Abstract
The X-linked gene encoding aristaless-related homeobox (ARX) is a bi-functional transcription factor capable of activating or repressing gene transcription, whose mutations have been found in a wide spectrum of neurodevelopmental disorders (NDDs); these include cortical malformations, paediatric epilepsy, intellectual disability (ID) and autism. In addition to point mutations, duplications of the ARX locus have been detected in male patients with ID. These rearrangements include telencephalon ultraconserved enhancers, whose structural alterations can interfere with the control of ARX expression in the developing brain. Here, we review the structural features of 15 gain copy-number variants (CNVs) of the ARX locus found in patients presenting wide-ranging phenotypic variations including ID, speech delay, hypotonia and psychiatric abnormalities. We also report on a further novel Xp21.3 duplication detected in a male patient with moderate ID and carrying a fully duplicated copy of the ARX locus and the ultraconserved enhancers. As consequences of this rearrangement, the patient-derived lymphoblastoid cell line shows abnormal activity of the ARX-KDM5C-SYN1 regulatory axis. Moreover, the three-dimensional (3D) structure of the Arx locus, both in mouse embryonic stem cells and cortical neurons, provides new insight for the functional consequences of ARX duplications. Finally, by comparing the clinical features of the 16 CNVs affecting the ARX locus, we conclude that—depending on the involvement of tissue-specific enhancers—the ARX duplications are ID-associated risk CNVs with variable expressivity and penetrance.
1. Introduction
The aristaless-related homeobox gene (ARX; MIM 300382), which belongs to the aristaless-related subset of the paired (Prd) class of homeodomain proteins, encodes an essential transcription factor (TF) involved in the development of the mammalian cortex [1]. In doing this, ARX controls numerous functions such as neuronal stem-cell proliferation, migration and differentiation, axonal guidance, and synaptic activity [2,3,4,5,6,7]. As ARX is located in Xp21.3, loss-of-function (LoF) and partial-LoF mutations contribute to a nearly continuous series of X-chromosome-linked neurodevelopmental disorders (NDDs) ranging from lissencephaly with abnormal genitalia (XLAG; MIM 300215), developmental and epileptic encephalopathy type 1 (DEE1; MIM 308350), X-linked intellectual disability (XLID; 300419), ID with hand dystonia (Partington syndrome; MIM 309510), and autism [8,9,10,11,12]. Of note, many target genes of this pleiotropic TF have been correlated with ID and autism, emphasizing the importance of ARX for brain development and functioning [7,9,13]. Indeed, as previously shown by us, ARX regulates the transcription of the X-linked NDD gene Lysine-specific demethylase 5C (KDM5C/JARID1C/SMCX; MIM 314690). We also established that ARX belongs to a convergent transcriptional axis that involves other NDD genes such as Synapsin I (SYN1; MIM 313440) and Sodium channel neuronal type II alpha subunit (SCN2A; MIM 182390) [9,14,15]. Most importantly, it has been suggested that the tissue-specific activities of ARX are determined by physical interactions with other transcription factors, such as MyoD and Mef2C [16]; with cofactor proteins, such as Groucho/transducin-like enhancer split protein (TLE); or with Wnt signaling [17,18]. In mice, the complete knockdown of Arx results in a severe defective corticogenesis with a phenotype similar to the clinical manifestations observed in XLAG individuals with ARX LoF mutations [19,20]. Unlike the effects caused by point mutations, it is unclear how duplications of the ARX locus can interfere with brain development and functioning. Some studies proposed that ARX duplications do not have detrimental effects on the brain. Thus, the abnormal neurological phenotype was ascribed to the presence of an extra copy resulting from the effect of other not-yet-identified chromosomal or molecular anomalies, alone or in association with the ARX duplication [21]. However, other studies proposed that the genomic duplications of the ARX locus could affect an auto-regulatory feedback loop of ARX mediated by ultraconserved long-range enhancers [22,23]. In mice, these regulatory regions are specifically active in the forebrain; furthermore, animals with single or pairwise deletions of these regions showed structural brain defects and alterations of specific neuron populations [24]. Based on these findings, we cannot exclude that copy-number gain may alter the expression level of ARX in specific brain areas during embryonic development. Furthermore, as witnessed by several chromosome conformation capture (3C) studies, genomic duplications alter the chromatin portioning units, leading to the formation of new topologically associated domains (neo-TAD) [25,26,27]. Mechanistically, changes in the genomic architecture of a developmentally regulated gene could disturb the activity of its regulatory elements and cause developmental disorders with variable expressivity. Remarkably, duplications of the Xp21.3 region, where ARX is located, were found in male patients presenting varying degrees of cognition defects [21,22,28,29,30,31]. Moreover, the correlation of genotype–phenotype was not fully resolved because, out of the 15 Xp21.3 duplications described in the literature, two were detected in two individuals with normal intelligence [21]. The unpredictable and variable phenotypic outcome associated with Xp21.3 duplications poses an enormous diagnostic difficulty to the clinician and, as such, it challenges the general paradigm according to which rearrangements involving dose-sensitive genes may induce a disease. In this framework, finding out whether ARX-target genes and related molecular pathways are sensitive to the amount of ARX would be an important step in understanding the molecular effects caused by an extra copy of this crucial brain TF. Here, we provide an overview of the 15 ARX duplications reported in the literature and one new case identified by us. We also propose that ARX is a dosage-sensitive regulator of the target genes involved in neuronal maturation [9,15], providing an entry point to study the molecular effects resulting from ARX duplication. We also reconstructed, for the first time, the three-dimensional (3D) structure of the Arx locus, both in mouse embryonic stem cells (ESC-46C) and murine cortical neurons (CNs); this allowed us to identify the TAD features underlining changes during neuronal cortical commitment. In the context of ID pathology, understanding the impact of genomic structural variants on the transcriptional regulation of ARX could aid in the interpretation of rare structural variants affecting this locus, and ameliorate the diagnosis and classification of ARX-related phenotypes.
2. Duplications of the p23.1 Region of the X Chromosome
Segmental duplications of the p21.3 region on the short arm of X chromosome—where ARX is located—have been identified in several patients with ID [32]. They are rare, vary in size (from a few kilobases to many megabases) and their clinical evaluation remains debated [32]. To date, 15 duplications of Xp23.1 have been reported (Figure 1 and Table 1). Whibley et al. [32] describe a 41 kb duplication, maternally inherited, identified in a male patient with moderate ID (family 505). This duplication, in direct tandem orientation, includes the terminal exon of Polymerase DNA alpha-1 (POLA1; MIM 312040), a ubiquitously expressed gene encoding the catalytic subunit of DNA polymerase, and across the entire coding sequence of ARX (Table 1). Of note, mutations in POLA1 have been found in patients presenting a multisystemic disorder without neurological symptoms, named X-linked reticulate pigmentary disorder (PDR; MIM 301220) [33].
