Pleiotropic Roles of Scavenger Receptors in Circadian Retinal Phagocytosis: A New Function for Lysosomal SR-B2/LIMP-2 at the RPE Cell Surface
Abstract
1. Introduction
2. Results
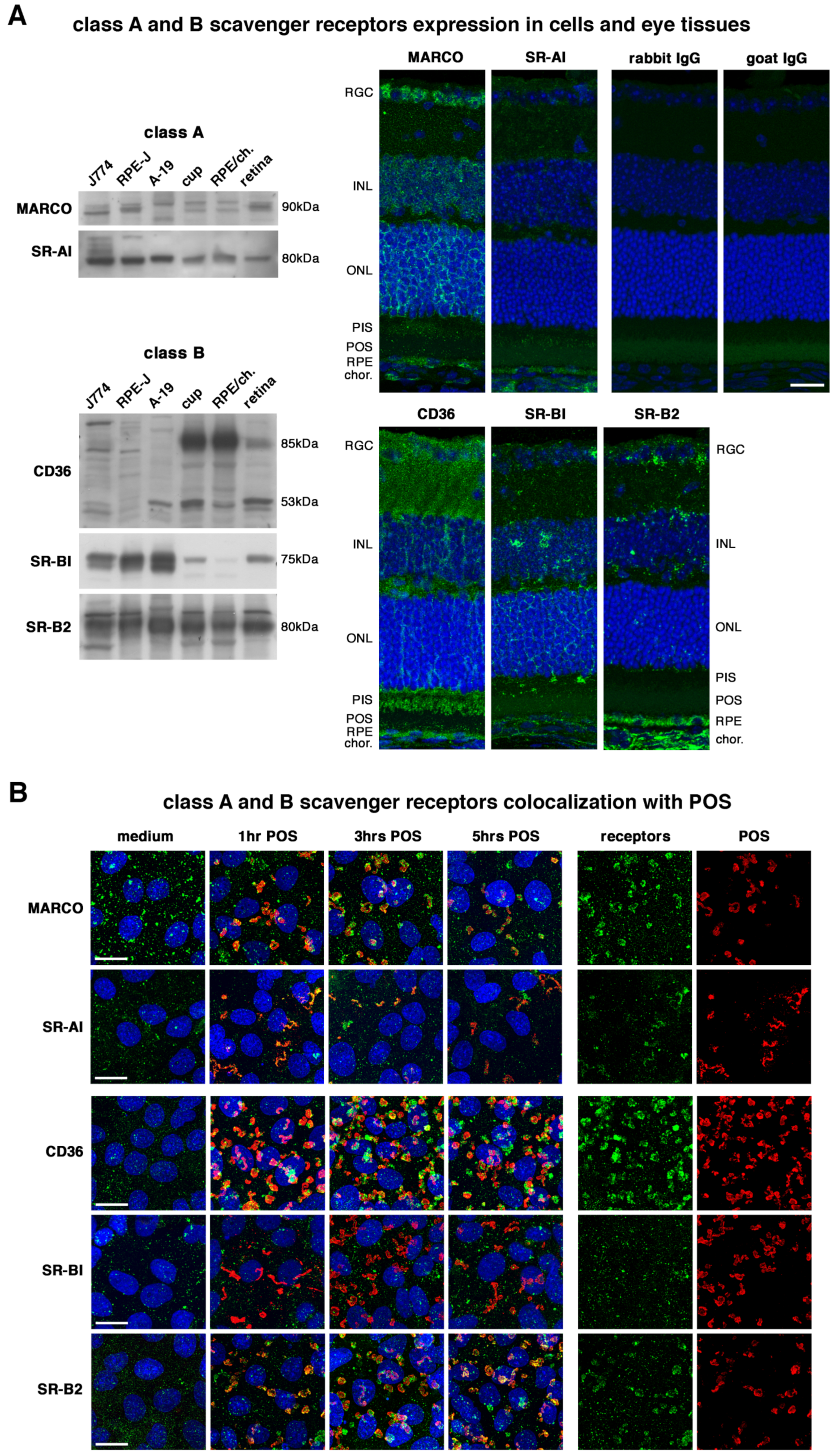
2.1. All Receptors Are Readily Expressed by RPE Cells and Colocalize with POS except SR-BI
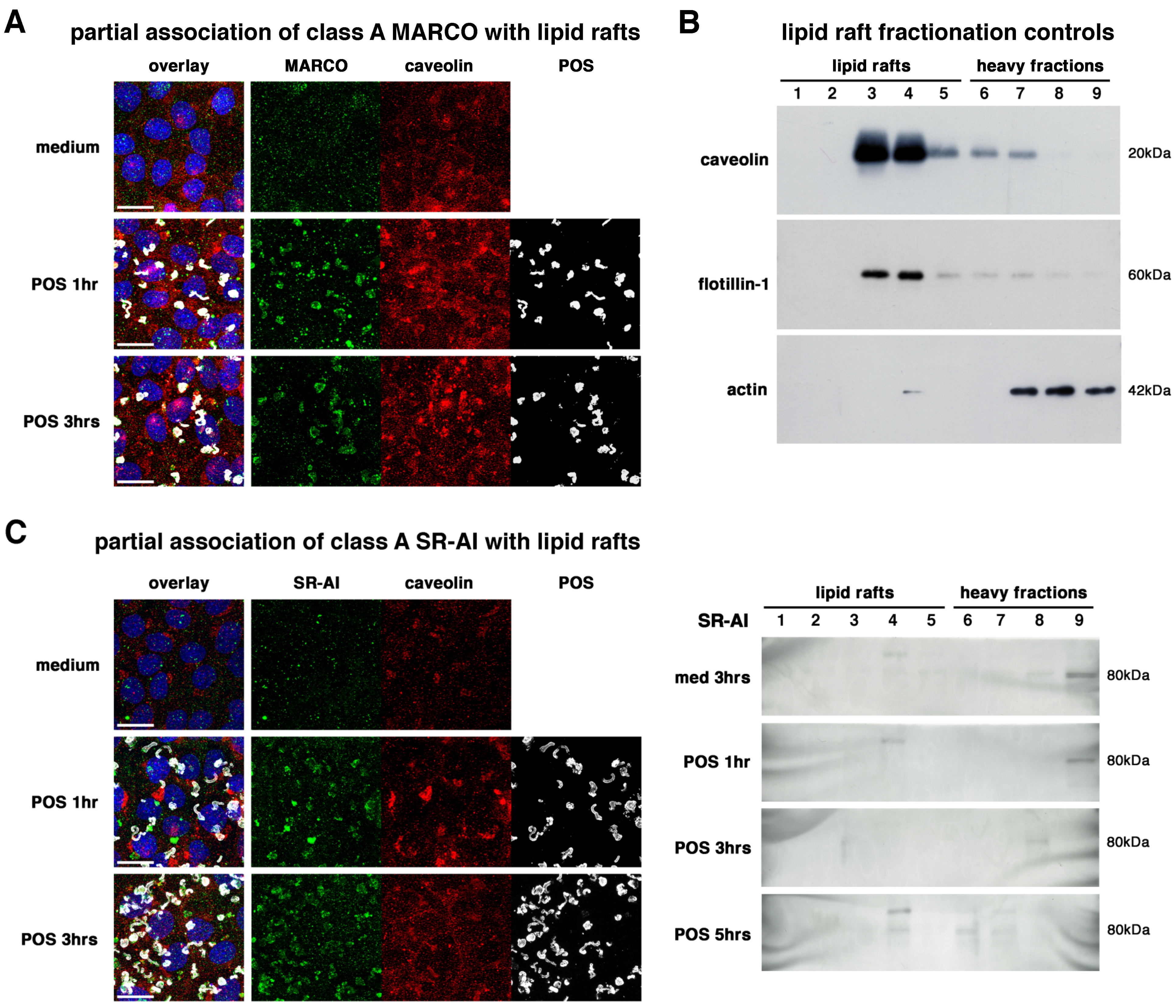
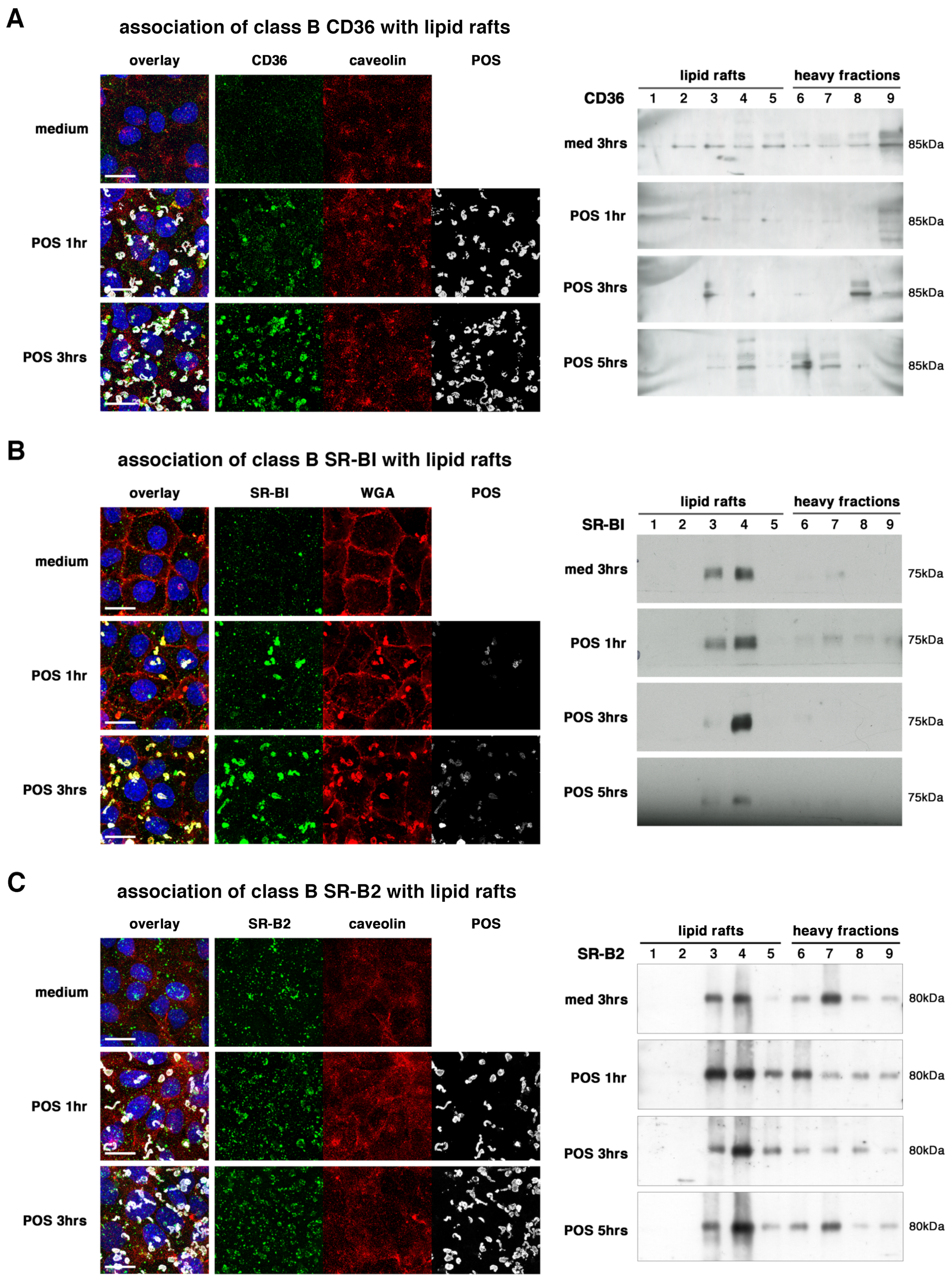
2.2. All Receptors Associate with Lipid Rafts
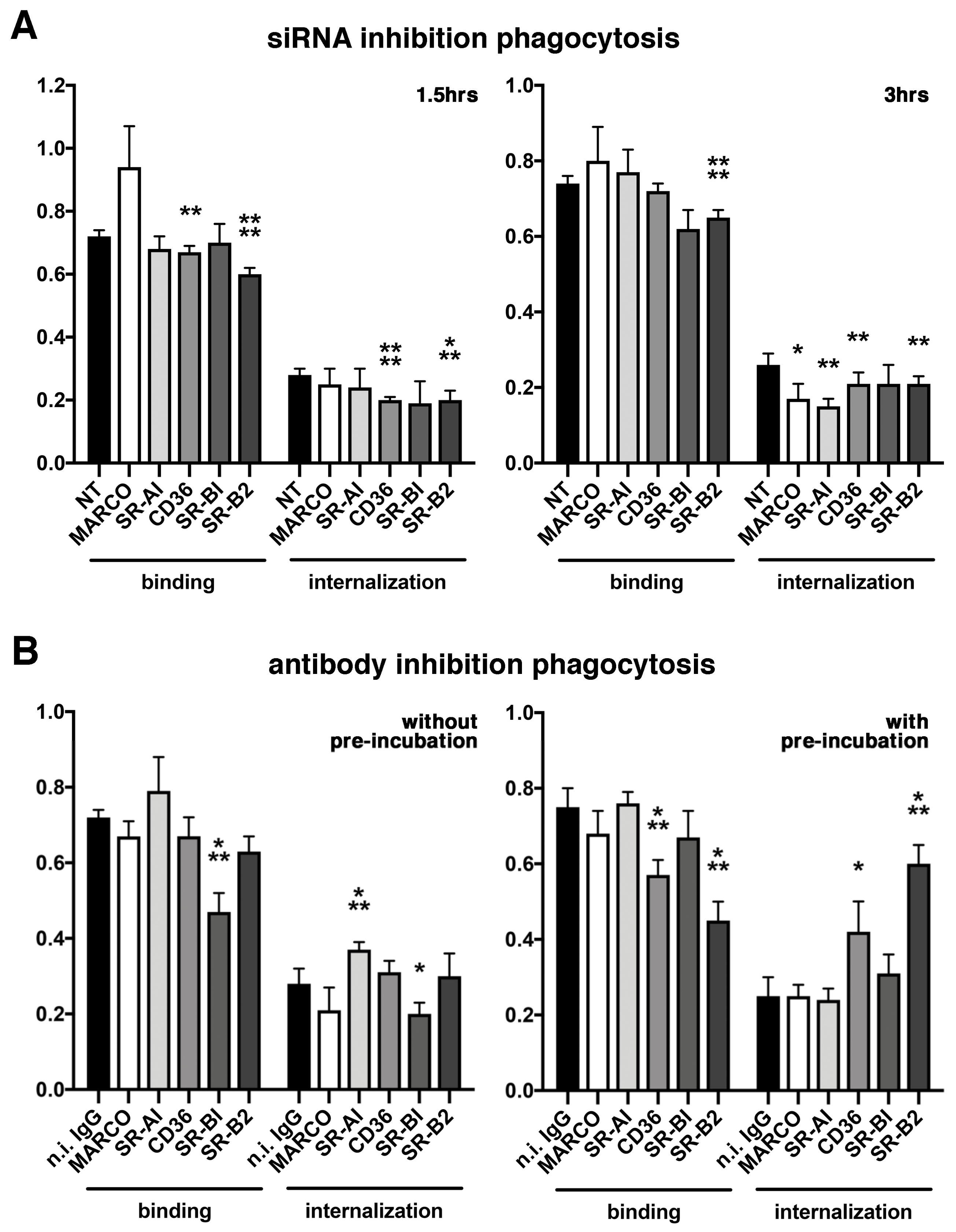
2.3. Various Effects of Receptors Inhibition on POS Phagocytosis
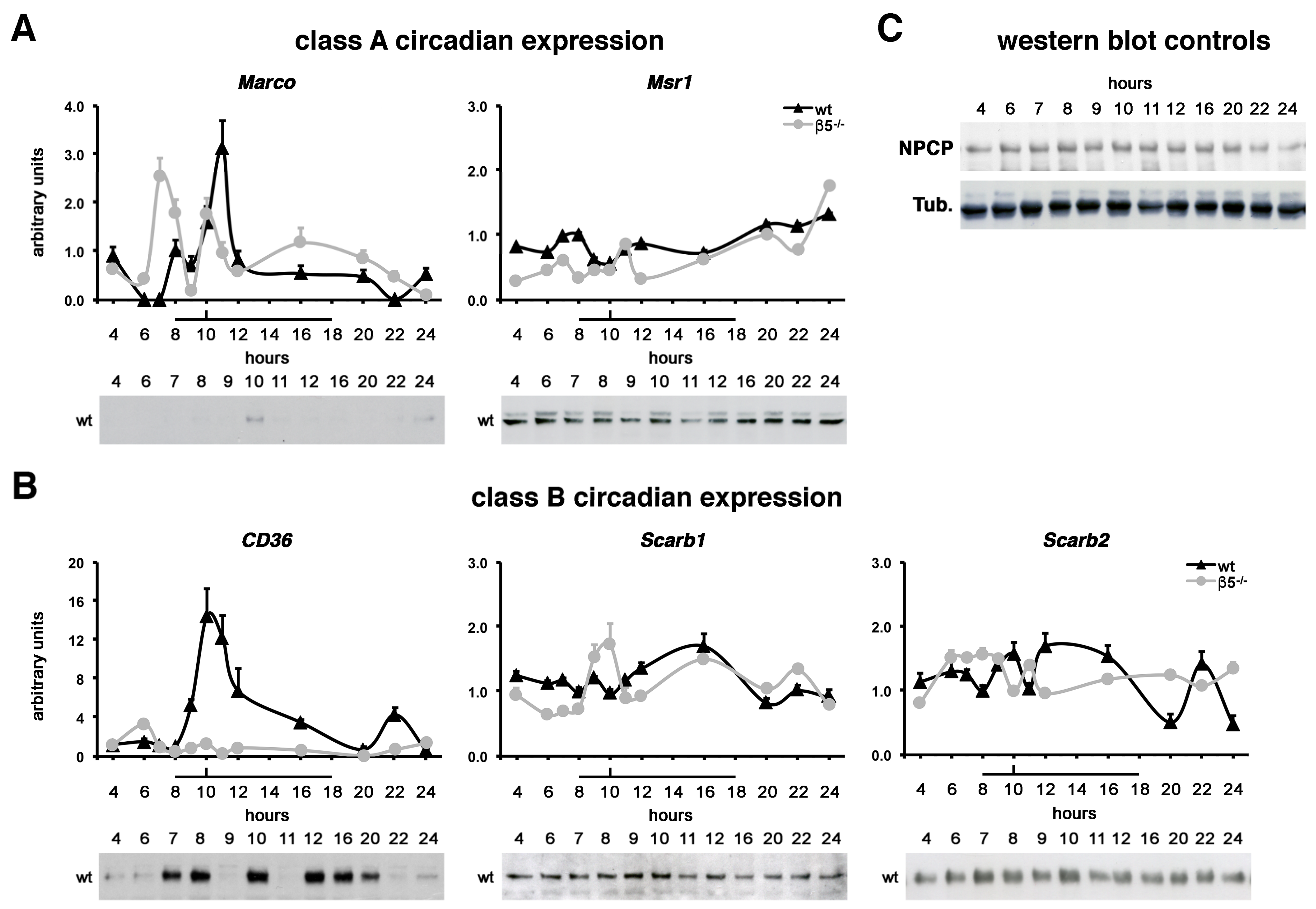
2.4. Class A MARCO and Class B CD36 Show Rhythmic Expression Patterns In Vivo
3. Discussion
4. Materials and Methods
4.1. Reagents, Antibodies and Cell Culture
4.2. Cell Culture and siRNA transfection
4.3. Animals and Tissue Collection
4.4. Sample Lysis and Immunoblotting
4.5. POS Isolation
4.6. POS Phagocytosis
4.7. Immunohistochemistry, Immunocytochemistry and Microscopy
4.8. Lipid Rafts Isolation by a Detergent-Free Method
4.9. Quantification of Gene Expression
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [PubMed]
- LaVail, M.M. Rod outer segment disk shedding in rat retina: Relationship to cyclic lighting. Science 1976, 194, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Bok, D.; Hall, M.O. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J. Cell Biol. 1971, 49, 664–682. [Google Scholar] [CrossRef]
- Nandrot, E.; Dufour, E.M.; Provost, A.C.; Péquignot, M.O.; Bonnel, S.; Gogat, K.; Marchant, D.; Rouillac, C.; Sépulchre de Condé, B.; Bihoreau, M.T.; et al. Homozygous deletion in the coding sequence of the c-mer gene in RCS rats unravels general mechanisms of physiological cell adhesion and apoptosis. Neurobiol. Dis. 2000, 7, 586–599. [Google Scholar] [CrossRef]
- Nandrot, E.F.; Kim, Y.; Brodie, S.E.; Huang, X.; Sheppard, D.; Finnemann, S.C. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J. Exp. Med. 2004, 200, 1539–1545. [Google Scholar] [CrossRef]
- Gal, A.; Li, Y.; Thompson, D.A.; Weir, J.; Orth, U.; Jacobson, S.G.; Apfelstedt-Sylla, E.; Vollrath, D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat. Genet. 2000, 26, 270–271. [Google Scholar] [CrossRef] [PubMed]
- Parinot, C.; Nandrot, E.F. A Comprehensive Review of Mutations in the MERTK Proto-Oncogene. Adv. Exp. Med. Biol. 2016, 854, 259–265. [Google Scholar]
