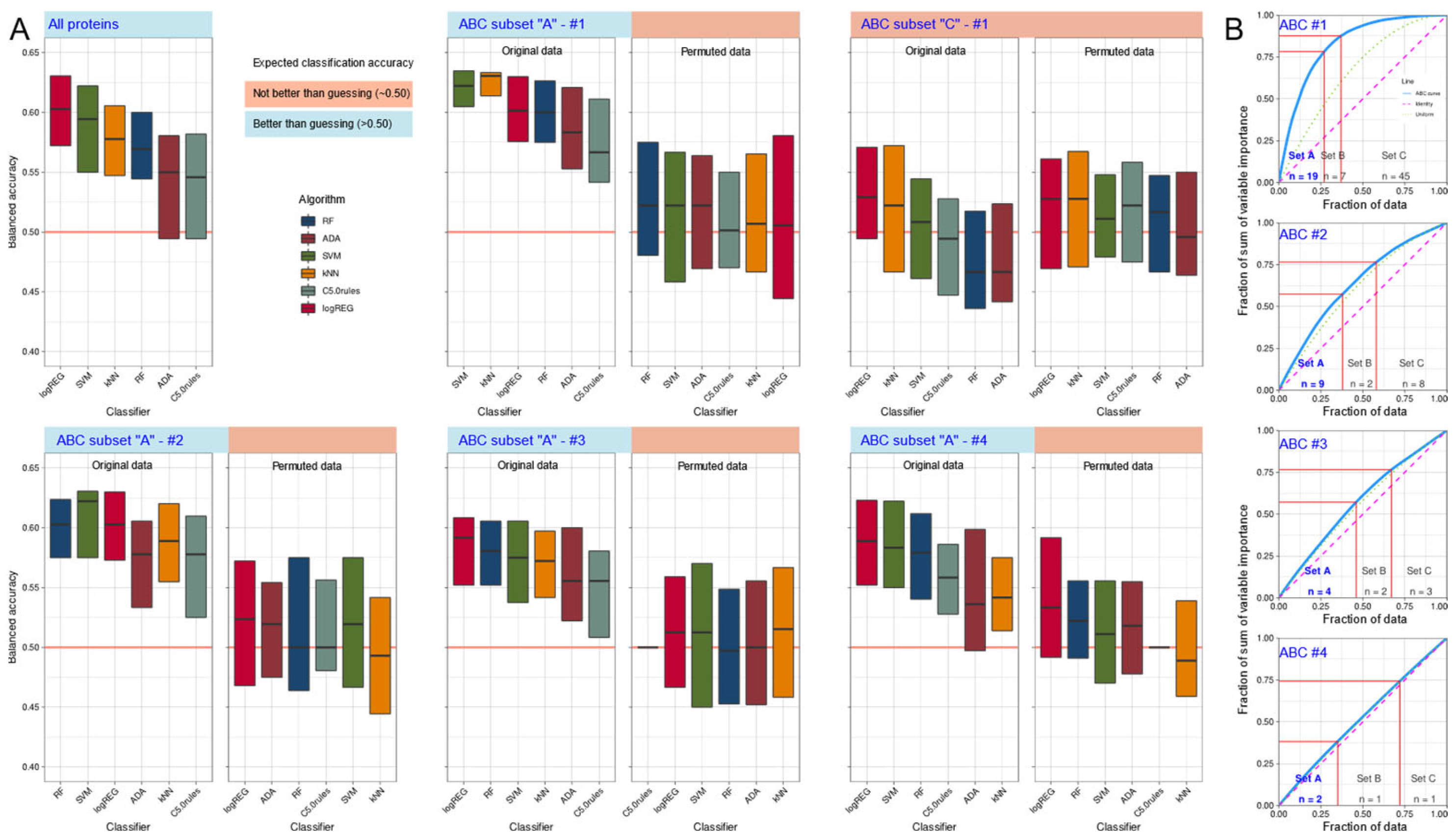
2.3. Supervised Machine Learning-Based Identification and Evaluation of Proteomic Markers Informative for Pain-Related Subgroup Segregation
3. Discussion
4. Methods
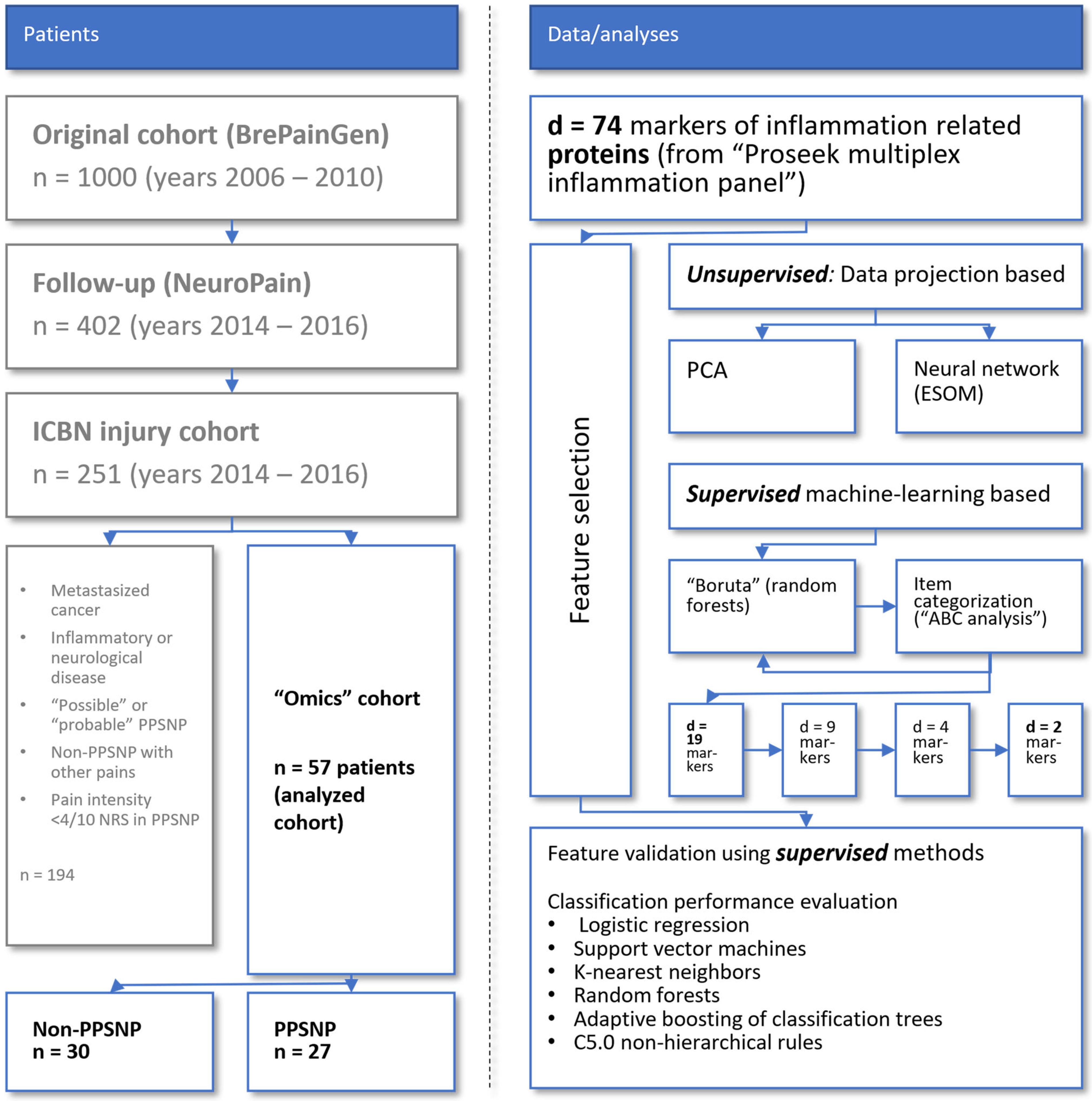
4.1. Patients and Study Design
4.2. Acquisition of Pain-Related Information
4.3. Blood Samples and Quantification of Serum Concentrations of Inflammatory Proteins
4.4. Data Analysis
4.4.1. Summary of the Concept of Data Analysis
4.4.2. Quantitative Information Analyzed
4.4.3. Data Projection-Based Assessment of Proteomics Data Structures Relevant to Pain-Related Subgroup Separation
4.4.4. Supervised Machine-Learning Based Assessment of Proteomics Data Structures Relevant to Pain-Related Subgroup Separation
4.4.5. Supervised Machine Learning-Based Evaluation of Identified Proteomic Markers to Distinguish Pain-Related Patient Subgroups
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Finnerup, N.B.; Haroutounian, S.; Kamerman, P.; Baron, R.; Bennett, D.L.H.; Bouhassira, D.; Cruccu, G.; Freeman, R.; Hansson, P.; Nurmikko, T.; et al. Neuropathic pain: An updated grading system for research and clinical practice. Pain 2016, 157, 1599–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilhan, E.; Chee, E.; Hush, J.; Moloney, N. The prevalence of neuropathic pain is high after treatment for breast cancer: A systematic review. Pain 2017, 158, 2082–2091. [Google Scholar] [CrossRef] [PubMed]
- Mustonen, L.; Aho, T.; Harno, H.; Sipila, R.; Meretoja, T.; Kalso, E. What makes surgical nerve injury painful? A 4-year to 9-year follow-up of patients with intercostobrachial nerve resection in women treated for breast cancer. Pain 2019, 160, 246–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazerani, P.; Vinterhøj, H.S.H. ‘Omics’: An emerging field in pain research and management. Future Neurol. 2016, 11, 255–265. [Google Scholar] [CrossRef]
- Calvo, M.; Davies, A.J.; Hébert, H.L.; Weir, G.A.; Chesler, E.J.; Finnerup, N.B.; Levitt, R.C.; Smith, B.H.; Neely, G.G.; Costigan, M.; et al. The Genetics of Neuropathic Pain from Model Organisms to Clinical Application. Neuron 2019, 104, 637–653. [Google Scholar] [CrossRef]
- Korczeniewska, O.A.; Katzmann Rider, G.; Gajra, S.; Narra, V.; Ramavajla, V.; Chang, Y.J.; Tao, Y.; Soteropoulos, P.; Husain, S.; Khan, J.; et al. Differential gene expression changes in the dorsal root versus trigeminal ganglia following peripheral nerve injury in rats. Eur. J. Pain 2020, 24, 967–982. [Google Scholar] [CrossRef]
- Møller Johansen, L.; Gerra, M.C.; Arendt-Nielsen, L. Time course of DNA methylation in pain conditions: From experimental models to humans. Eur. J. Pain 2021, 25, 296–312. [Google Scholar] [CrossRef]
- Bohren, Y.; Timbolschi, D.I.; Muller, A.; Barrot, M.; Yalcin, I.; Salvat, E. Platelet-rich plasma and cytokines in neuropathic pain: A narrative review and a clinical perspective. Eur. J. Pain 2021, 26, 43–60. [Google Scholar] [CrossRef]
- Teckchandani, S.; Nagana Gowda, G.A.; Raftery, D.; Curatolo, M. Metabolomics in chronic pain research. Eur. J. Pain 2021, 25, 313–326. [Google Scholar] [CrossRef]
- Niederberger, E.; Geisslinger, G. Proteomics in neuropathic pain research. Anesthesiology 2008, 108, 314–323. [Google Scholar] [CrossRef] [Green Version]
- Gerdle, B.; Ghafouri, B. Proteomic studies of common chronic pain conditions-a systematic review and associated network analyses. Expert Rev. Proteom. 2020, 17, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Gineste, C.; Ho, L.; Pompl, P.; Bianchi, M.; Pasinetti, G.M. High-throughput proteomics and protein biomarker discovery in an experimental model of inflammatory hyperalgesia: Effects of nimesulide. Drugs 2003, 63 (Suppl. S1), 23–29. [Google Scholar] [CrossRef] [PubMed]