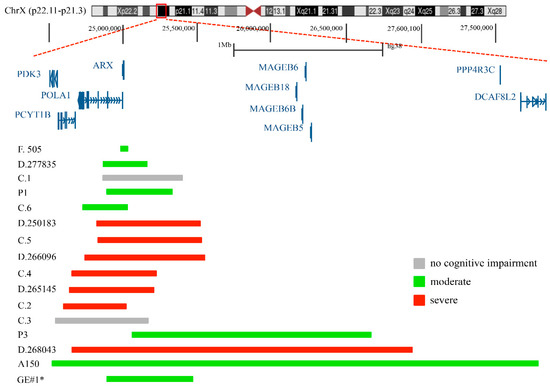
Figure 1.
Genomic mapping of duplications encompassing the ARX locus. Of them, 15 have already been described in the literature, and one, GE#1*, is described in this report for the first time.
Popovici et al. [21] reports on four duplications of Xp21.3. Two of them, a 440 Kb and a 580 Kb duplication, included the full copies of POLA1 and ARX genes and were found in two males with syndromic ID (case 2 and case 4; Table 1); meanwhile, the remaining two—a 540 Kb duplication, which included a full copy of the ARX gene, and a 630 Kb duplication, which included full copies of Phosphate cytidyltransferase 1 choline beta isoform (PCYT1B; MIM 300948), POLA1 and ARX—were found in two males with diagnoses of normal intelligence (case 1 and case 3; Table 1). In addition, the authors define two other duplications reported in public databases: a 720 Kb de novo duplication that contained a full copy of PCYT1B, POLA1 and ARX genes, identified in a boy with syndromic ID (case 5, reported by DECIPHER https://www.deciphergenomics.org/, accessed on 31 January 2022); and a 290 Kb duplication covering partial POLA1 and ARX genes, identified in a patient with developmental delay (reported by the ISCA consortium; case 6; Table 1).

Table 1.
ARX duplications and clinical spectrum.
Table 1.
ARX duplications and clinical spectrum.
| Patient | Extent (kb) | ChrX Genomic Coordinates (UCSC Release) | Duplicated Genes | Duplicated Enhancers | Clinical Signs/Reference |
|---|---|---|---|---|---|
| 505 | 41.1 | 24,992,915–25,033,979 (GRCh37/hg19) 24,974,798–25,015,862 (GRCh38/hg38) | ARX (full gene) | hs121, hs122, hs145 | Moderate Syndromic ID [32] |
| Case 1 | 540 | 24,861,402–25,398,496 (GRCh37/hg19) 24,843,285–25,380,379 (GRCh38/hg38) | ARX (full gene) | hs118, hs119, hs121, hs122, hs145 | No clinical signs [21] |
| Case 2 | 440.5 | 24,593,306–25,033,770 (GRCh37/hg19) 24,575,189–25,015,653 (GRCh38/hg38) | POLA1 (full gene) ARX (exons 2-5) | hs118, hs119, hs121, hs122, hs145 | Severe ID, Microphthalmia, growth retardation [21] |
| Case 3 | 630 | 24,537,027–25,163,704 (GRCh37/hg19) 24,518,910–25,145,587 (GRCh38/hg38) | PCYT1B (full gene); POLA1 (full gene); ARX (full gene), | hs118, hs119, hs121, hs122, hs145 | No clinical signs [21] |
| Case 4 | 580 | 24,650,157–25,230,368 (GRCh37/hg19) 24,632,040–25,212,251 (GRCh38/hg38) | POLA1 (full gene), ARX (full gene) | hs118, hs119, hs121, hs122, hs145 | autism, hyperactivity, delayed speech [21] |
| Case 5 | 720 | 24,542,008–25,542,728 (GRCh37/hg19) 24,523,891–25,524,611 (GRCh38/hg38) | PCYT1B (full gene); POLA1 (full gene); ARX (full gene), | hs118, hs119, hs121, hs122, hs145, hs123 | ID, psychiatric abnormalities, delayed speech [21] |
| Case 6 | 290 | 24,733,304–25,022,540 (GRCh37/hg19) 24,715,187–25,004,423 (GRCh38/hg38) | POLA1 (partial), ARX (partial) | hs118, hs119, hs121, hs122, hs145 | Developmental delay [21] |
| A150 | 3350 | 24,513,979–27,864,451 (GRCh37/hg19) 24,495,862–27,846,334 (GRCh38/hg38) | PCYT1B (full gene), POLA1 (full gene); ARX (full gene) & others | hs118, hs119, hs121, hs122, hs145, hs123 | Autism [30] |
| DECIPHER 277835 | 302 | 24,843,484–25,145,646 (GRCh38/hg38) | ARX (full gene) | hs118, hs119, hs121, hs122, hs145, hs123 | Moderate ID [30] |
| DECIPHER 268043 | 2300 | 24,810,754–27,125,219 (GRCh37/hg19) 24,792,637–27,107,102 (GRCh38/hg38) | ARX (full gene) &others | hs118, hs119, hs121, hs122, hs145, hs123 | ID; short stature [30] |
| DECIPHER 250183 | 717 | 24,807,990–25,524,611 (GRCh37/hg19) 24,789,873–25,506,494 (GRCh38/hg38) | ARX (full coding region) | hs118, hs119, hs121, hs122, hs123, hs145 | Behavioral abnormality; ID; delayed speech [30] |
| DECIPHER 265145 | 580 | 24,632,040–25,212,251 (GRCh37/hg19) 24,613,923–25,194,134 (GRCh38/hg38) | POLA1 (full coding region) ARX (full coding region) | hs118, hs119, hs121, hs122, hs145 | attention deficit; hyperactivity; autism; delayed speech [30] |
| P1 | 438 | 24,887,676–25,325,777 (GRCh37/hg19) 24,869,559–25,307,660 (GRCh38/hg38) | ARX (full coding region) | hs118, hs119, hs121, hs122, hs145 | mild ID, speech delay and hypotonia [22] |
| P3 | 377 | 24,677,441–25,054,698 (GRCh37/hg19) 24,659,324–25,036,581 (GRCh38/hg38) | POLA1 (full coding region) ARX (full coding region) | hs118, hs119, hs121, hs122, hs145 | Developmental delay, growth retardation, delayed speech [22] |
| DECIPHER 266096 DP3 | 813 | 24,741,372–25,554,818 (GRCh37/hg19) 24,723,255–25,536,701 (GRCh38/hg38) | ARX (full coding region) | hs118, hs119, hs121, hs122, hs145, hs123 | hypotonia, microcephaly [22] |