- Finnemann, S.C.; Rodriguez-Boulan, E. Macrophage and retinal pigment epithelium phagocytosis: Apoptotic cells and photoreceptors compete for alphavbeta3 and alphavbeta5 integrins, and protein kinase C regulates alphavbeta5 binding and cytoskeletal linkage. J. Exp. Med. 1999, 190, 861–874. [Google Scholar] [CrossRef]
- Nandrot, E.F.; Anand, M.; Almeida, D.; Atabai, K.; Sheppard, D.; Finnemann, S.C. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci. USA 2007, 104, 12005–12010. [Google Scholar] [CrossRef] [PubMed]
- Nandrot, E.F.; Silva, K.E.; Scelfo, C.; Finnemann, S.C. Retinal pigment epithelial cells use a MerTK-dependent mechanism to limit the phagocytic particle binding activity of αvβ5 integrin. Biol. Cell 2012, 104, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Savill, J.S.; Haslett, C.; Bratton, D.L.; Doherty, D.E.; Campbell, P.A.; Henson, P.M. Different populations of macrophages use either the vitronectin receptor or the phosphatidylserine receptor to recognize and remove apoptotic cells. J. Immunol. 1992, 149, 4029–4035. [Google Scholar] [PubMed]
- Ruggiero, L.; Connor, M.P.; Chen, J.; Langen, R.; Finnemann, S.C. Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. Proc. Natl. Acad. Sci. USA 2012, 109, 8145–8148. [Google Scholar] [CrossRef]
- Hanayama, R.; Tanaka, M.; Miwa, K.; Shinohara, A.; Iwamatsu, A.; Nagata, S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002, 417, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Kawamoto, K.; Higashino, K.; Arita, H. Prevention of growth arrest-induced cell death of vascular smooth muscle cells by a product of growth arrest-specific gene, gas6. FEBS Lett. 1996, 387, 78–80. [Google Scholar] [CrossRef]
- Law, A.L.; Parinot, C.; Chatagnon, J.; Gravez, B.; Sahel, J.A.; Bhattacharya, S.S.; Nandrot, E.F. Cleavage of Mer tyrosine kinase (MerTK) from the cell surface contributes to the regulation of retinal phagocytosis. J. Biol. Chem. 2015, 290, 4941–4952. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Finnemann, S.C. Tetraspanin CD81 is required for the alphavbeta5-integrin-dependent particle-binding step of RPE phagocytosis. J. Cell Sci. 2007, 120, 3053–3063. [Google Scholar] [CrossRef]
- Ravichandran, K.S. Beginnings of a good apoptotic meal: The find-me and eat-me signaling pathways. Immunity 2011, 35, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Prabhudas, M.; Bowdish, D.; Drickamer, K.; Febbraio, M.; Herz, J.; Kobzik, L.; Krieger, M.; Loike, J.; Means, T.K.; Moestrup, S.K.; et al. Standardizing scavenger receptor nomenclature. J. Immunol. 2014, 192, 1997–2006. [Google Scholar] [CrossRef]
