RNA Sequencing of Arabidopsis thaliana Seedlings after Non-Thermal Plasma-Seed Treatment Reveals Upregulation in Plant Stress and Defense Pathways
Abstract
:1. Introduction
2. Results
2.1. Germination Rate of Plasma-Treated A. thaliana Seeds
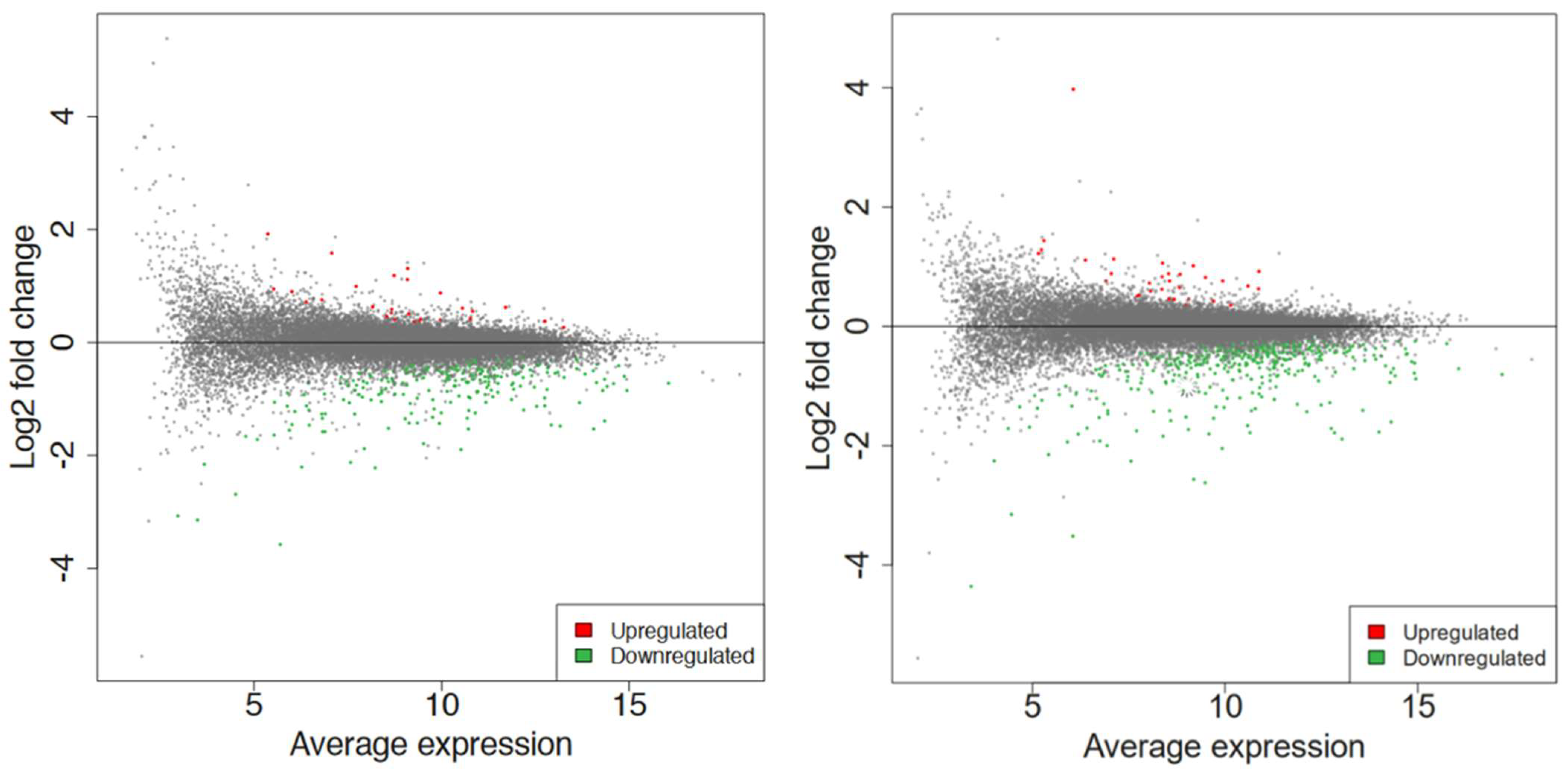
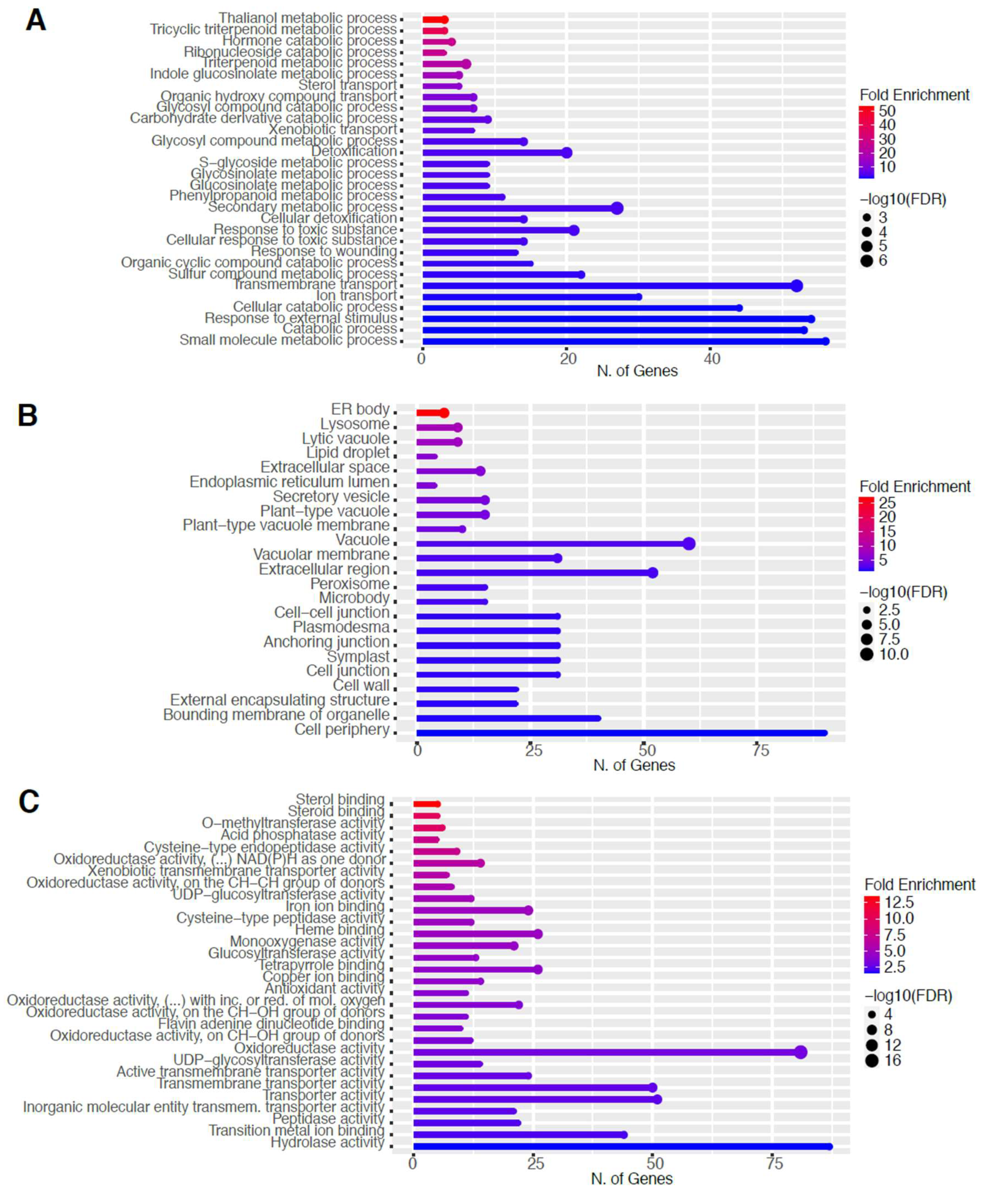
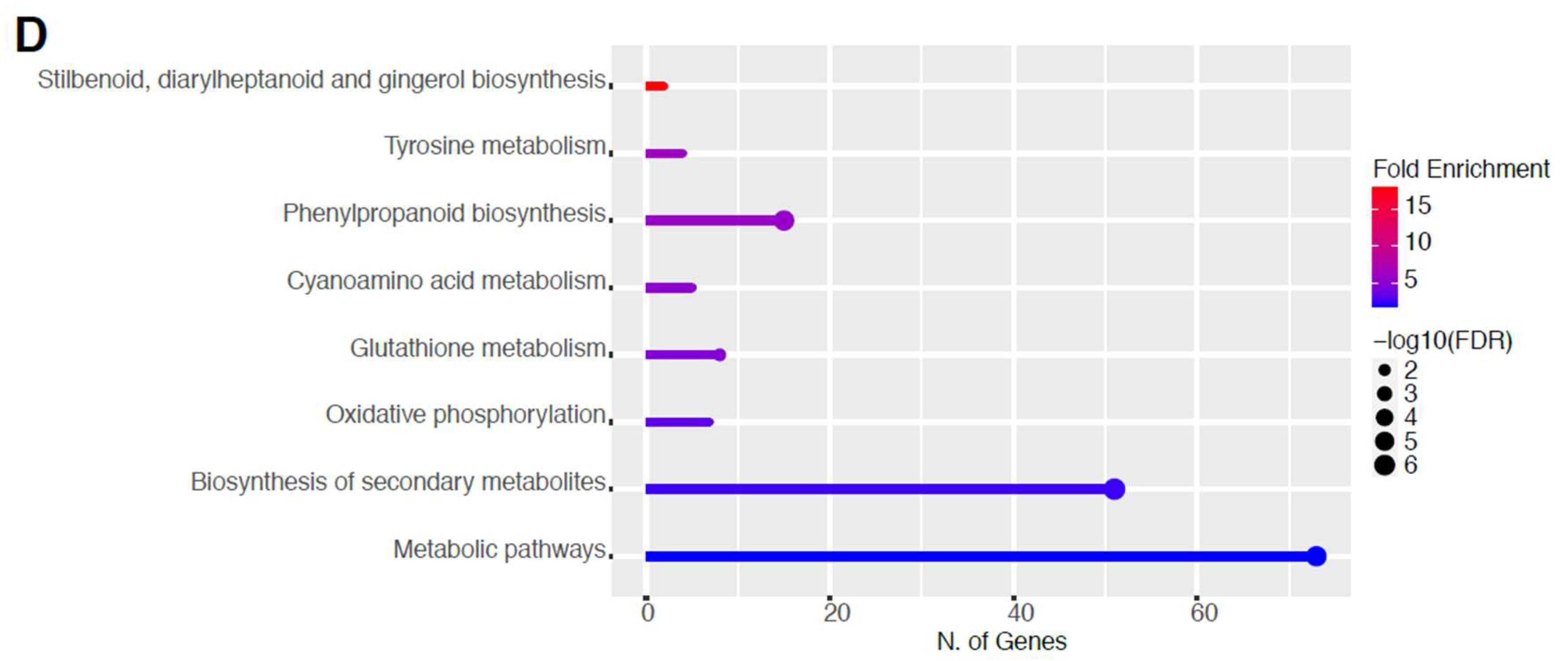