| GE#1 | 803 | 24,828,871–25,631,863 (GRCh38/hg38) | ARX (full coding region) | hs118, hs119, hs121, hs122, hs145, hs123 | Moderate ID; psychomotor retardation, behavioural and developmental disorder [this report] |
Egger et al. [30] describe a de novo 3.3 Mb duplication containing the full copy of ARX and other genes, detected in a male patient with autism (A150; Table 1). They also report on four gain CNVs that are present in the DECIPHER database: a 302 Kb duplication (DECIPHER 277835) including the full copy of the ARX locus, detected in a male patient with moderate ID; a 2.3 Mb duplication (DECIPHER 268043), identified in a male patient with ID; a 717 Kb duplication (DECIPHER 250183), detected in a male patient with ID and behavioral abnormality; and a 580 Kb duplication (DECIPHER 265145), found in a male patient with autism and delayed speech and language development (Table 1). In Ishibashi et al. [22], three gain CNVs including the ARX locus were described: a 438 Kb duplication (P1) including the full copy of ARX, detected in a male patient with mild ID, speech delay and hypotonia; a 377 Kb duplication (P3) including a full copy of POLA1 and ARX, found in a male patient with developmental delay, growth retardation, and delayed speech; and a 813 Kb duplication (DECIPHER 266096 DP3), detected in a male patient with hypotonia and microcephaly (Table 1). Additionally, we identified, in a male patient with moderate ID and psychomotor retardation (GE#1; see Supplementary Materials), a new interstitial duplication of Xp22.11-p21.3 with an estimated size of at least 803 Kb including the full copy of ARX (Table 1). Summarizing what is described above, except for two cases (case 1 and case 3), all the published duplications and the new one reported here were identified in patients with various neurodevelopmental problems, intellectual disabilities and autism (Table 1) [22,30,32]. Furthermore, all these duplications show different breakpoint positions around the ARX locus, and in several cases, they include a gene desert area (1.1 Mb length) located between ARX (hg38 chrX: 25,003,694–25,015,965) and the MAGE family member B18 (MAGEB18; hg38 chrX:26,138,343–26,140,736).
3. Identification of a Novel Xp22.11-p21.3—Duplication in a Male Child with Moderate ID and Analysis of Functional Implications
A new interstitial duplication of Xp22.11-p21.3 was detected by CGH array in a male child with moderate ID and his asymptomatic mother (family GE#1; Figure 2A). As determined by the UCSC Genome database (GRCh38/hg38 release), this rearrangement has an estimate size at least of 803 Kb, from nucleotide position 24.828.871 (Xp22.11) to 25.631.863 (Xp21.3) (Figure 2A,B). By inspecting the sequence content, we established that this interstitial duplication contained a partially duplicated POLA1 locus, an additional copy of the ARX locus (including intact duplicated promoter sequence and all enhancer elements) and a non-actively transcribed gene region (Figure 2B). We analyzed the effect of this rearrangement in the lymphoblastoid cell line derived from the ID patient (ID-LCLs) compared to LCLs derived from the control XY individual. Accordingly, with the presence of two active gene copies of ARX, we observed about a 3,5-fold increased expression of the ARX transcript with respect to control cells (Figure 2C). On the contrary, no difference in the level of POLA1 expression was found between the control and patient cells (Supplementary Figure S1A), indicating that this gene is not involved in the disease phenotype.
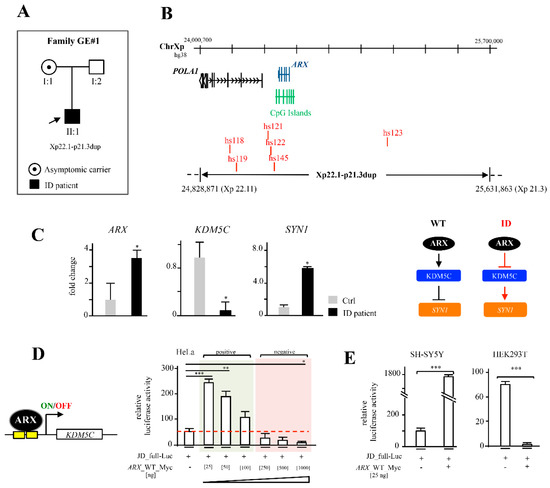
Figure 2.
Identification of a novel ARX duplication and its functional implications. Genealogical tree of family GE#1 and segregation of Xp22.1-p21.3dup: (A) Physical map of the ARX duplication on the human X chromosome (ChrX). (B) The involved genes POLA1 and ARX, the CpG Islands and the ultraconserved enhancers expressed in embryonic mouse brain (VISTA Enhancer Browser at https://enhancer.lbl.gov/, accessed on 24 January 2022) are shown. ARX, KDM5C and SYN1 expression levels in GE#1 ID patient’s lymphoblastoid cell lines (LCLs) and XYWT LCLs (C). Results were normalized using HPRT1 as reference gene and shown as mean ± SD from three independent experiments. p < 0.05. Schematic representation of ARX-KDM5C-SYN1 regulatory path is shown. Dose- and cell-type-dependent activity of ARX on 5′ KDM5C regulatory region (D,E). Luciferase assay in HeLa cells at increasing concentrations of ARX and Luciferase assays in HEK293T and SHSY5Y cell lines. Data were normalized to relative luciferase activity of JD-full-Luc. Results are shown as mean ± SD from three independent experiments. p < 0.05 *; p < 0.005 **; p < 0.0005 ***.