- Sommer, C.; Leinders, M.; Üçeyler, N. Inflammation in the pathophysiology of neuropathic pain. Pain 2018, 159, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Backonja, M.M.; Coe, C.L.; Muller, D.A.; Schell, K. Altered cytokine levels in the blood and cerebrospinal fluid of chronic pain patients. J. Neuroimmunol. 2008, 195, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Bäckryd, E.; Lind, A.L.; Thulin, M.; Larsson, A.; Gerdle, B.; Gordh, T. High levels of cerebrospinal fluid chemokines point to the presence of neuroinflammation in peripheral neuropathic pain: A cross-sectional study of 2 cohorts of patients compared with healthy controls. Pain 2017, 158, 2487–2495. [Google Scholar] [CrossRef]
- Uçeyler, N.; Schäfers, M.; Sommer, C. Mode of action of cytokines on nociceptive neurons. Exp. Brain Res. 2009, 196, 67–78. [Google Scholar] [CrossRef]
- Calvo, M.; Bennett, D.L. The mechanisms of microgliosis and pain following peripheral nerve injury. Exp. Neurol. 2012, 234, 271–282. [Google Scholar] [CrossRef]
- Uçeyler, N.; Rogausch, J.P.; Toyka, K.V.; Sommer, C. Differential expression of cytokines in painful and painless neuropathies. Neurology 2007, 69, 42–49. [Google Scholar] [CrossRef]
- Kringel, D.; Lippmann, C.; Parnham, M.J.; Kalso, E.; Ultsch, A.; Lötsch, J. A machine-learned analysis of human gene polymorphisms modulating persisting pain points to major roles of neuroimmune processes. Eur. J. Pain 2018, 22, 1735–1756. [Google Scholar] [CrossRef] [Green Version]
- Kaunisto, M.A.; Jokela, R.; Tallgren, M.; Kambur, O.; Tikkanen, E.; Tasmuth, T.; Sipilä, R.; Palotie, A.; Estlander, A.-M.; Leidenius, M.; et al. Pain in 1000 women treated for breast cancer: A prospective study of pain sensitivity and postoperative pain. Anesthesiology 2013, 119, 1410–1421. [Google Scholar] [CrossRef] [Green Version]
- Klevebro, S.; Björkander, S.; Ekström, S.; Merid, S.K.; Gruzieva, O.; Mälarstig, A.; Johansson, Å.; Kull, I.; Bergström, A.; Melén, E. Inflammation-related plasma protein levels and association with adiposity measurements in young adults. Sci. Rep. 2021, 11, 11391. [Google Scholar] [CrossRef] [PubMed]
- Solheim, N.; Östlund, S.; Gordh, T.; Rosseland, L.A. Women report higher pain intensity at a lower level of inflammation after knee surgery compared with men. Pain Rep. 2017, 2, e595. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Song, M.; Ito, H.; Oze, I.; Koyanagi, Y.N.; Kasugai, Y.; Rabkin, C.S.; Matsuo, K. Associations of circulating mediators of inflammation, cell regulation and immune response with esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2021, 147, 2885–2892. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, A.M.; Stewart, R.E.; Köke, A.J.A.; Oosterwijk, R.F.A.; Swaan, J.L.; Schreurs, K.M.G.; Schiphorst Preuper, H.R. Cut-Off Points for Mild, Moderate, and Severe Pain on the Numeric Rating Scale for Pain in Patients with Chronic Musculoskeletal Pain: Variability and Influence of Sex and Catastrophizing. Front. Psychol. 2016, 7, 1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerbershagen, H.J.; Rothaug, J.; Kalkman, C.J.; Meissner, W. Determination of moderate-to-severe postoperative pain on the numeric rating scale: A cut-off point analysis applying four different methods. Br. J. Anaesth. 2011, 107, 619–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual comparisons by ranking methods. Biometrics 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2011, 39, D52–D57. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Gentleman, R. Annotate. Annotation for Microarrays. Available online: https://www.bioconductor.org/packages/annotate/ (accessed on 14 March 2022).