2.2. Global RNA-Seq Analysis of Young Seedlings after Non-Thermal Plasma-Seed Treatment
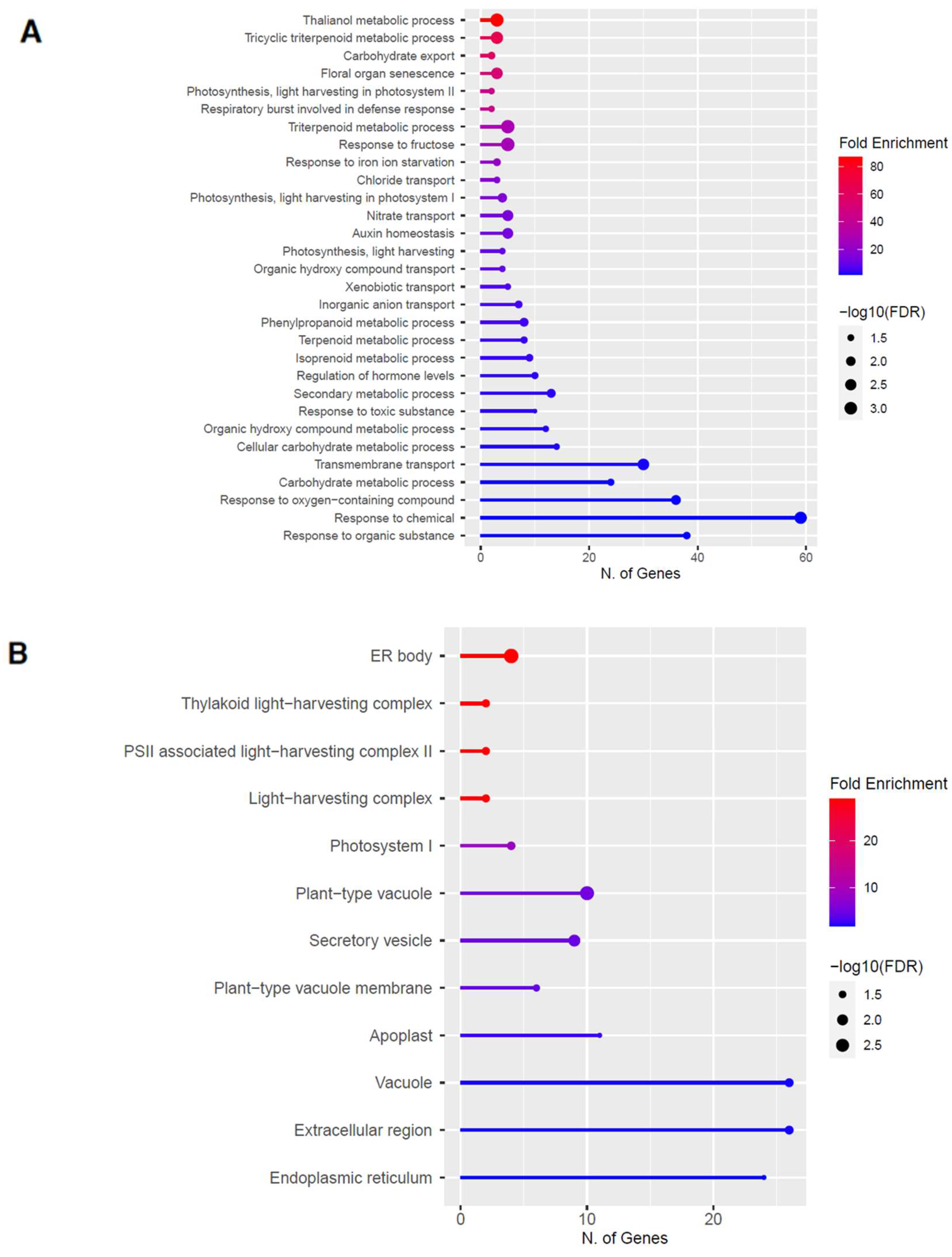
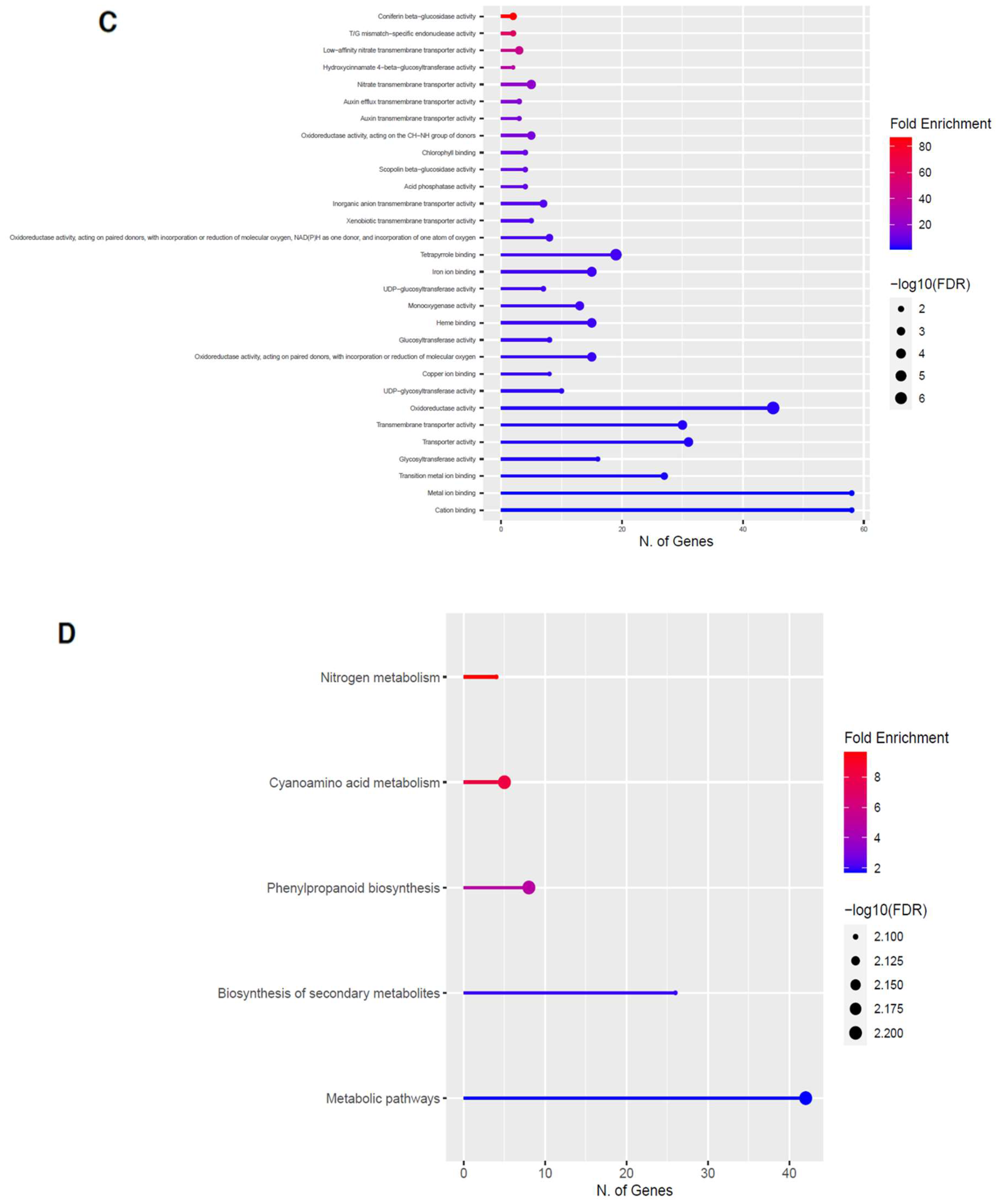
2.3. Upregulation of Genes in the Phenylpropanoid Pathway
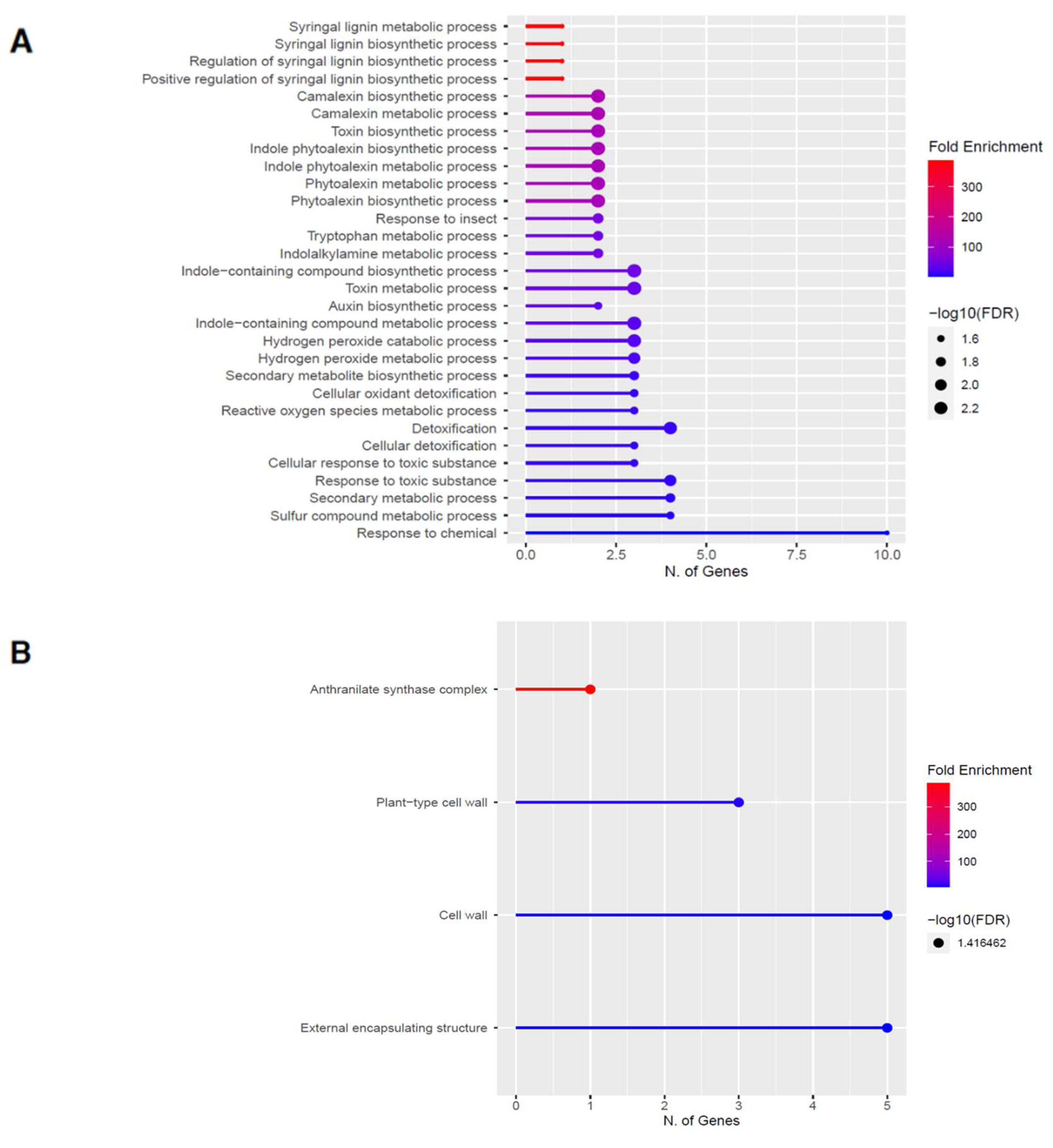
2.4. Downregulation of Other Secondary Metabolic Pathways
2.5. Upregulation of Genes in the Glucosinolate Pathway
2.6. Downregulation of Other Secondary Metabolic Pathways
3. Discussion and Conclusions
3.1. Gene Enrichment as a Result of Short Non-Thermal Air Plasma Treatment
3.2. Comparison with Other Transcriptomic Studies
3.3. Transcriptomic Plant Response to Plasma Treatment and Its Limitations
3.4. Conclusions
4. Materials and Methods
4.1. Seed Material
4.2. Germination Rate Measurements
4.3. Surface Dielectric Barrier Discharge Description
4.4. Plasma Parameters for the Seed Treatment
4.5. Germination Statistics
4.6. RNA Isolation, Library Construction, and RNA Sequencing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waskow, A.; Howling, A.A.; Furno, I. Mechanisms of plasma-seed treatments as a potential seed processing technology. Front. Phys. 2021, 9, 174. [Google Scholar] [CrossRef]
- FAO. News Article: Pollutants from Agriculture a Serious Threat to World’s Water. Available online: https://www.fao.org/news/story/en/item/1141534/icode/ (accessed on 10 January 2022).