- Alquraini, A.; El Khoury, J. Scavenger receptors. Curr. Biol. 2020, 30, R790–R795. [Google Scholar] [CrossRef]
- Plüddemann, A.; Neyen, C.; Gordon, S. Macrophage scavenger receptors and host-derived ligands. Methods 2007, 43, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Ryeom, S.W.; Silverstein, R.L.; Scotto, A.; Sparrow, J.R. Binding of anionic phospholipids to retinal pigment epithelium may be mediated by the scavenger receptor CD36. J. Biol. Chem. 1996, 271, 20536–20539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tait, J.F.; Smith, C. Phosphatidylserine receptors: Role of CD36 in binding of anionic phospholipid vesicles to monocytic cells. J. Biol. Chem. 1999, 274, 3048–3054. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.E.; Sun, M.; Zhang, R.; Febbraio, M.; Silverstein, R.; Hazen, S.L. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J. Exp. Med. 2006, 203, 2613–2625. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.E.; Tacon, D.; Tedbury, P.R.; Hadden, J.M.; Knowling, S.; Sawamura, T.; Peckham, M.; Phillips, S.E.; Walker, J.H.; Ponnambalam, S. LOX-1 scavenger receptor mediates calcium-dependent recognition of phosphatidylserine and apoptotic cells. Biochem. J. 2006, 393, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Ryeom, S.W.; Sparrow, J.R.; Silverstein, R.L. CD36 participates in the phagocytosis of rod outer segments by retinal pigment epithelium. J. Cell Sci. 1996, 109, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Finnemann, S.C.; Silverstein, R.L. Differential roles of CD36 and alphavbeta5 integrin in photoreceptor phagocytosis by the retinal pigment epithelium. J. Exp. Med. 2001, 194, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, J.R.; Ryeom, S.W.; Abumrad, N.A.; Ibrahimi, A.; Silverstein, R.L. CD36 expression is altered in retinal pigment epithelial cells of the RCS rat. Exp. Eye Res. 1997, 64, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Finnemann, S.C.; Febbraio, M.; Shan, L.; Annangudi, S.P.; Podrez, E.A.; Hoppe, G.; Darrow, R.; Organisciak, D.T.; Salomon, R.G.; et al. Light-induced oxidation of photoreceptor outer segment phospholipids generates ligands for CD36-mediated phagocytosis by retinal pigment epithelium: A potential mechanism for modulating outer segment phagocytosis under oxidant stress conditions. J. Biol. Chem. 2006, 281, 4222–4230. [Google Scholar] [CrossRef]