It has been estimated that gene dosage alterations caused by structural variations are responsible for ~15% of NDD cases [34]. Given the extra dosage of ARX transcript, we evaluated the expression levels of two ARX-related disease genes involved in ID and epilepsy: KDM5C, a direct target of ARX—as already established by us in both human and murine cell systems [9,14,35]—and SYN1, a gene found physiologically repressed by KDM5C [14,15]. A marked downregulation of KDM5C/KDM5C, both at mRNA and protein levels (Figure 2C and Supplementary Figure S1B), coupled with the overexpression of SYN1, was observed in ID-LCLs with respect to the control (Figure 2C). As ARX can activate or repress target promoters [7,9], we next analyzed whether the transcriptional activity of ARX operates in a dosage-dependent manner. By means of a luciferase assay performed in HeLa cells, we tested the promoter response of KDM5C, varying the amount of the transfected construct expressing the ARX WT cDNA (ARX_Myc-WT). Thus, an inverse correlation between the reporter response of the KDM5C promoter (JD_full_Luc) and the ARX dosage was found (Figure 2D). This evidence suggests that ARX may act on the KDM5C promoter as repressor at a high concentration, and as activator at a low concentration (Figure 2D). Next, we verified whether this double function of ARX on KDM5C is cell-context dependent by testing the transient activity of ARX on the KDM5C promoter in two different human cell lines. We found that a low concentration of ARX [25 ng] transiently transfected in HEK293T and SH-SY5Y induces the repression or the stimulation of KDM5C promoter, respectively (Figure 2E), indicating that the bi-functional activity of ARX is dose- and cell-context dependent.
4. Ultraconserved ARX/Arx Brain Enhancers
Similarly to other developmentally expressed TF loci, the ARX locus has a high density of multiple and very long human/rodent ultraconserved enhancers [23,24,36,37,38,39]. As annotated in the VISTA enhancer database, all these regulatory elements, except one, are located downstream of the coding sequence of ARX, between the ARX and POLA1 loci (Figure 3A) [36]. In the mouse forebrain, four of them (hs119, hs121, hs122 and hs123) showed enhancer activity similar to the Arx gene expression (Figure 3B). Of note, mice with single or pairwise deletions of ultraconserved Arx enhancers presented neurological or growth abnormalities (Figure 3C) [24]. In particular, the loss of enhancer hs121 (hs121-KO) caused altered densities of specific cortical interneuron types, including cholinergic neurons; meanwhile the loss of enhancer hs122 (hs122-KO), which is active in the dorsal forebrain, caused a reduced expression of Arx in the dorsal forebrain and changes in the size and morphology of the hippocampus. Regarding hs119, its deletion caused growth abnormalities and reduced body mass (Figure 3C) [24]. Very interestingly, hemizygous double-knockout mice for hs119 and hs121 (hs119/hs121-KO) displayed a more severe combination of defects when each enhancer was deleted individually (Figure 3C). Furthermore, several of the neuronal alterations observed in single- and double-knockout mice were very similar to a subset of defects observed in Arx KO mice [3,19,38,39]. These findings sustain not only the crucial role of these enhancers in the regulation of Arx, but also reinforce the concept that rearrangements involving the ARX locus could interfere with their regulatory activity. Moreover, a functional analysis carried out in zebrafish revealed that three of the ultraconserved enhancers mediate negative and positive autoregulation of ARX in specific brain regions [22]. The authors proposed a model, explaining how breakpoints in long-range enhancers might alter the expression levels of Arx in specific brain regions, and how this effect can lead to subtle neuronal phenotypes [22]. With this in mind, we therefore conclude that perturbations of long-range activity of ARX/Arx ultraconserved enhancers could have dosage-sensitivity-driven effects, and thus, constitute a risk for neurodevelopmental disorders.
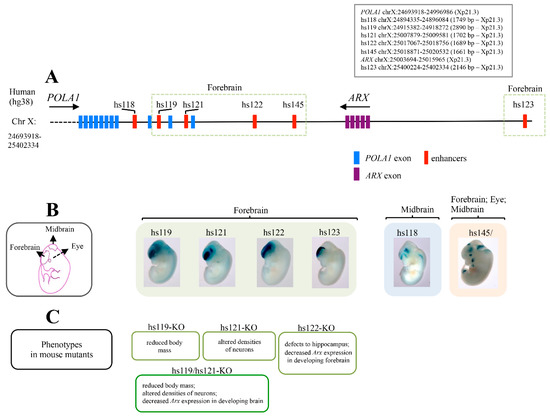
Figure 3.
The ARX locus has a high density of forebrain ultraconserved sequences. Schematic representation of Xp region containing the multiple ultraconserved sequences located near the ARX locus, created on the annotation of UCSC hg38 assembly. Nucleotide positions are shown in box (A). Whole-mount staining images, obtained freely from VISTA Enhancer Browser (https://enhancer.lbl.gov/, accessed on 24 January 2022), showing the expression profiles of each enhancer in E11.5 embryos. Schematic representation of E11.5 brain subregions is shown in (B). Summary of phenotypic and molecular features of knockout mice lacking individual, or combinations of, ultraconserved Arx enhancers in shown in (C).
5. Three-Dimensional (3D) Structure of the Arx Locus in Embryonic Stem Cells (ESs) and Cortical Neurons (CNs)
The alteration of the 3D structure of chromosomes is emerging as one of the causes of ID [25]. Mechanistically, when chromosomal rearrangement events affect TAD boundaries, they alter the interactions between genes and enhancers, leading to an abnormal expression of genes. Given the complexity of the ARX/Arx locus, we investigate its architecture, analyzing published murine HiC data in embryonic stem cells (ESs) and cortical neurons (CNs) [40]. Thus, we focused on a 5 Mb genomic region (chrX:91,000,000–96,000,000, UCSC mm10) encompassing the Arx gene and its ultraconserved enhancers [23]. From the analysis of the contact maps, it emerges that the locus undergoes structural rearrangements involving the Arx gene and the enhancer elements, highlighting how architectural features have a strong functional impact on the regulation of this gene (Figure 4). In ESs, the Arx gene tends to interact locally with its most proximal enhancers (hs119, hs121 and hs122) and interacts weakly with its flanking regions (Figure 4B, left matrix). In CNs, in addition to the contact with the most proximal enhancers (hs119, hs121 and hs122), we observed long-range contacts with the upstream region involving the most distant hs123 ultraconserved enhancer, located in a large, well-defined interaction domain (Figure 4B, right matrix). These features are compatible with a transcriptional activity change in Arx; it is inactive in ESs, and active in CNs in which all four of the ultraconserved enhancers cooperate to control the Arx expression [24]. Next, we investigated the 3D structure of the locus in both cell lines by employing the Strings and Binder Switch polymer model [41,42,43,44]. The model assumes that the chromatin filament (the string) has a specific binding site that interacts with molecular factors (the binders), which are naturally present in the nuclear environment. Moreover, the interaction between binding sites and binders forms stable loops, and shapes the locus organization (see Supplementary Materials). The contact maps obtained from the model effectively recapitulate the interaction pattern of experimental data (Pearson r = 0.98, Figure 4A, bottom matrices). Regarding the distribution of the binding sites, the studied region (Figure 4B) reveals specific architectural features: in ESs, the Arx gene is associated to a binding domain (number 9) containing the hs119, hs121 and hs122 enhancers, but not the hs123; in CNs, this binding domain changes and spreads, involving the hs123 enhancer, in agreement with HiC data. In order to give a structural rationale to these findings, we performed Molecular Dynamics (MD) simulations and reproduced the 3D architecture in both ES and CN cells (Figure 4C). For visualization purposes, we divided the polymer in three consecutive regions; then, we colored them according to the binding domain distributions. We found that in ESs the three regions tend to form locally self-interacting domains, weakly interacting with each other. Conversely, in CNs, we found that the green and blue regions interact strongly in order to bring the enhancer hs123 into close proximity with Arx, ensuring its activity (Figure 4C). Based on this evidence, we therefore proposed that the long-range interactions between the regulatory elements of Arx and its promoter could differently regulate the expression of this pleiotropic TF during embryonic stem-cell proliferation and neuron differentiation. Very interestingly, comparison of ARX expression data across brain development in mice, humans and human-derived brain organoids, available in the Human and Mouse Development atlas (http://www.humous.org/, accessed on 2 February 2022) [45], revealed a distinctive time- and cell-dependent expression pattern in the mouse neocortex, human neocortex and human-derived brain organoids (Figure 5A–C). In particular, in the murine cortex, Arx is strongly expressed in apical radial glia cells (aRGs) at E12, and is decreased in immature neurons (iNs) and mature neurons (mNs), along the following corticogenesis stages (E15; Figure 5A). Conversely in the human cortex, ARX is strongly expressed in basal radial glia cells (bRGs) and iNs at gestation week 8, and decreased along the following embryonic stages (at week 20; Figure 5B); meanwhile in human-derived brain organoids ARX is expressed in RG cells at the early phase of in vitro culture (at week 3; Figure 5C). Based on these findings, we therefore conclude that the fine regulation of ARX expression in time and space mirrors the architectural features of its genetic locus and the relative regulatory elements.