- Carlson, M. org.Hs.eg.db: Genome Wide Annotation for Human. Available online: https://bioconductor.org/packages/org.Hs.eg.db/ (accessed on 14 March 2022).
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Ciucci, S.; Ge, Y.; Duran, C.; Palladini, A.; Jimenez-Jimenez, V.; Martinez-Sanchez, L.M.; Wang, Y.; Sales, S.; Shevchenko, A.; Poser, S.W.; et al. Enlightening discriminative network functional modules behind Principal Component Analysis separation in differential-omic science studies. Sci. Rep. 2017, 7, 43946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ultsch, A. Pareto Density Estimation: A Density Estimation for Knowledge Discovery. In Proceedings of the Innovations in Classification, Data Science, and Information Systems-Proceedings 27th Annual Conference of the German Classification Society (GfKL), Technical University Cottbus, Cottbus, Germany, 12–14 March 2003. [Google Scholar]
- Fisher, R.A. On the Interpretation of Chi Square from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Available online: https://CRAN.R-project.org/ (accessed on 14 March 2022).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Ultsch, A.; Lötsch, J. Machine-learned cluster identification in high-dimensional data. J. Biomed. Inform. 2017, 66, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Lötsch, J.; Ultsch, A. Exploiting the structures of the U-matrix. In Advances in Intelligent Systems and Computing; Villmann, T., Schleif, F.-M., Kaden, M., Lange, M., Eds.; Springer: Heidelberg, Germany, 2014; Volume 295, pp. 248–257. [Google Scholar]
- Jeppson, H.; Hofmann, H.; Cook, D. Ggmosaic: Mosaic Plots in the ‘ggplot2′ Framework. Available online: https://cran.r-project.org/package=ggmosaic (accessed on 14 March 2022).
- Lötsch, J.; Lerch, F.; Djaldetti, R.; Tegeder, I.; Ultsch, A. Identification of disease-distinct complex biomarker patterns by means of unsupervised machine-learning using an interactive R toolbox (Umatrix). BMC Big Data Anal. 2018, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Tillé, Y.; Matei, A. Sampling: Survey Sampling. 2016. Available online: https://cran.r-project.org/package=ABCanalysis (accessed on 14 March 2022).
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 13. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. A power primer. Psych. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef]
- Lötsch, J.; Kringel, D.; Ultsch, A. Explainable artificial intelligence (XAI) in biomedicine. Mak-ing AI decisions trust-worthy for physicians and patients. BioMedInformatics 2022, 2, 1–17. [Google Scholar] [CrossRef]
- Datta, A.; Matlock, M.K.; Le Dang, N.; Moulin, T.; Woeltje, K.F.; Yanik, E.L.; Joshua Swamidass, S. ‘Black Box’ to ‘Conversational’ Machine Learning: Ondansetron Reduces Risk of Hospital-Acquired Venous Thromboembolism. IEEE J. Biomed. Health Inform. 2021, 25, 2204–2214. [Google Scholar] [CrossRef]
- Maxwell, M.M.; Tomkinson, E.M.; Nobles, J.; Wizeman, J.W.; Amore, A.M.; Quinti, L.; Chopra, V.; Hersch, S.M.; Kazantsev, A.G. The Sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Hum. Mol. Genet. 2011, 20, 3986–3996. [Google Scholar] [CrossRef]
- Werner, H.B.; Kuhlmann, K.; Shen, S.; Uecker, M.; Schardt, A.; Dimova, K.; Orfaniotou, F.; Dhaunchak, A.; Brinkmann, B.G.; Möbius, W.; et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J. Neurosci. 2007, 27, 7717–7730. [Google Scholar] [CrossRef]