- Courtois, Y. The role of CD36 receptor in the phagocytosis of oxidized lipids and AMD. Aging 2010, 2, 888–889. [Google Scholar] [CrossRef] [PubMed]
- Rigotti, A.; Acton, S.L.; Krieger, M. The class B scavenger receptors SR-BI and CD36 are receptors for anionic phospholipids. J. Biol. Chem. 1995, 270, 16221–16224. [Google Scholar] [CrossRef] [PubMed]
- Picard, E.; Houssier, M.; Bujold, K.; Sapieha, P.; Lubell, W.; Dorfman, A.; Racine, J.; Hardy, P.; Febbraio, M.; Lachapelle, P.; et al. CD36 plays an important role in the clearance of oxLDL and associated age-dependent sub-retinal deposits. Aging 2010, 2, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Dufour, E.M.; Nandrot, E.; Marchant, D.; Van Den Berghe, L.; Gadin, S.; Issilame, M.; Dufier, J.L.; Marsac, C.; Carper, D.; Menasche, M.; et al. Identification of novel genes and altered signaling pathways in the retinal pigment epithelium during the Royal College of Surgeons rat retinal degeneration. Neurobiol. Dis. 2003, 14, 166–180. [Google Scholar] [CrossRef]
- Duncan, K.G.; Bailey, K.R.; Kane, J.P.; Schwartz, D.M. Human retinal pigment epithelial cells express scavenger receptors BI and BII. Biochem. Biophys. Res. Commun. 2002, 292, 1017–1022. [Google Scholar] [CrossRef]
- Provost, A.C.; Péquignot, M.O.; Sainton, K.M.; Gadin, S.; Sallé, S.; Marchant, D.; Hales, D.B.; Abitbol, M. Expression of SR-BI receptor and StAR protein in rat ocular tissues. C. R. Biol. 2003, 326, 841–851. [Google Scholar] [CrossRef]
- Duncan, K.G.; Hosseini, K.; Bailey, K.R.; Yang, H.; Lowe, R.J.; Matthes, M.T.; Kane, J.P.; LaVail, M.M.; Schwartz, D.M.; Duncan, J.L. Expression of reverse cholesterol transport proteins ATP-binding cassette A1 (ABCA1) and scavenger receptor BI (SR-BI) in the retina and retinal pigment epithelium. Br. J. Ophthalmol. 2009, 93, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Provost, A.C.; Vede, L.; Bigot, K.; Keller, N.; Tailleux, A.; Jaïs, J.P.; Savoldelli, M.; Ameqrane, I.; Lacassagne, E.; Legeais, J.M.; et al. Morphologic and electroretinographic phenotype of SR-BI knockout mice after a long-term atherogenic diet. Invest. Ophthalmol. Vis. Sci. 2009, 50, 3931–3942. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Feng, Y.; Sun, H.; Yang, S.; Sun, T.; Guo, X.; Ji, F.; Wu, B.; Zhou, D. Scavenger receptor MARCO contributes to macrophage phagocytosis and clearance of tumor cells. Exp. Cell Res. 2021, 408, 112862. [Google Scholar] [CrossRef]