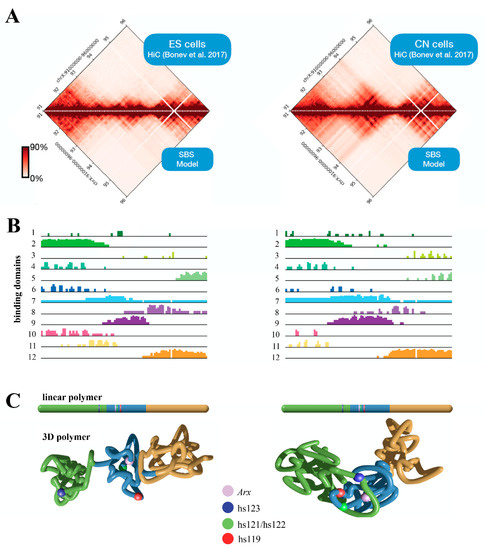
Figure 4.
Polymer physics description of the murine Arx locus. Bottom matrices and contact maps predicted from the SBS polymer model by analysing the genomic region chrX:91,000,000–95,000,000 (UCSC, mm10). HiC data were obtained from a previous study [20] (A). Binding domain distributions for ESs (left) and CNs (right) (B). Snapshots from real MD simulations describing the 3D structure in ESs (left) and CNs (right). The colour scheme used is reported in the linear bar above (C).
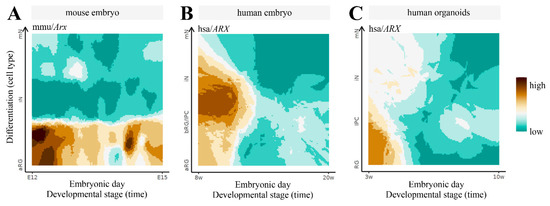
Figure 5.
Spatiotemporal gene expression trajectories of Arx/ARX across the corticogenesis: in mouse, from the embryonic stage E12 to E15 (A); in humans, from gestation week 8 to 20 (B); and in human-derived brain organoids, from in vitro developmental week 3 to 10 (C). RG, radial glia; aRG, apical radial glia; vRG, ventral radial glia; oRG, outer radial glia; IPC, intermediate progenitor cell; BP, basal progenitor; N, neuron; iN, immature neuron; mN, mature neuron. Data can be accessed freely on http://www.humous.org/, accessed on 2 February 2022.
6. Genotype-Phenotype Correlation
Sixteen distinct Xp21.3 duplications with different breakpoints, ranging in size from 41.1 kb to 3.3 Mb, have been identified to date (Table 1). Clinical findings in Xp21.3 duplication include syndromic ID (from mild–moderate to severe), global developmental delay, hypotonia, autism, and hyperactivity (Table 1). The majority of these rearrangements include a full copy of ARX with the telencephalon enhancers, and a full copy or break of further genes. Remarkably, the smallest one—identified in a patient with moderate syndromic ID (Patient 505) [32]—includes the full copy of ARX and the enhancer elements hs121, hs122 and hs145 (Table 1). In contrast, large duplications include full copies or breakages of PCYT1B and/or POLA1, two genes not associated with central nervous system disorders [33]. The molecular characterization of these rearrangements suggests that the core of ID syndrome is largely due to the duplication of the ARX locus. In this context, the activity of the duplicated telencephalon enhancers could contribute to the neurological features. Indeed, similarly to other developmentally expressed genes with pleiotropic activity, ARX has multiple enhancers orchestrating its regulation in time and space in the developing brain [23,24,36,37,38,39]. Thus, a structural alteration within these regions can specifically disturb the transcriptional regulation of ARX. Starting with these considerations, we propose that the ID variability observed in patients with Xp21.3 duplication could be due to disturbed cis-regulatory mechanisms of ARX expression that, in turn, lead to different syndromic conditions. Particular consideration should be given to the two duplications reported as asymptomatic by Popovici et al. [21] (case 1 and case 3), which raised the general question of whether the duplications of Xp21.3 are pathogenetic. As already described, two male individuals with normal IQ, carrying two different duplications of 540 kb (case 1) and 630 kb (case 3), were identified 21]. Both rearrangements enclosed the ARX gene and the enhancer elements hs118, hs119, hs121, hs122 and hs145 [21]. Since this region has been found duplicated in several male patients with varying degrees of ID and autism, it does not seem plausible that these rearrangements are completely asymptomatic. As previously commented by others, the presence of these two duplications could be attributable to an untested somatic mosaicism condition [22]. Consequently, the two rearrangements could be absent in the brain tissues or present in a small fraction of cells, and thus, could not disturb the control of ARX expression in the developing brain. As an alternative explanation, we cannot rule out that the non-ID phenotypes associated with these two duplications could be the result of variable phenotypic penetrance, a feature often occurring in syndromic ID conditions. Altogether, these considerations strongly support the concept that Xp21.3 duplications are ID-associated risk CNVs with variable expressivity and incomplete penetrance. Certainly, all patients with Xp21.3 duplication should receive a thorough genetic and neurological evaluation in order to avoid neglecting early treatments that could help manage the disease. In this regard, it should be reiterated that the close alliance between clinicians, scientists and families could improve both the characterization of the ID phenotypes and the management of patients.