- Rothgiesser, K.M.; Erener, S.; Waibel, S.; Lüscher, B.; Hottiger, M.O. SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci. 2010, 123, 4251–4258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.; Jung, Y.J.; Kim, D.; Nguyen-Thanh, T.; Kang, K.P.; Lee, S.; Park, S.K.; Kim, W. SIRT2 ameliorates lipopolysaccharide-induced inflammation in macrophages. Biochem. Biophys. Res. Commun. 2014, 450, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.A.; Ma, X.J.; Huang, S.B.; Hao, X.R.; Li, D.M.; Feng, K.Y.; Wang, W.M. SIRT2 inhibits oxidative stress and inflammatory response in diabetic osteoarthritis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2855–2864. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Wang, X.; Fang, N.; Xu, A.; Lin, Y.; Zhao, X.; Nazarali, A.J.; Ji, S. SIRT2 suppresses expression of inflammatory factors via Hsp90-glucocorticoid receptor signalling. J. Cell Mol. Med. 2020, 24, 7439–7450. [Google Scholar] [CrossRef] [PubMed]
- Pais, T.F.; Szegő, É.M.; Marques, O.; Miller-Fleming, L.; Antas, P.; Guerreiro, P.; de Oliveira, R.M.; Kasapoglu, B.; Outeiro, T.F. The NAD-dependent deacetylase sirtuin 2 is a suppressor of microglial activation and brain inflammation. EMBO J. 2013, 32, 2603–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Sandoval, A.; Nutile-McMenemy, N.; DeLeo, J.A. Spinal microglial and perivascular cell cannabinoid receptor type 2 activation reduces behavioral hypersensitivity without tolerance after peripheral nerve injury. Anesthesiology 2008, 108, 722–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taves, S.; Berta, T.; Chen, G.; Ji, R.-R. Microglia and spinal cord synaptic plasticity in persistent pain. Neural Plast. 2013, 2013, 753656. [Google Scholar] [CrossRef]
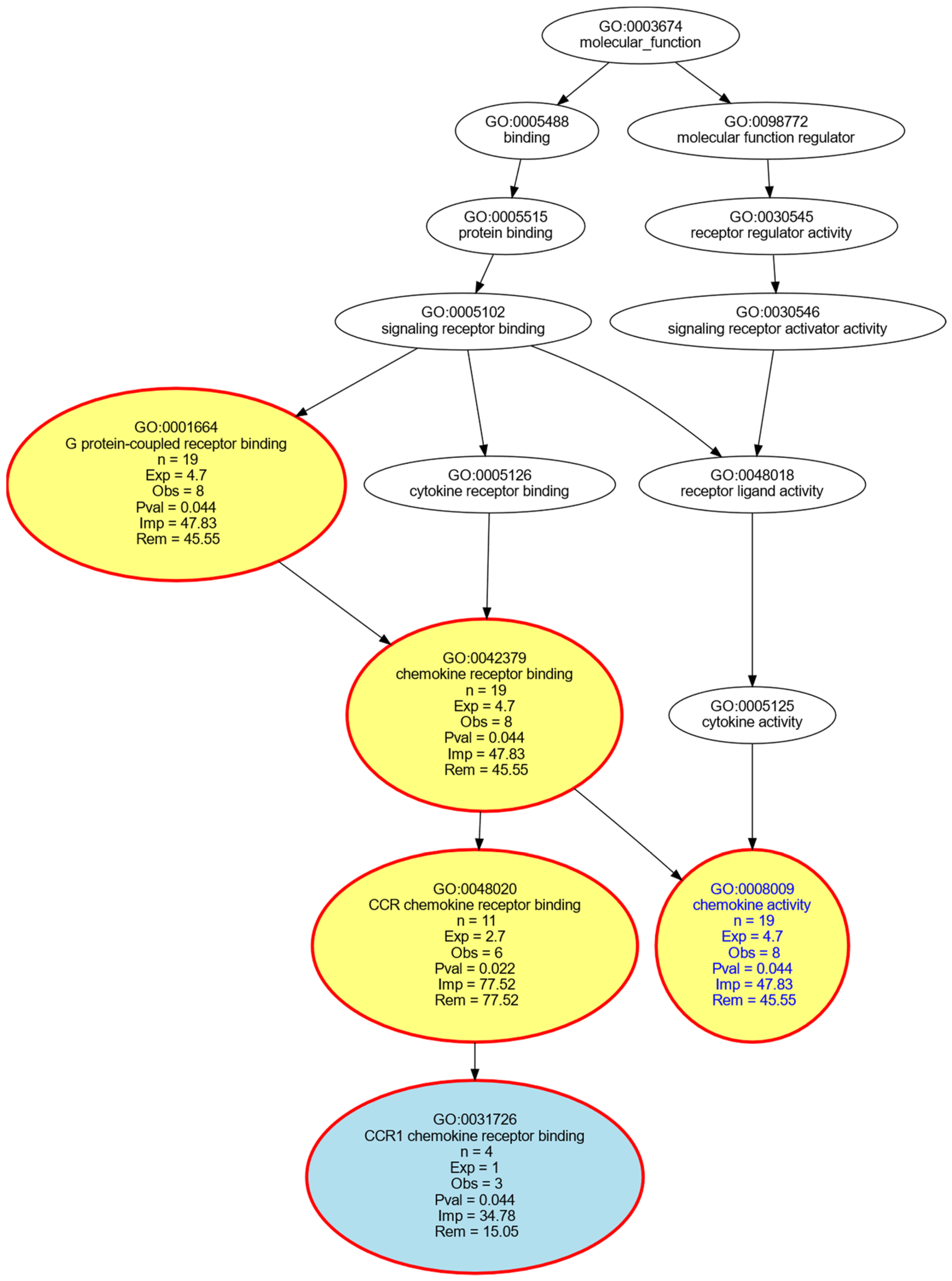
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Ultsch, A.; Kringel, D.; Kalso, E.; Mogil, J.S.; Lötsch, J. A data science approach to candidate gene selection of pain regarded as a process of learning and neural plasticity. Pain 2016, 157, 2747–2757. [Google Scholar] [CrossRef]
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front. Pharmacol. 2019, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-H.; Zhu, Y.; Ozden, O.; Kim, H.-S.; Jiang, H.; Deng, C.-X.; Gius, D.; Vassilopoulos, A. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl. Cancer Res. 2012, 1, 15–21. [Google Scholar] [PubMed]
- Kozako, T.; Mellini, P.; Ohsugi, T.; Aikawa, A.; Uchida, Y.-i.; Honda, S.-i.; Suzuki, T. Novel small molecule SIRT2 inhibitors induce cell death in leukemic cell lines. BMC Cancer 2018, 18, 791. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, L.M.; Zino, S.; MacDonald, A.I.; Curle, J.; Reilly, J.E.; Mohammed, Z.M.; McMillan, D.C.; Mallon, E.; Payne, A.P.; Edwards, J.; et al. SIRT2: Tumour suppressor or tumour promoter in operable breast cancer? Eur. J. Cancer 2014, 50, 290–301. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, R.M.; Sarkander, J.; Kazantsev, A.G.; Outeiro, T.F. SIRT2 as a Therapeutic Target for Age-Related Disorders. Front. Pharmacol. 2012, 3, 82. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Huang, P.; Hu, C. The role of SIRT2 in cancer: A novel therapeutic target. Int. J. Cancer 2020, 147, 3297–3304. [Google Scholar] [CrossRef]