- Platt, N.; Suzuki, H.; Kurihara, Y.; Kodama, T.; Gordon, S. Role for the class A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes in vitro. Proc. Natl. Acad. Sci. USA 1996, 93, 12456–12460. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.M.; Murao, K.; Imachi, H.; Sato, M.; Nakano, T.; Kodama, T.; Sasaguri, Y.; Wong, N.C.; Takahara, J.; Ishida, T. Phosphatidylinositol 3-OH kinase-Akt/protein kinase B pathway mediates Gas6 induction of scavenger receptor a in immortalized human vascular smooth muscle cell line. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1592–1597. [Google Scholar]
- Todt, J.C.; Hu, B.; Curtis, J.L. The scavenger receptor SR-A I/II (CD204) signals via the receptor tyrosine kinase Mertk during apoptotic cell uptake by murine macrophages. J. Leukoc. Biol. 2008, 84, 510–518. [Google Scholar] [CrossRef]
- Araki, N.; Higashi, T.; Mori, T.; Shibayama, R.; Kawabe, Y.; Kodama, T.; Takahashi, K.; Shichiri, M.; Horiuchi, S. Macrophage scavenger receptor mediates the endocytic uptake and degradation of advanced glycation end products of the Maillard reaction. Eur. J. Biochem. 1995, 230, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Platt, N.; Suzuki, H.; Kodama, T.; Gordon, S. Apoptotic thymocyte clearance in scavenger receptor class A-deficient mice is apparently normal. J. Immunol. 2000, 164, 4861–4867. [Google Scholar] [CrossRef] [PubMed]
- Heybrock, S.; Kanerva, K.; Meng, Y.; Ing, C.; Liang, A.; Xiong, Z.J.; Weng, X.; Ah Kim, Y.; Collins, R.; Trimble, W.; et al. Lysosomal integral membrane protein-2 (LIMP-2/SCARB2) is involved in lysosomal cholesterol export. Nat. Commun. 2019, 10, 3521. [Google Scholar] [CrossRef] [PubMed]
- Neculai, D.; Schwake, M.; Ravichandran, M.; Zunke, F.; Collins, R.F.; Peters, J.; Neculai, M.; Plumb, J.; Loppnau, P.; Pizarro, J.C.; et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature 2013, 504, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Marín, E.; Fernández-Prieto, L.; Rodriguez-Del Rio, E.; Madrazo-Toca, F.; Reinheckel, T.; Saftig, P.; Alvarez-Dominguez, C. LIMP-2 links late phagosomal trafficking with the onset of the innate immune response to Listeria monocytogenes: A role in macrophage activation. J. Biol. Chem. 2011, 286, 3332–3341. [Google Scholar] [CrossRef]