- IPCC. Land Is a Critical Resource, IPCC Report Says. Available online: https://www.ipcc.ch/2019/08/08/land-is-a-critical-resource_srccl/ (accessed on 10 January 2022).
- Guragain, R.P.; Baniya, H.B.; Dhungana, S.; Chhetri, G.K.; Sedhai, B.; Basnet, N.; Shakya, A.; Pandey, B.P.; Pradhan, S.P.; Joshi, U.M.; et al. Effect of plasma treatment on the seed germination and seedling growth of radish (Raphanus sativus). Plasma Sci. Technol. 2021, 24, 015502. [Google Scholar] [CrossRef]
- Li, K.; Zhong, C.; Shi, Q.; Bi, H.; Gong, B. Cold plasma seed treatment improves chilling resistance of tomato plants through hydrogen peroxide and abscisic acid signaling pathway. Free Radic. Biol. Med. 2021, 172, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Waskow, A.; Avino, F.; Howling, A.; Furno, I. Entering the plasma agriculture field: An attempt to standardize protocols for plasma treatment of seeds. Plasma Processes Polym. 2021, 19, e2100152. [Google Scholar] [CrossRef]
- Suriyasak, C.; Hatanaka, K.; Tanaka, H.; Okumura, T.; Yamashita, D.; Attri, P.; Koga, K.; Shiratani, M.; Hamaoka, N.; Ishibashi, Y. Alterations of DNA Methylation Caused by Cold Plasma Treatment Restore Delayed Germination of Heat-Stressed Rice (Oryza sativa L.) Seeds. ACS Agric. Sci. Technol. 2021, 1, 5–10. [Google Scholar] [CrossRef]
- Tamošiūnė, I.; Gelvonauskienė, D.; Haimi, P.; Mildažienė, V.; Koga, K.; Shiratani, M.; Baniulis, D. Cold plasma treatment of sunflower seeds modulates plant-associated microbiome and stimulates root and lateral organ growth. Front. Plant Sci. 2020, 11, 1347. [Google Scholar] [CrossRef] [PubMed]
- Tamošiūnė, I.; Gelvonauskienė, D.; Ragauskaitė, L.; Koga, K.; Shiratani, M.; Baniulis, D. Cold plasma treatment of Arabidopsis thaliana (L.) seeds modulates plant-associated microbiome composition. Appl. Phys. Express 2020, 13, 076001. [Google Scholar] [CrossRef]
- Mildaziene, V.; Aleknavičiūtė, V.; Žūkienė, R.; Paužaitė, G.; Naučienė, Z.; Filatova, I.; Lyushkevich, V.; Haimi, P.; Tamošiūnė, I.; Baniulis, D. Treatment of Common Sunflower (Helianthus annus L.) Seeds with Radio-frequency Electromagnetic Field and Cold Plasma Induces Changes in Seed Phytohormone Balance, Seedling Development and Leaf Protein Expression. Sci. Rep. 2019, 9, 6437. [Google Scholar] [CrossRef]
- Ji, S.H.; Ki, S.H.; Kang, M.H.; Choi, J.S.; Park, Y.; Oh, J.; Kim, S.B.; Yoo, S.J.; Choi, E.H.; Park, G. Characterization of physical and bio-chemical changes in plasma treated spinach seed during germination. J. Phys. D Appl. Phys. 2018, 51, 145205. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, Y.; Zhang, H.; Qu, G.; Wang, T.; Sun, Q.; Liang, D. Alleviation of adverse effects of drought stress on wheat seed germination using atmospheric dielectric barrier discharge plasma treatment. Sci. Rep. 2017, 7, 16680. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.M.; Sajib, S.A.; Rahi, M.S.; Tahura, S.; Roy, N.C.; Parvez, S.; Reza, A.; Talukder, M.R.; Kabir, A.H. Mechanisms and signaling associated with LPDBD plasma mediated growth improvement in wheat. Sci. Rep. 2018, 8, 10498. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, B.; Adhikari, M.; Ghimire, B.; Park, G.; Choi, E.H. Cold atmospheric plasma-activated water irrigation induces defense hormone and gene expression in tomato seedlings. Sci. Rep. 2019, 9, 16080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhikari, B.; Adhikari, M.; Ghimire, B.; Adhikari, B.C.; Park, G.; Choi, E.H. Cold plasma seed priming modulates growth, redox homeostasis and stress response by inducing reactive species in tomato (Solanum lycopersicum). Free Radic. Biol. Med. 2020, 156, 57–69. [Google Scholar] [CrossRef]
- Li, K.; Zhang, L.; Shao, C.; Zhong, C.; Cao, B.; Shi, Q.; Gong, B. Utilising cold plasma seed treatment technologies to delay cotyledon senescence in tomato seedlings. Sci. Hortic. 2021, 281, 109911. [Google Scholar] [CrossRef]