- Zhang, Y.; Chi, D. Overexpression of SIRT2 Alleviates Neuropathic Pain and Neuroinflammation Through Deacetylation of Transcription Factor Nuclear Factor-Kappa B. Inflammation 2018, 41, 569–578. [Google Scholar] [CrossRef]
- Palada, V.; Ahmed, A.S.; Freyhult, E.; Hugo, A.; Kultima, K.; Svensson, C.I.; Kosek, E. Elevated inflammatory proteins in cerebrospinal fluid from patients with painful knee osteoarthritis are associated with reduced symptom severity. J. Neuroimmunol. 2020, 349, 577391. [Google Scholar] [CrossRef]
- Almeida-Souza, L.; Timmerman, V.; Janssens, S. Microtubule dynamics in the peripheral nervous system: A matter of balance. Bioarchitecture 2011, 1, 267–270. [Google Scholar] [CrossRef] [Green Version]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- D’Ydewalle, C.; Krishnan, J.; Chiheb, D.M.; Van Damme, P.; Irobi, J.; Kozikowski, A.P.; Vanden Berghe, P.; Timmerman, V.; Robberecht, W.; Van Den Bosch, L. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat. Med. 2011, 17, 968–974. [Google Scholar] [CrossRef]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaidya, S.V.; Mathew, P.A. Of Mice and Men: Different Functions of the Murine and Human 2B4 (CD244) Receptor on NK Cells. Immunol. Lett. 2006, 105, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Agresta, L.; Hoebe, K.H.N.; Janssen, E.M. The Emerging Role of CD244 Signaling in Immune Cells of the Tumor Microenvironment. Front. Immunol. 2018, 9, 2809. [Google Scholar] [CrossRef] [PubMed]
- Maleki, F.; Ovens, K.; Hogan, D.J.; Kusalik, A.J. Gene Set Analysis: Challenges, Opportunities, and Future Research. Front. Genet. 2020, 11, 654. [Google Scholar] [CrossRef]
- Camon, E.; Magrane, M.; Barrell, D.; Lee, V.; Dimmer, E.; Maslen, J.; Binns, D.; Harte, N.; Lopez, R.; Apweiler, R. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004, 32, D262–D266. [Google Scholar] [CrossRef] [Green Version]
- Camon, E.; Magrane, M.; Barrell, D.; Binns, D.; Fleischmann, W.; Kersey, P.; Mulder, N.; Oinn, T.; Maslen, J.; Cox, A.; et al. The Gene Ontology Annotation (GOA) project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 2003, 13, 662–672. [Google Scholar] [CrossRef] [Green Version]
- Thulasiraman, K.; Swamy, M.N.S. Graphs: Theory and Algorithms; Wiley: New York, NY, USA, 1992; p. XV, 460 S. [Google Scholar]
- Kringel, D.; Malkusch, S.; Lötsch, J. Drugs and Epigenetic Molecular Functions. A Pharmacological Data Scientometric Analysis. Int. J. Mol. Sci. 2021, 22, 7250. [Google Scholar] [CrossRef]
- Lippmann, C.; Kringel, D.; Ultsch, A.; Lotsch, J. Computational functional genomics-based approaches in analgesic drug discovery and repurposing. Pharmacogenomics 2018, 19, 783–797. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M. GO.db: A Set of Annotation Maps Describing the Entire Gene Ontology. Available online: https://bioconductor.org/packages/release/data/annotation/html/GO.db.html (accessed on 14 March 2022).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate-a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ultsch, A.; Lötsch, J. Functional abstraction as a method to discover knowledge in gene ontologies. PLoS ONE 2014, 9, e90191. [Google Scholar] [CrossRef] [Green Version]
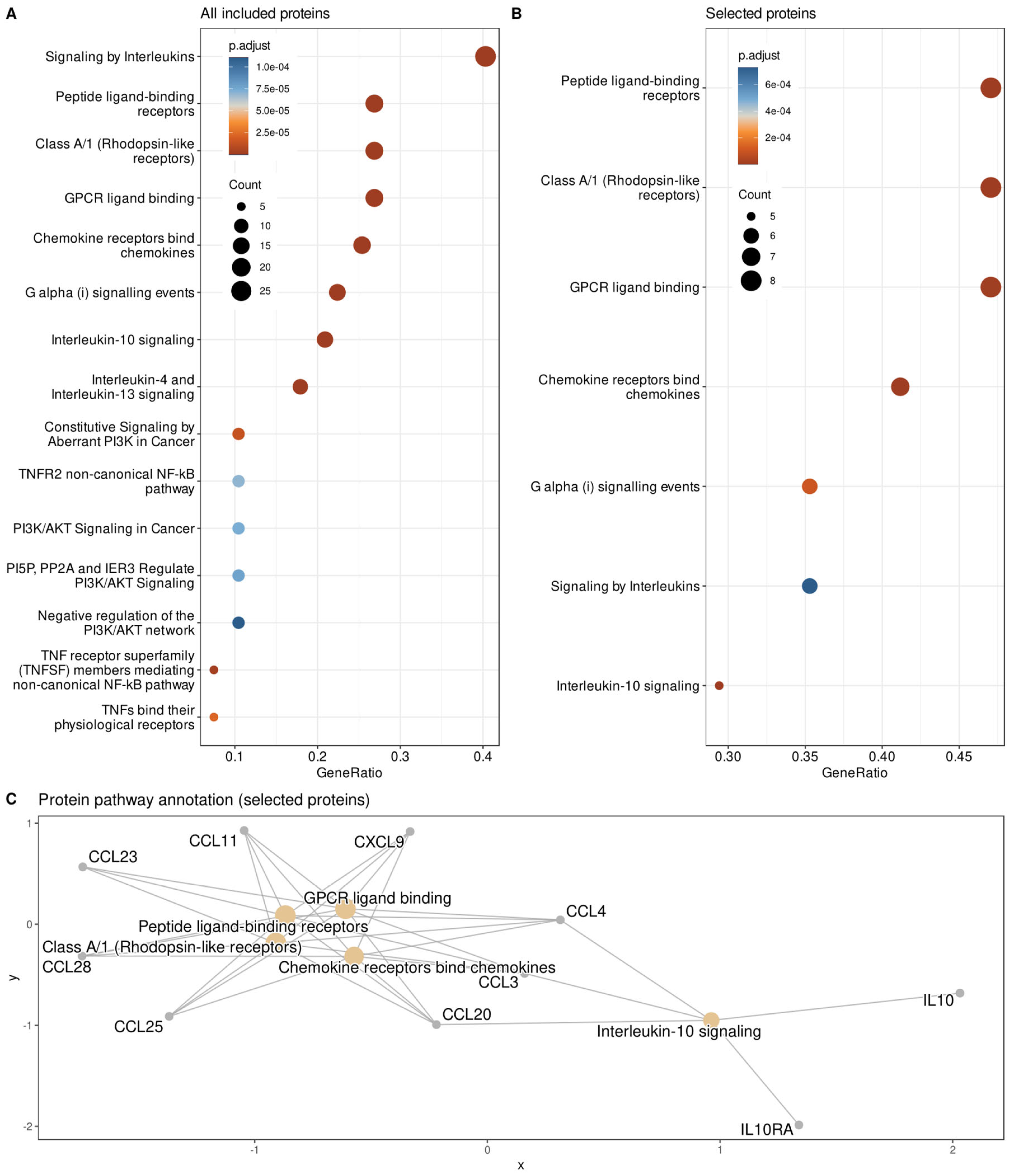
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Ezzat, A. DrugBank Database XML Parser. 2020. Available online: https://cran.r-project.org/package=dbparser (accessed on 14 March 2022).