- Gamp, A.C.; Tanaka, Y.; Lüllmann-Rauch, R.; Wittke, D.; D’Hooge, R.; De Deyn, P.P.; Moser, T.; Maier, H.; Hartmann, D.; Reiss, K.; et al. LIMP-2/LGP85 deficiency causes ureteric pelvic junction obstruction, deafness and peripheral neuropathy in mice. Hum. Mol. Genet. 2003, 12, 631–646. [Google Scholar] [CrossRef]
- Pike, L.J. Lipid rafts: Bringing order to chaos. J. Lipid Res. 2003, 44, 655–667. [Google Scholar] [CrossRef]
- Head, B.P.; Patel, H.H.; Insel, P.A. Interaction of membrane/lipid rafts with the cytoskeleton: Impact on signaling and function: Membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta 2014, 1838, 532–545. [Google Scholar] [CrossRef]
- El Biari, K.; Gaudioso, A.; Fernández-Alonso, M.C.; Jiménez-Barbero, J.; Cañada, F.J. Peptidoglycan Recognition by Wheat Germ Agglutinin. A View by NMR. Nat. Prod. Comm. 2019, 14, 1–10. [Google Scholar]
- Thorne, R.F.; Meldrum, C.J.; Harris, S.J.; Dorahy, D.J.; Shafren, D.R.; Berndt, M.C.; Burns, G.F.; Gibson, P.G. CD36 forms covalently associated dimers and multimers in platelets and transfected COS-7 cells. Biochem. Biophys. Res. Commun. 1997, 240, 812–818. [Google Scholar] [CrossRef]
- Reaven, E.; Cortez, Y.; Leers-Sucheta, S.; Nomoto, A.; Azhar, S. Dimerization of the scavenger receptor class B type I: Formation, function, and localization in diverse cells and tissues. J. Lipid Res. 2004, 45, 513–528. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, D.L.; Bhattacharya, A.; Sha, Y.; Xu, Y.; Xiang, Q.; Kan, A.; Jagannath, C.; Komatsu, M.; Eissa, N.T. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity 2013, 39, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Ferracini, M.; Rios, F.J.; Pecenin, M.; Jancar, S. Clearance of apoptotic cells by macrophages induces regulatory phenotype and involves stimulation of CD36 and platelet-activating factor receptor. Mediat. Inflamm. 2013, 2013, 950273. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, Z.; Hu, R.; Bo, L.; Minshall, R.D.; Malik, A.B.; Hu, G. Inactivation of Rab11a GTPase in Macrophages Facilitates Phagocytosis of Apoptotic Neutrophils. J. Immunol. 2017, 198, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Averaimo, S.; Assali, A.; Ros, O.; Couvet, S.; Zagar, Y.; Genescu, I.; Rebsam, A.; Nicol, X. A plasma membrane microdomain compartmentalizes ephrin-generated cAMP signals to prune developing retinal axon arbors. Nat. Commun. 2016, 7, 12896. [Google Scholar] [CrossRef]