- Ghasempour, M.; Iranbakhsh, A.; Ebadi, M.; Oraghi Ardebili, Z. Seed priming with cold plasma improved seedling performance, secondary metabolism, and expression of deacetylvindoline O-acetyltransferase gene in Catharanthus roseus. Contrib. Plasma Phys. 2020, 60, e201900159. [Google Scholar] [CrossRef]
- Islam, S.; Omar, F.B.; Sajib, S.A.; Roy, N.C.; Reza, A.; Hasan, M.; Talukder, M.R.; Kabir, A.H. Effects of LPDBD Plasma and Plasma Activated Water on Germination and Growth in Rapeseed (Brassica napus). Gesunde Pflanz 2019, 71, 175–185. [Google Scholar] [CrossRef]
- Iranbakhsh, A.; Ardebili, Z.O.; Molaei, H.; Ardebili, N.O.; Amini, M. Cold plasma up-regulated expressions of WRKY1 transcription factor and genes involved in biosynthesis of cannabinoids in Hemp (Cannabis sativa L.). Plasma Chem. Plasma Process. 2020, 40, 527–537. [Google Scholar] [CrossRef]
- Ghaemi, M.; Majd, A.; Iranbakhsh, A. Transcriptional responses following seed priming with cold plasma and electromagnetic field in Salvia nemorosa L. J. Theor. Appl. Phys. 2020, 14, 323–328. [Google Scholar] [CrossRef]
- Ebrahimibasabi, E.; Ebrahimi, A.; Momeni, M.; Reza Amerian, M. Elevated expression of diosgenin-related genes and stimulation of the defense system in Trigonella foenum-graecum (Fenugreek) by cold plasma treatment. Sci. Hortic. 2020, 271, 109494. [Google Scholar] [CrossRef]
- Sajib, S.A.; Billah, M.; Mahmud, S.; Miah, M.; Hossain, F.; Omar, F.B.; Roy, N.C.; Hoque, K.M.F.; Talukder, M.R.; Kabir, A.H.; et al. Plasma activated water: The next generation eco-friendly stimulant for enhancing plant seed germination, vigor and increased enzyme activity, a study on black gram (Vigna mungo L.). Plasma Chem. Plasma Process. 2020, 40, 119–143. [Google Scholar] [CrossRef]
- Perez, S.M.; Biondi, E.; Laurita, R.; Proto, M.; Sarti, F.; Gherardi, M.; Bertaccini, A.; Colombo, V. Plasma activated water as resistance inducer against bacterial leaf spot of tomato. PLoS ONE 2019, 14, e0217788. [Google Scholar] [CrossRef] [PubMed]
- Ka, D.H.; Priatama, R.A.; Park, J.Y.; Park, S.J.; Kim, S.B.; Lee, I.A.; Lee, Y.K. Plasma-Activated Water Modulates Root Hair Cell Density via Root Developmental Genes in Arabidopsis thaliana L. Appl. Sci. 2021, 11, 2240. [Google Scholar] [CrossRef]
- Tong, J.; He, R.; Tang, X.; Li, M.; Wan, J. Transcriptomic analysis of seed germination improvement of Andrographis paniculata responding to air plasma treatment. PLoS ONE 2020, 15, e0240939. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Yin, Y.; Li, H.; Hu, X.; Zhuang, J.; Ma, R.; Jiao, Z. Comparative transcriptome analysis of atmospheric pressure cold plasma enhanced early seedling growth in Arabidopsis thaliana. Plasma Sci. Technol. 2021, 23, 085502. [Google Scholar] [CrossRef]
- Han, B.; Yu, N.-N.; Zheng, W.; Zhang, L.-N.; Liu, Y.; Yu, J.-B.; Zhang, Y.-Q.; Park, G.; Sun, H.-N.; Kwon, T. Effect of non-thermal plasma (NTP) on common sunflower (Helianthus annus L.) seed growth via upregulation of antioxidant activity and energy metabolism-related gene expression. Plant Growth Regul. 2021, 95, 271–281. [Google Scholar] [CrossRef]
- Wang, P.; Chen, F.; Huang, Y.; Sun, H.; Fan, M.; Ning, D.; Wang, T.; Wang, H.; Liu, M. New insights of low-temperature plasma effects on seeds germination of Platycodon grandiflorum. Isr. J. Plant Sci. 2021, 68, 306–318. [Google Scholar] [CrossRef]
- Waskow, A.; Ibba, L.; Leftley, M.; Howling, A.; Ambrico, P.F.; Furno, I. An In Situ FTIR Study of DBD Plasma Parameters for Accelerated Germination of Arabidopsis thaliana Seeds. Int. J. Mol. Sci. 2021, 22, 11540. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis. In International Encyclopedia of Statistical Science; Lovric, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar] [CrossRef]
- Jin, X.; Han, J. K-Means Clustering. In Encyclopedia of Machine Learning; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2011. [Google Scholar] [CrossRef]