7. Discussion & Conclusions
Patients with various neurodevelopmental problems, intellectual disabilities and autism, were identified to have duplications encompassing the ARX gene locus. Although ARX rearrangements are rare, thus making access to a wide number of cases difficult, the identification of new cases with CNVs in Xp21.3 allows us to improve our knowledge on ARX and to stimulate the pursuit of further functional studies. Indeed, in spite of the progress made over the years on the functional impacts of point mutations in ARX, the need remains to clarify how duplications of specific ARX enhancers can alter its bi-functional dose-dependent transcriptional activity. First evidence suggests that these rearrangements may disturb the pleiotropic tunable bifunctional activity of ARX, and thus, may cause neurodevelopmental defects. Indeed, these gain CNVs seem to affect both the protein-coding portion of ARX and the copy number and/or the position of the telencephalon enhancers. As a result, they can reshuffle the higher-order chromatin structure of the ARX locus. Remarkably, although enhancer duplications may impact gene expression in unpredictable ways [46], structural variants can disrupt the 3D structure of a gene locus, modifying the spacing between enhancers and their position relative to the promoter [47]. Since the level of expression of ARX is highly sensitive to the genomic architecture, as observed in mice [24] and GE#1 patient LCLs, we hypothesize that changes in the genomic architecture of the ARX locus may dramatically impact its expression levels and, in turn, on the activity of its protein product. Moreover, the 3D structure of the Arx locus revealed differences between mouse ESs and cortical neurons in the long-range interactions of the enhancer elements with its promoter. Given the multiple activities of ARX in the brain [48], perturbations of its transcriptional regulation during the critical phases of embryonic development—such as RG proliferation, and neuron differentiation and maturation—could alter the cortical multilayers [1,2,3,4,5,6,7].
In conclusion, duplications of the Xp21.3 region can lead to cognition defects by disrupting the 3D structure of the ARX locus and altering the dose-sensitivity action of ARX, which is crucial for corticogenesis [1,2,48]. Very importantly, further studies aimed at integrating clinical data with genetic and molecular findings are required to dissect the specific functional processes that are damaged in patients with Xp21.3 duplication, providing prognostic indicators and therapeutic perspectives.
Supplementary Materials
Supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23063084/s1.
Author Contributions
Writing—Original Draft Preparation, L.P.; Methodology and Validation, M.M. and A.P.; Data Curation, D.D. and L.V.; Methodology and Data Curation, A.M.C., S.B. and M.N.; Resources, M.B.L.; Data Curation, M.P., E.S. and D.C.; Conceptualization, Writing—Original Draft Preparation, Review and Editing, M.G.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Jerome Lejeune Foundation grant (1372-MM2015A), the Telethon Foundation grant (GGP14198), and the Italian Ministry of Economic Development grant (F/050011/02/X32) to M.G.M. D.D. was partially supported by a grant from the Maria Rosaria Maglione Onlus Foundation; D.C. and M.M. were partially supported by the grants MSALRC21 (Ministero Salute Ricerca Corrente) and ALBURLORF, DLDG514/20 (Progetto Ricerca collaborative Rete IRCSS IDE).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Regione Liguria (Comitato Etico Regione Liguria) (protocol code 5/2014 and date of approval 13 May 2014) and the Ethics Committee of Istituto Neurologico Mediterraneo Neuromed (protocol 10/2018 in date 29 October 2018) for studies involving humans.
Informed Consent Statement
Informed consent was obtained from the subjects involved in the study.
Data Availability Statement
Publicly archived datasets analyzed: DECIPHER database https://www.deciphergenomics.org/, accessed on 31 January 2022; Human and Mouse Development atlas http://www.humous.org/, accessed on 2 February 2022.
Acknowledgments
We would like to thank the family GE#1 and the healthy control individuals and “SPECIALmente Noi Onlus Foundation” for promoting research into autism spectrum disorder (ASD). We also thank the Genetic Bank of Laboratory of Human Genetics at Gaslini Institute, and members of the Telethon Network of Genetic Biobanks and of the EuroBioBank network, who provided us with specimens.
Conflicts of Interest
The authors declare that they have no conflict of interests.