- Stuart, L.M.; Bell, S.A.; Stewart, C.R.; Silver, J.M.; Richard, J.; Goss, J.L.; Tseng, A.A.; Zhang, A.; Khoury, J.B.E.; Moore, K.J. CD36 signals to the actin cytoskeleton and regulates microglial migration via a p130Cas complex. J. Biol. Chem. 2007, 282, 27392–27401. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Yancey, P.G.; Babaev, V.R.; Blakemore, J.L.; Zhang, Y.; Ding, L.; Fazio, S.; Linton, M.F. Macrophage SR-BI mediates efferocytosis via Src/PI3K/Rac1 signaling and reduces atherosclerotic lesion necrosis. J. Lipid Res. 2015, 56, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Seixas, E.; Escrevente, C.; Seabra, M.C.; Barral, D.C. Rab GTPase regulation of bacteria and protozoa phagocytosis occurs through the modulation of phagocytic receptor surface expression. Sci. Rep. 2018, 8, 12998. [Google Scholar] [CrossRef]
- Ren, Y.; Silverstein, R.L.; Allen, J.; Savill, J. CD36 gene transfer confers capacity for phagocytosis of cells undergoing apoptosis. J. Exp. Med. 1995, 181, 1857–1862. [Google Scholar] [CrossRef]
- Conrad, K.S.; Cheng, T.W.; Ysselstein, D.; Heybrock, S.; Hoth, L.R.; Chrunyk, B.A.; Am Ende, C.W.; Krainc, D.; Schwake, M.; Saftig, P.; et al. Lysosomal integral membrane protein-2 as a phospholipid receptor revealed by biophysical and cellular studies. Nat. Commun. 2017, 8, 1908. [Google Scholar] [CrossRef]
- Sun, B.; Qi, N.; Shang, T.; Wu, H.; Deng, T.; Han, D. Sertoli cell-initiated testicular innate immune response through toll-like receptor-3 activation is negatively regulated by Tyro3, Axl, and mer receptors. Endocrinology 2010, 151, 2886–2897. [Google Scholar] [CrossRef]
- Stewart, C.R.; Stuart, L.M.; Wilkinson, K.; van Gils, J.M.; Deng, J.; Halle, A.; Rayner, K.J.; Boyer, L.; Zhong, R.; Frazier, W.A.; et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 2010, 11, 155–161. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Varin, A.; Chen, Y.; Liu, B.; Tryggvason, K.; Gordon, S. SR-A/MARCO-mediated ligand delivery enhances intracellular TLR and NLR function, but ligand scavenging from cell surface limits TLR4 response to pathogens. Blood 2011, 117, 1319–1328. [Google Scholar] [CrossRef]
- Sigola, L.B.; Fuentes, A.-L.; Millis, L.M.; Vapenik, J.; Murira, A. Effects of Toll-like receptor ligands on RAW 264.7 macrophage morphology and zymosan phagocytosis. Tissue Cell 2016, 48, 389–396. [Google Scholar] [CrossRef]
- Cao, D.; Luo, J.; Chen, D.; Xu, H.; Shi, H.; Jing, X.; Zang, W. CD36 regulates lipopolysaccharide-induced signaling pathways and mediates the internalization of Escherichia coli in cooperation with TLR4 in goat mammary gland epithelial cells. Sci. Rep. 2016, 6, 23132. [Google Scholar] [CrossRef] [PubMed]