- Lötsch, J.; Oertel, B.G.; Ultsch, A. Human models of pain for the prediction of clinical analgesia. Pain 2014, 155, 2014–2021. [Google Scholar] [CrossRef] [PubMed]
- Oertel, B.G.; Lötsch, J. Clinical pharmacology of analgesics assessed with human experimental pain models: Bridging basic and clinical research. Br. J. Pharmacol. 2013, 168, 534–553. [Google Scholar] [CrossRef] [Green Version]
- Staahl, C.; Olesen, A.E.; Andresen, T.; Arendt-Nielsen, L.; Drewes, A.M. Assessing analgesic actions of opioids by experimental pain models in healthy volunteers-an updated review. Br. J. Clin. Pharmacol. 2009, 68, 149–168. [Google Scholar] [CrossRef]
- Lötsch, J.; Walter, C.; Zunftmeister, M.; Zinn, S.; Wolters, M.; Ferreiros, N.; Rossmanith, T.; Oertel, B.G.; Geisslinger, G. A data science approach to the selection of most informative readouts of the human intradermal capsaicin pain model to assess pregabalin effects. Basic Clin. Pharmacol. Toxicol. 2020, 126, 318–331. [Google Scholar] [CrossRef] [Green Version]
- Sisignano, M.; Lotsch, J.; Parnham, M.J.; Geisslinger, G. Potential biomarkers for persistent and neuropathic pain therapy. Pharmacol. Ther. 2019, 199, 16–29. [Google Scholar] [CrossRef]
- Sisignano, M.; Angioni, C.; Park, C.K.; Meyer Dos Santos, S.; Jordan, H.; Kuzikov, M.; Liu, D.; Zinn, S.; Hohman, S.W.; Schreiber, Y.; et al. Targeting CYP2J to reduce paclitaxel-induced peripheral neuropathic pain. Proc. Natl. Acad. Sci. USA 2016, 113, 12544–12549. [Google Scholar] [CrossRef] [Green Version]
- Cleeland, C.S.; Ryan, K.M. Pain assessment: Global use of the Brief Pain Inventory. Ann. Acad. Med. Singap. 1994, 23, 129–138. [Google Scholar]
- Wiberg, A.; Olsson-Strömberg, U.; Herman, S.; Kultima, K.; Burman, J. Profound but Transient Changes in the Inflammatory Milieu of the Blood during Autologous Hematopoietic Stem Cell Transplantation. Biol. Blood Marrow Transpl. 2020, 26, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Saeys, Y.; Inza, I.; Larranaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 498–520. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Santosa, F.; Symes, W.W. Linear Inversion of Band-Limited Reflection Seismograms. SIAM J. Sci. Stat. Comput. 1986, 7, 1307–1330. [Google Scholar] [CrossRef]
- Lötsch, J.; Ultsch, A. Random Forests Followed by Computed ABC Analysis as a Feature Selection Method for Machine Learning in Biomedical Data; Springer: Singapore, 2020; pp. 57–69. [Google Scholar]
- Ho, T.K. Random decision forests. In Proceedings of the Third International Conference on Document Analysis and Recognition, Montreal, QC, Canada , 14–16 August 1995; p. 278. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Lötsch, J.; Schiffmann, S.; Schmitz, K.; Brunkhorst, R.; Lerch, F.; Ferreiros, N.; Wicker, S.; Tegeder, I.; Geisslinger, G.; Ultsch, A. Machine-learning based lipid mediator serum concentration patterns allow identification of multiple sclerosis patients with high accuracy. Sci. Rep. 2018, 8, 14884. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Bolstad, B. Preprocesscore: A Collection of Pre-Processing Functions. Available online: https://www.bioconductor.org/packages/release/bioc/html/preprocessCore.html (accessed on 14 March 2022).
- Ultsch, A.; Thrun, M.C.; Hansen-Goos, O.; Lötsch, J. Identification of Molecular Fingerprints in Human Heat Pain Thresholds by Use of an Interactive Mixture Model R Toolbox (AdaptGauss). Int. J. Mol. Sci. 2015, 16, 25897–25911. [Google Scholar] [CrossRef]
- Smirnov, N. Table for Estimating the Goodness of Fit of Empirical Distributions. Ann. Math. Stat. 1948, 19, 279–281. [Google Scholar] [CrossRef]
- Bayes, M.; Price, M. An Essay towards Solving a Problem in the Doctrine of Chances. By the Late Rev. Mr. Bayes, F.R.S. Communicated by Mr. Price, in a Letter to John Canton, A.M.F.R.S. Philos. Trans. 1763, 53, 370–418. [Google Scholar] [CrossRef]
- Kohonen, T. Self-organized formation of topologically correct feature maps. Biol. Cybernet. 1982, 43, 59–69. [Google Scholar] [CrossRef]
- Ultsch, A. Maps for Visualization of High-Dimensional Data Spaces. Available online: https://www.researchgate.net/profile/Alfred-Ultsch/publication/228706090_Maps_for_the_visualization_of_high-dimensional_data_spaces/links/544652950cf2f14fb80f3134/Maps-for-the-visualization-of-high-dimensional-data-spaces.pdf (accessed on 14 March 2022).
- Ultsch, A.; Sieman, H.P. Kohonen’s self organizing feature maps for exploratory data analysis. In Proceedings of the INNC’90, Int. Neural Network Conference, Dordrecht, The Netherlands, 9–13 July 1990; pp. 305–308. [Google Scholar]
- Good, P.I. Resampling Methods: A Practical Guide to Data Analysis; Birkhäuser: Boston, MA, USA, 2006. [Google Scholar]
- Ultsch, A.; Lötsch, J. Computed ABC Analysis for Rational Selection of Most Informative Variables in Multivariate Data. PLoS ONE 2015, 10, e0129767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juran, J.M. The non-Pareto principle; Mea culpa. Qual. Prog. 1975, 8, 8–9. [Google Scholar]
- Brodersen, K.H.; Ong, C.S.; Stephan, K.E.; Buhmann, J.M. The Balanced Accuracy and Its Posterior Distribution. In Proceedings of the Pattern Recognition (ICPR), 2010 20th International Conference on, Istanbul, Turkey, 23–26 August 2010; pp. 3121–3124. [Google Scholar]
- Peterson, W.; Birdsall, T.; Fox, W. The theory of signal detectability. Trans. IRE Prof. Group Inf. Theory 1954, 4, 171–212. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Diagnostic tests 2: Predictive values. BMJ 1994, 309, 102. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.G.; Bland, J.M. Diagnostic tests. 1: Sensitivity and specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, T.J. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Its Application to Analyses of the Vegetation on Danish Commons; I Kommission Hos E. Munksgaard: København, Denmark, 1948. [Google Scholar]
- Jardine, N.; van Rijsbergen, C.J. The use of hierarchic clustering in information retrieval. Inf. Storage Retr. 1971, 7, 217–240. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and Regression Training. Available online: https://cran.r-project.org/package=caret (accessed on 14 March 2022).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Schapire, R.E.; Freund, Y. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 771–780. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Fix, E.; Hodges, J.L. Discriminatory Analysis. Nonparametric Discrimination: Consistency Properties. Int. Stat. Rev. /Rev. Int. De Stat. 1951, 57, 238–247. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Berkson, J. Application of the Logistic Function to Bio-Assay. J. Am. Stat. Assoc. 1944, 39, 357–365. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Press, S.J.; Wilson, S. Choosing between Logistic Regression and Discriminant Analysis. J. Am. Stat. Assoc. 1978, 73, 699–705. [Google Scholar] [CrossRef]
- Antonogeorgos, G.; Panagiotakos, D.B.; Priftis, K.N.; Tzonou, A. Logistic Regression and Linear Discriminant Analyses in Evaluating Factors Associated with Asthma Prevalence among 10- to 12-Years-Old Children: Divergence and Similarity of the Two Statistical Methods. Int. J. Pediatrics 2009, 2009, 952042. [Google Scholar] [CrossRef]
- Lotsch, J.; Ultsch, A. Machine learning in pain research. Pain 2017, 159, 623–630. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme Gradient Boosting. 2020. Available online: https://cran.r-project.org/package=xgboost (accessed on 14 March 2022).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071). Available online: https://cran.r-project.org/package=e1071 (accessed on 14 March 2022).