References
- Strømme, P.; Mangelsdorf, M.E.; Shaw, M.A.; Lower, K.M.; Lewis, S.M.; Bruyere, H.; Lütcherath, V.; Gedeon, A.K.; Wallace, R.H.; Scheffer, I.E.; et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 2002, 30, 441–445. [Google Scholar] [CrossRef]
- Colombo, E.; Galli, R.; Cossu, G.; Gécz, J.; Broccoli, V. Mouse orthologue of ARX; a gene mutated in several X-linked forms of mental retardation and epilepsy; is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev. Dyn. 2004, 231, 631–639. [Google Scholar] [CrossRef]
- Colombo, E.; Collombat, P.; Colasante, G.; Bianchi, M.; Long, J.; Mansouri, A.; Rubenstein, J.L.; Broccoli, V. Inactivation of Arx; the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene; leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J. Neurosci. 2007, 27, 4786–4798. [Google Scholar] [CrossRef]
- Friocourt, G.; Poirier, K.; Rakić, S.; Parnavelas, J.G.; Chelly, J. The role of ARX in cortical development. Eur J. Neurosci. 2006, 23, 869–876. [Google Scholar] [CrossRef]
- Friocourt, G.; Kanatani, S.; Tabata, H.; Yozu, M.; Takahashi, T.; Antypa, M.; Raguénès, O.; Chelly, J.; Férec, C.; Nakajima, K.; et al. Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J. Neurosci. 2008, 28, 5794–5805. [Google Scholar] [CrossRef]
- Seufert, D.W.; Prescott, N.L.; El-Hodiri, H.M. Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev. Dyn. 2005, 232, 313–324. [Google Scholar] [CrossRef]
- Fulp, C.T.; Cho, G.; Marsh, E.D.; Nasrallah, I.M.; Labosky, P.A.; Golden, J.A. Identification of Arx transcriptional targets in the developing basal forebrain. Hum. Mol. Genet. 2008, 17, 3740–3760. [Google Scholar] [CrossRef]
- Laperuta, C.; Spizzichino, L.; D′Adamo, P.; Monfregola, J.; Maiorino, A.; D′Eustacchio, A.; Ventruto, V.; Neri, G.; D’Urso, M.; Chiurazzi, P.; et al. MRX87 family with Aristaless X dup24bp mutation and implication for polyalanine expansions. BMC Med. Genet. 2007, 8, 25. [Google Scholar] [CrossRef]
- Poeta, L.; Fusco, F.; Drongitis, D.; Shoubridge, C.; Manganelli, G.; Filosa, S.; Paciolla, M.; Courtney, M.; Collombat, P.; Lioi, M.B.; et al. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 2013, 92, 114–125. [Google Scholar] [CrossRef]
- Shoubridge, C.; Tan, M.H.; Seiboth, G.; Gécz, J. ARX homeodomain mutations abolish DNA binding and lead to a loss of transcriptional repression. Hum. Mol. Genet. 2012, 21, 1639–1647. [Google Scholar] [CrossRef]
- Fullston, T.; Brueton, L.; Willis, T.; Philip, S.; MacPherson, L.; Finnis, M.; Gecz, J.; Morton, J. Ohtahara syndrome in a family with an ARX protein truncation mutation (c.81C>G/p.Y27X). Eur. J. Hum. Genet. 2010, 18, 157–162. [Google Scholar] [CrossRef]
- Kato, M.; Saitoh, S.; Kamei, A.; Shiraishi, H.; Ueda, Y.; Akasaka, M.; Tohyama, J.; Akasaka, N.; Hayasaka, K.A. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome). Am. J. Hum. Genet. 2007, 81, 361–366. [Google Scholar] [CrossRef]
- Quillé, M.L.; Carat, S.; Quéméner-Redon, S.; Hirchaud, E.; Baron, D.; Benech, C.; Guihot, J.; Placet, M.; Mignen, O.; Férec, C.; et al. High-throughput analysis of promoter occupancy reveals new targets for Arx; a gene mutated in mental retardation and interneuronopathies. PLoS ONE 2011, 6, e25181. [Google Scholar] [CrossRef]
- Poeta, L.; Padula, A.; Attianese, B.; Valentino, M.; Verrillo, L.; Filosa, S.; Shoubridge, C.; Barra, A.; Schwartz, C.E.; Christensen, J.; et al. Histone demethylase KDM5C is a SAHA-sensitive central hub at the crossroads of transcriptional axes involved in multiple neurodevelopmental disorders. Hum. Mol. Genet. 2019, 28, 4089–4102. [Google Scholar] [CrossRef]
- Poeta, L.; Padula, A.; Lioi, M.B.; van Bokhoven, H.; Miano, M.G. Analysis of a Set of KDM5C Regulatory Genes Mutated in Neurodevelopmental Disorders Identifies Temporal Coexpression Brain Signatures. Genes 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Biressi, S.; Messina, G.; Collombat, P.; Tagliafico, E.; Monteverde, S.; Benedetti, L.; Cusella De Angelis, M.G.; Mansouri, A.; Ferrari, S.; Tajbakhsh, S.; et al. The homeobox gene Arx is a novel positive regulator of embryonic myogenesis. Cell Death Differ. 2008, 15, 94–104. [Google Scholar] [CrossRef][Green Version]
- McKenzie, O.; Ponte, I.; Mangelsdorf, M.; Finnis, M.; Colasante, G.; Shoubridge, C.; Stifani, S.; Gécz, J.; Broccoli, V. Aristaless-related homeobox gene; the gene responsible for West syndrome and related disorders; is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience 2007, 146, 236–247. [Google Scholar] [CrossRef]
- Cho, I.T.; Lim, Y.; Golden, J.A.; Cho, G. Aristaless Related Homeobox (ARX) Interacts with β-Catenin; BCL9; and P300 to Regulate Canonical Wnt Signaling. PLoS ONE 2017, 12, e0170282. [Google Scholar] [CrossRef]
- Kitamura, K.; Yanazawa, M.; Sugiyama, N.; Miura, H.; Iizuka-Kogo, A.; Kusaka, M.; Omichi, K.; Suzuki, R.; Kato-Fukui, Y.; Kamiirisa, K.; et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002, 32, 359–369. [Google Scholar] [CrossRef]
- Poirier, K.; Van Esch, H.; Friocourt, G.; Saillour, Y.; Bahi, N.; Backer, S.; Souil, E.; Castelnau-Ptakhine, L.; Beldjord, C.; Francis, F.; et al. Neuroanatomical distribution of ARX in brain and its localisation in GABAergic neurons. Brain Res. Mol. Brain Res. 2004, 122, 35–46. [Google Scholar] [CrossRef]
- Popovici, C.; Busa, T.; Boute, O.; Thuresson, A.C.; Perret, O.; Sigaudy, S.; Södergren, T.; Andrieux, J.; Moncla, A.; Philip, N. Whole ARX gene duplication is compatible with normal intellectual development. Am. J. Med. Genet. A 2014, 164A, 2324–2327. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Manning, E.; Shoubridge, C.; Krecsmarik, M.; Hawkins, T.A.; Giacomotto, J.; Zhao, T.; Mueller, T.; Bader, P.I.; Cheung, S.W.; et al. Copy number variants in patients with intellectual disability affect the regulation of ARX transcription factor gene. Hum. Genet. 2015, 134, 1163–1182. [Google Scholar] [CrossRef] [PubMed]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef] [PubMed]
- Dickel, D.E.; Ypsilanti, A.R.; Pla, R.; Zhu, Y.; Barozzi, I.; Mannion, B.J.; Khin, Y.S.; Fukuda-Yuzawa, Y.; Plajzer-Frick, I.; Pickle, C.S.; et al. Ultraconserved Enhancers Are Required for Normal Development. Cell 2018, 172, 491–499. [Google Scholar] [CrossRef]