- Yamada, C.; Beron-Pelusso, C.; Algazzaz, N.; Heidari, A.; Luz, D.; Rawas-Qalaji, M.; Toderas, I.; Mascarenhas, A.K.; Kawai, T.; Movila, A. Age-dependent effect between MARCO and TLR4 on PMMA particle phagocytosis by macrophages. J. Cell Mol. Med. 2016, 23, 5827–5831. [Google Scholar] [CrossRef] [PubMed]
- Parinot, C.; Rieu, Q.; Chatagnon, J.; Finnemann, S.C.; Nandrot, E.F. Large-scale purification of porcine or bovine photoreceptor outer segments for phagocytosis assays on retinal pigment epithelial cells. J. Vis. Exp. 2014, 94, 52100. [Google Scholar] [CrossRef] [PubMed]
- Finnemann, S.C.; Bonilha, V.L.; Marmorstein, A.D.; Rodriguez-Boulan, E. Phagocytosis of rod outer segments by retinal pigment epithelial cells requires alpha(v)beta5 integrin for binding but not for internalization. Proc. Natl. Acad. Sci. USA 1997, 94, 12932–12937. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rieu, Q.; Bougoüin, A.; Zagar, Y.; Chatagnon, J.; Hamieh, A.; Enderlin, J.; Huby, T.; Nandrot, E.F. Pleiotropic Roles of Scavenger Receptors in Circadian Retinal Phagocytosis: A New Function for Lysosomal SR-B2/LIMP-2 at the RPE Cell Surface. Int. J. Mol. Sci. 2022, 23, 3445. https://doi.org/10.3390/ijms23073445
Rieu Q, Bougoüin A, Zagar Y, Chatagnon J, Hamieh A, Enderlin J, Huby T, Nandrot EF. Pleiotropic Roles of Scavenger Receptors in Circadian Retinal Phagocytosis: A New Function for Lysosomal SR-B2/LIMP-2 at the RPE Cell Surface. International Journal of Molecular Sciences. 2022; 23(7):3445. https://doi.org/10.3390/ijms23073445
Chicago/Turabian StyleRieu, Quentin, Antoine Bougoüin, Yvrick Zagar, Jonathan Chatagnon, Abdallah Hamieh, Julie Enderlin, Thierry Huby, and Emeline F. Nandrot. 2022. "Pleiotropic Roles of Scavenger Receptors in Circadian Retinal Phagocytosis: A New Function for Lysosomal SR-B2/LIMP-2 at the RPE Cell Surface" International Journal of Molecular Sciences 23, no. 7: 3445. https://doi.org/10.3390/ijms23073445
APA StyleRieu, Q., Bougoüin, A., Zagar, Y., Chatagnon, J., Hamieh, A., Enderlin, J., Huby, T., & Nandrot, E. F. (2022). Pleiotropic Roles of Scavenger Receptors in Circadian Retinal Phagocytosis: A New Function for Lysosomal SR-B2/LIMP-2 at the RPE Cell Surface. International Journal of Molecular Sciences, 23(7), 3445. https://doi.org/10.3390/ijms23073445