- Kuhn, M.; Quinlan, R. C50: C5.0 Decision Trees and Rule-Based Models. Available online: https://CRAN.R-project.org/package=C50 (accessed on 14 March 2022).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S; Springer: New York, NY, USA, 2002. [Google Scholar]
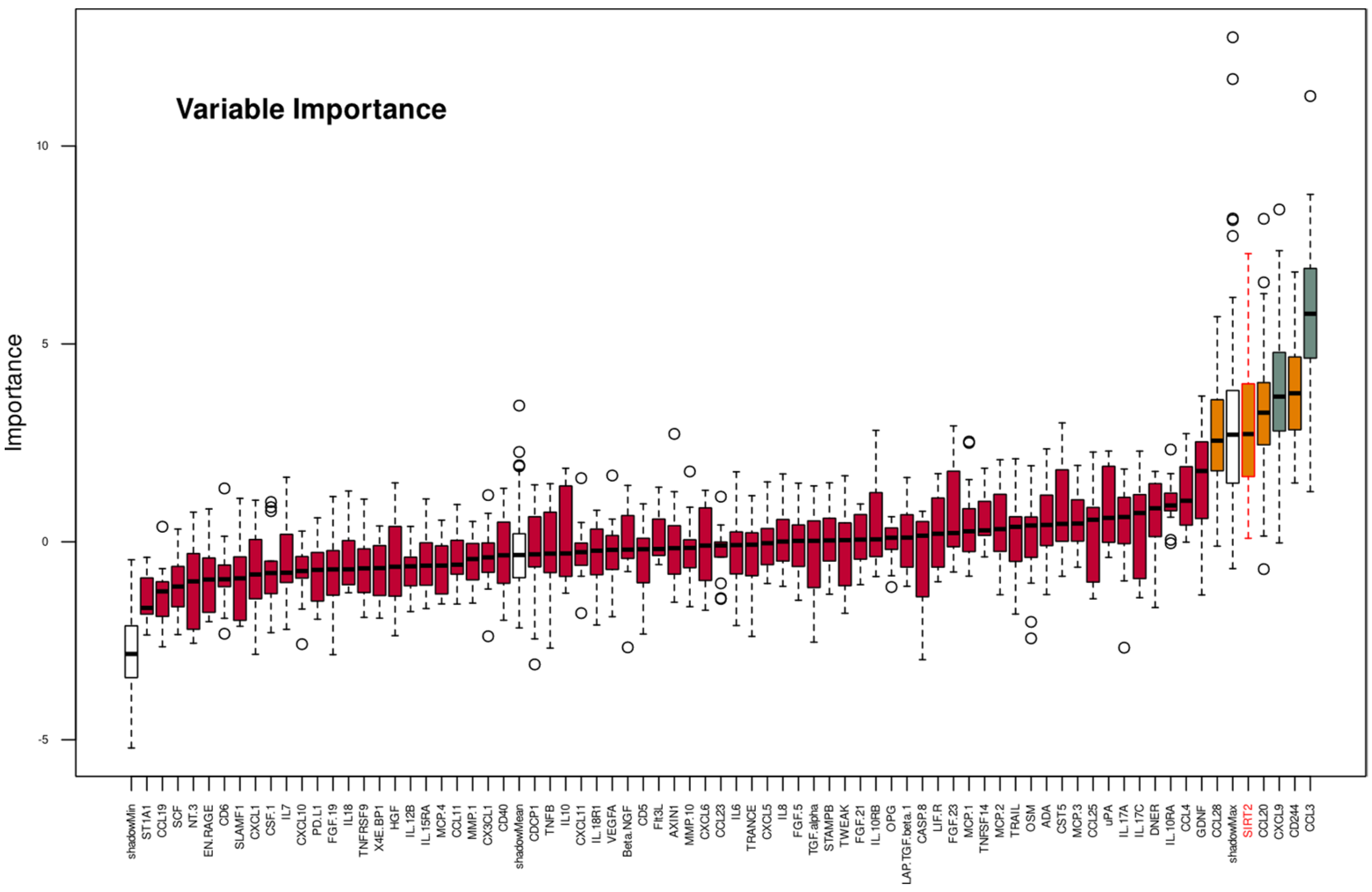
| Parameter | Full Feature Set | Un-Selected Features | Reduced Set #1 | Reduced Set #2 | Reduced Set #3 | Reduced Set #4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein # | 74 | 45 | 19 | 9 | 4 | 2 | ||||||
| Data | Original | Permuted | Original | Permuted | Original | Permuted | Original | Permuted | Original | Permuted | Original | Permuted |
| Sensitivity | 65 (60–75) | 65 (58.75–75) | 57.5 (50–65) | 60 (53.75–70) | 70 (65–80) | 60 (50–65) | 70 (65–75) | 55 (50–65) | 65 (60–75) | 65 (58.75–75) | 70 (60–75) | 55 (50–65) |
| Specificity | 50 (44.44–61.11) | 38.89 (27.78–44.44) | 33.33 (27.78–38.89) | 38.89 (27.78–44.44) | 66.67 (55.56–72.22) | 44.44 (33.33–50) | 66.67 (55.56–72.22) | 44.44 (33.33–55.56) | 50 (44.44–61.11) | 38.89 (27.78–44.44) | 47.22 (38.89–61.11) | 44.44 (33.33–55.56) |
| Pos Pred Value | 60 (56.52–64.78) | 54.01 (48.28–58.33) | 48.15 (45.83–52.29) | 52.51 (49.57–56.52) | 68.83 (63.52–73.91) | 53.39 (46.07–59.09) | 68.42 (63.64–72.22) | 52.63 (47.96–59.32) | 60 (56.52–64.78) | 54.01 (48.28–58.33) | 60 (56–64) | 53.85 (48.11–57.89) |
| Neg Pred Value | 58.11 (52.86–65.42) | 50 (40.88–58.33) | 40 (35.71–46.84) | 47.21 (41.18–53.33) | 66.67 (62.35–71.63) | 48.81 (38.37–55.73) | 64.85 (61.05–70.15) | 47.37 (40.88–55) | 58.11 (52.86–65.42) | 50 (40.88–58.33) | 58.11 (53.24–63.8) | 49 (40.91–53.33) |
| Precision | 60 (56.52–64.78) | 54.01 (48.28–58.33) | 48.15 (45.83–52.29) | 52.51 (49.57–56.52) | 68.83 (63.52–73.91) | 53.39 (46.07–59.09) | 68.42 (63.64–72.22) | 52.63 (47.96–59.32) | 60 (56.52–64.78) | 54.01 (48.28–58.33) | 60 (56–64) | 53.85 (48.11–57.89) |
| Recall | 65 (60–75) | 65 (58.75–75) | 57.5 (50–65) | 60 (53.75–70) | 70 (65–80) | 60 (50–65) | 70 (65–75) | 55 (50–65) | 65 (60–75) | 65 (58.75–75) | 70 (60–75) | 55 (50–65) |
| F1 | 63.29 (59.09–68.66) | 59.09 (54.04–65.22) | 53.2 (48.86–57.14) | 56.52 (51.16–60.57) | 69.77 (65.12–74.32) | 56.47 (47.77–61.3) | 68.36 (65.09–72.73) | 55.16 (50–61.22) | 63.29 (59.09–68.66) | 59.09 (54.04–65.22) | 63.53 (57.87–68.11) | 55.68 (49.72–60) |
| Prevalence | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) | 52.63 (52.63–52.63) |
| Detection Rate | 34.21 (31.58–39.47) | 34.21 (30.92–39.47) | 30.26 (26.32–34.21) | 31.58 (28.29–36.84) | 36.84 (34.21–42.11) | 31.58 (26.32–34.21) | 36.84 (34.21–39.47) | 28.95 (26.32–34.21) | 34.21 (31.58–39.47) | 34.21 (30.92–39.47) | 36.84 (31.58–39.47) | 28.95 (26.32–34.21) |
| Detection Prevalence | 60.53 (52.63–65.79) | 65.79 (57.89–71.71) | 61.84 (57.24–65.79) | 60.53 (55.26–68.42) | 55.26 (50–60.53) | 57.89 (52.63–63.16) | 52.63 (47.37–60.53) | 57.89 (50–63.16) | 60.53 (52.63–65.79) | 65.79 (57.89–71.71) | 60.53 (52.63–65.79) | 55.26 (50–63.82) |
| Balanced Accuracy | 57.92 (54.72–64.24) | 51.81 (44.44–57.01) | 44.44 (41.39–49.58) | 49.86 (45.56–54.72) | 68.33 (62.5–71.67) | 50.97 (43.33–57.5) | 66.25 (61.94–70.56) | 50 (44.38–57.57) | 57.92 (54.72–64.24) | 51.81 (44.44–57.01) | 59.17 (54.44–62.57) | 51.25 (44.1–55.35) |