- Li, R.; Liu, Y.; Li, T.; Li, C. 3Disease Browser: A Web server for integrating 3D genome and disease-associated chromosome rearrangement data. Sci. Rep. 2016, 6, 34651. [Google Scholar] [CrossRef] [PubMed]
- Franke, M.; Ibrahim, D.M.; Andrey, G.; Schwarzer, W.; Heinrich, V.; Schöpflin, R.; Kraft, K.; Kempfer, R.; Jerković, I.; Chan, W.L.; et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 2016, 538, 265–269. [Google Scholar] [CrossRef]
- Bianco, S.; Lupiáñez, D.G.; Chiariello, A.M.; Annunziatella, C.; Kraft, K.; Schöpflin, R.; Wittler, L.; Andrey, G.; Vingron, M.; Pombo, A.; et al. Polymer physics predicts the effects of structural variants on chromatin architecture. Nat. Genet. 2018, 50, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Thorson, L.; Bryke, C.; Rice, G.; Artzer, A.; Schilz, C.; Israel, J.; Huber, S.; Laffin, J.; Raca, G. Clinical and molecular characterization of overlapping interstitial Xp21-p22 duplications in two unrelated individuals. Am. J. Med. Genet. Part A 2010, 152A, 904–915. [Google Scholar] [CrossRef]
- Wu, L.; Liu, J.; Lv, W.; Wen, J.; Xia, Y.; Liang, D. An Xp21.3p11.4 duplication observed in a boy with intellectual deficiency and speech delay and his asymptomatic mother. Birth Defects Res. A Clin. Mol. Teratol. 2013, 97, 467–470. [Google Scholar] [CrossRef]
- Egger, G.; Roetzer, K.M.; Noor, A.; Lionel, A.C.; Mahmood, H.; Schwarzbraun, T.; Boright, O.; Mikhailov, A.; Marshall, C.R.; Windpassinger, C.; et al. Identification of risk genes for autism spectrum disorder through copy number variation analysis in Austrian families. Neurogenetics 2014, 15, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Piccione, M.; Sanfilippo, C.; Cavani, S.; Salatiello, P.; Malacarne, M.; Pierluigi, M.; Fichera, M.; Luciano, D.; Corsello, G. Molecular and clinical characterization of a small duplication Xp in a human female with psychiatric disorders. J. Genet. 2011, 90, 473–477. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abidi, F.; Fullston, T.; Choma, M.K.; Boucher, C.A.; Shepherd, L.; Willatt, L.; Parkin, G.; Smith, R.; Futreal, P.A.; Shaw, M.; et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am. J. Hum. Genet. 2010, 87, 173–188. [Google Scholar]
- Starokadomskyy, P.; Gemelli, T.; Rios, J.J.; Xing, C.; Wang, R.C.; Li, H.; Pokatayev, V.; Dozmorov, I.; Khan, S.; Miyata, N.; et al. DNA polymerase-α regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat. Immunol. 2016, 17, 495–504. [Google Scholar] [CrossRef] [PubMed]
- D’haene, E.; Vergult, S. Interpreting the impact of noncoding structural variation in neurodevelopmental disorders. Genet. Med. 2021, 23, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Shulha, H.; Weng, Z.; Akbarian, S. Regulation of histone H3K4 methylation in brain development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130514. [Google Scholar] [CrossRef] [PubMed]
- Visel, A.; Taher, L.; Girgis, H.; May, D.; Golonzhka, O.; Hoch, R.V.; McKinsey, G.L.; Pattabiraman, K.; Silberberg, S.N.; Blow, M.J.; et al. A high-resolution enhancer atlas of the developing telencephalon. Cell 2013, 152, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A.; Ahituv, N.; Moses, A.M.; Prabhakar, S.; Nobrega, M.A.; Shoukry, M.; Minovitsky, S.; Dubchak, I.; Holt, A.; Lewis, K.D.; et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 2006, 444, 499–502. [Google Scholar] [CrossRef]
- Snetkova, V.; Ypsilanti, A.R.; Akiyama, J.A.; Mannion, B.J.; Plajzer-Frick, I.; Novak, C.S.; Harrington, A.N.; Pham, Q.T.; Kato, M.; Zhu, Y.; et al. Ultraconserved enhancer function does not require perfect sequence conservation. Nat. Genet. 2021, 53, 521–528. [Google Scholar] [CrossRef]
- Snetkova, V.; Pennacchio, L.A.; Visel, A.; Dickel, D.E. Perfect and imperfect views of ultraconserved sequences. Nat. Rev. Genet. 2021, 23, 182–194. [Google Scholar] [CrossRef]
- Bonev, B.; Mendelson Cohen, N.; Szabo, Q.; Fritsch, L.; Papadopoulos, G.L.; Lubling, Y.; Xu, X.; Lv, X.; Hugnot, J.P.; Tanay, A.; et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 2017, 171, 557–572. [Google Scholar] [CrossRef]
- Barbieri, M.; Chotalia, M.; Fraser, J.; Lavitas, L.M.; Dostie, J.; Pombo, A.; Nicodemi, M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA 2012, 109, 16173–16178. [Google Scholar] [CrossRef] [PubMed]
- Chiariello, A.M.; Esposito, A.; Annunziatella, C.; Bianco, S.; Fiorillo, L.; Prisco, A.; Nicodemi, M. A Polymer Physics Investigation of the Architecture of the Murine Orthologue of the 7q11.23 Human Locus. Front. Neurosci. 2017, 11, 559. [Google Scholar] [CrossRef] [PubMed]
- Chiariello, A.M.; Bianco, S.; Oudelaar, A.M.; Esposito, A.; Annunziatella, C.; Fiorillo, L.; Conte, M.; Corrado, A.; Prisco, A.; Larke, M.; et al. A Dynamic Folded Hairpin Conformation Is Associated with α-Globin Activation in Erythroid Cells. Cell Rep. 2020, 30, 2125–2135. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, L.; Musella, F.; Conte, M.; Kempfer, R.; Chiariello, A.M.; Bianco, S.; Kukalev, A.; Irastorza-Azcarate, I.; Esposito, A.; Abraham, A.; et al. Comparison of the Hi-C, GAM and SPRITE methods using polymer models of chromatin. Nat. Methods 2021, 18, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Klingler, E.; Francis, F.; Jabaudon, D.; Cappello, S. Mapping the molecular and cellular complexity of cortical malformations. Science 2021, 371, eaba4517. [Google Scholar] [CrossRef] [PubMed]
- Klopocki, E.; Ott, C.E.; Benatar, N.; Ullmann, R.; Mundlos, S.; Lehmann, K. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J. Med. Genet. 2008, 45, 370–375. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.; Ong, C.T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Drongitis, D.; Caterino, M.; Verrillo, L.; Santonicola, P.; Costanzo, M.; Poeta, L.; Attianese, B.; Barra, A.; Terrone, G.; Lioi, M.B.; et al. Deregulation of microtubule organization and RNA metabolism in Arx models for Lissencephaly and developmental epileptic encephalopathy. Hum. Mol. Genet. 2022, ddac028. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).