| ROC-AUC | 57.92 (54.72–64.24) | 54.44 (49.17–59.44) | 49.72 (44.17–56.04) | 52.5 (46.94–57.29) | 68.33 (62.5–71.67) | 55.56 (51.94–62.57) | 66.25 (61.94–70.56) | 54.72 (49.17–60.35) | 57.92 (54.72–64.24) | 54.44 (49.17–59.44) | 59.17 (54.44–62.57) | 54.86 (50–58.06) |
| ABC Subsets “A” | Proteomic Marker | Name | Gene Symbol | Frequency of Selection | Group Difference p-Value | Group Difference Cohen’s d | |||
|---|---|---|---|---|---|---|---|---|---|
| ABC subset A” #1 | ABC subset A” #2 | ABC subset A” #3 | ABC subset A” #4 | CD244 | CD244 molecule | CD244 | 477 | 0.124 | 0.288 |
| SIRT2 | Sirtuin 2 | SIRT2 | 424 | 0.0119 | 0.49 | ||||
| CCL28 | C-C motif chemokine ligand 28 | CCL28 | 409 | 0.203 | −0.399 | ||||
| CXCL9 | C-X-C motif chemokine ligand 9 | CXCL9 | 389 | 0.0229 | −0.383 | ||||
| CCL20 | C-C motif chemokine ligand 20 | CCL20 | 339 | 0.0115 | −0.312 | ||||
| CCL3 | C-C motif chemokine ligand 3 | CCL3 | 297 | 0.194 | 0.323 | ||||
| IL.10RA | Interleukin 10 receptor subunit alpha | IL10RA | 243 | 0.0647 | 0.037 | ||||
| MCP.1 | C-C motif chemokine ligand 2 | CCL2 | 241 | 0.0371 | 0.452 | ||||
| TRAIL | TNF superfamily member 10 | TNFSF10 | 241 | 0.0131 | −0.532 | ||||
| CCL25 | C-C motif chemokine ligand 25 | CCL25 | 237 | 0.027 | −0.469 | ||||
| IL10 | Interleukin 10 | IL10 | 200 | 0.0814 | −0.39 | ||||
| uPA | Plasminogen activator, urokinase | PLAU | 181 | 0.0474 | −0.42 | ||||
| CCL4 | C-C motif chemokine ligand 4 | CCL4 | 176 | 0.036 | 0.439 | ||||
| DNER | Delta/notch like EGF repeat containing | DNER | 146 | 0.0205 | −0.392 | ||||
| STAMPB | STAM binding protein | STAMBP | 137 | 0.0803 | 0.394 | ||||
| CCL23 | C-C motif chemokine ligand 23 | CCL23 | 113 | 0.0929 | −0.339 | ||||
| CST5 | Cystatin D | CST5 | 111 | 0.788 | 0.123 | ||||
| CCL11 | C-C motif chemokine ligand 11 | CCL11 | 108 | 0.275 | 0.252 | ||||
| FGF.23 | Fibroblast growth factor 23 | FGF23 | 108 | 0.0676 | −0.304 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lötsch, J.; Mustonen, L.; Harno, H.; Kalso, E. Machine-Learning Analysis of Serum Proteomics in Neuropathic Pain after Nerve Injury in Breast Cancer Surgery Points at Chemokine Signaling via SIRT2 Regulation. Int. J. Mol. Sci. 2022, 23, 3488. https://doi.org/10.3390/ijms23073488
Lötsch J, Mustonen L, Harno H, Kalso E. Machine-Learning Analysis of Serum Proteomics in Neuropathic Pain after Nerve Injury in Breast Cancer Surgery Points at Chemokine Signaling via SIRT2 Regulation. International Journal of Molecular Sciences. 2022; 23(7):3488. https://doi.org/10.3390/ijms23073488
Chicago/Turabian StyleLötsch, Jörn, Laura Mustonen, Hanna Harno, and Eija Kalso. 2022. "Machine-Learning Analysis of Serum Proteomics in Neuropathic Pain after Nerve Injury in Breast Cancer Surgery Points at Chemokine Signaling via SIRT2 Regulation" International Journal of Molecular Sciences 23, no. 7: 3488. https://doi.org/10.3390/ijms23073488
APA StyleLötsch, J., Mustonen, L., Harno, H., & Kalso, E. (2022). Machine-Learning Analysis of Serum Proteomics in Neuropathic Pain after Nerve Injury in Breast Cancer Surgery Points at Chemokine Signaling via SIRT2 Regulation. International Journal of Molecular Sciences, 23(7), 3488. https://doi.org/10.3390/ijms23073488