- Hinneburg, A. Visualizing Clustering Results. In Encyclopedia of Database Systems; Liu, L., Özsu, M.T., Eds.; Springer: Boston, MA, USA, 2009. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. Int. J. Mol. Sci. 2021, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Amen, T.; Guihur, A.; Zelent, C.; Ursache, R.; Wilting, J.; Kaganovich, D. Resveratrol and related stilbene derivatives induce stress granules with distinct clearance kinetics. Mol. Biol. Cell 2021, 32, ar18. [Google Scholar] [CrossRef] [PubMed]
- Bourgine, B.; Guihur, A. Heat shock signaling in land plants: From plasma membrane sensing to the transcription of small heat shock proteins. Front. Plant Sci. 2021, 12, 710801. [Google Scholar] [CrossRef]
- Guihur, A.; Fauvet, B.; Finka, A.; Quadroni, M.; Goloubinoff, P. Quantitative proteomic analysis to capture the role of heat-accumulated proteins in moss plant acquired thermotolerance. Plant Cell Environ. 2021, 44, 2117–2133. [Google Scholar] [CrossRef] [PubMed]
- Waskow, A.; Butscher, D.; Oberbossel, G.; Klöti, D.; Rudolf von Rohr, P.; Büttner-Mainik, A.; Schuppler, M. Low-energy electron beam has severe impact on seedling development compared to cold atmospheric pressure plasma. Sci. Rep. 2021, 11, 16373. [Google Scholar] [CrossRef]
- Wijesooriya, K.; Jadaan, S.A.; Perera, K.L.; Kaur, T.; Ziemann, M. Guidelines for reliable and reproducible functional enrichment analysis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Koga, K.; Thapanut, S.; Amano, T.; Seo, H.; Itagaki, N.; Hayashi, N.; Shiratani, M. Simple method of improving harvest by nonthermal air plasma irradiation of seeds of Arabidopsis thaliana (L.). Appl. Phys. Express 2015, 9, 16201. [Google Scholar] [CrossRef]
- Bafoil, M.; Le Ru, A.; Merbahi, N.; Eichwald, O.; Dunand, C.; Yousfi, M. New insights of low-temperature plasma effects on germination of three genotypes of Arabidopsis thaliana seeds under osmotic and saline stresses. Sci. Rep. 2019, 9, 8649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrico, P.F.; Simek, M.; Ambrico, M.; Morano, M.; Prukner, V.; Minafra, A.; Allegretta, I.; Porfido, C.; Senesi, G.S.; Terzano, R. On the air atmospheric pressure plasma treatment effect on the physiology, germination and seedlings of basil seeds. J. Phys. D Appl. Phys. 2020, 53, 104001. [Google Scholar] [CrossRef]
- Waskow, A.; Howling, A.; Furno, I. Advantages and limitations of surface analysis techniques on plasma-treated Arabidopsis thaliana seeds. Front. Mater. 2021, 8, 642099. [Google Scholar] [CrossRef]
- Butscher, D.; Zimmermann, D.; Schuppler, M.; von Rohr, P.R. Plasma inactivation of bacterial endospores on wheat grains and polymeric model substrates in a dielectric barrier discharge. Food Control 2016, 60, 636–645. [Google Scholar] [CrossRef]
- Butscher, D.; van Loon, H.; Waskow, A.; von Rohr, P.R.; Schuppler, M. Plasma inactivation of microorganisms on sprout seeds in a dielectric barrier discharge. Int. J. Food Microbiol. 2016, 238, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Mildaziene, V.; Ivankov, A.; Pauzaite, G.; Naucienė, Z.; Zukiene, R.; Degutyte-Fomins, L.; Lyushkevich, V. Seed treatment with cold plasma and electromagnetic field induces changes in red clover root growth dynamics, flavonoid exudation, and activates nodulation. Plasma Processes Polym. 2021, 18, 2000160. [Google Scholar] [CrossRef]
- Huang, A.C.; Jiang, T.; Liu, Y.-X.; Bai, Y.-C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.-W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Sandhu, K.; Bisht, N.; Nailwal, T.; Saini, I.; Kaushik, P. Ascertaining the Paradigm of Secondary Metabolism Enhancement through Gene Level Modification in Therapeutic Plants. J. Young Pharm. 2019, 11, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Binder, S. Branched-Chain Amino Acid Metabolism in Arabidopsis thaliana. Arab. Book 2010, 8, e0137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subki, A.; Abidin, A.A.Z.; Yusof, Z.N.B. The role of thiamine in plants and current perspectives in crop improvement. B Group Vitam.-Curr. Uses Perspect. 2018, 5, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Ahn, I.P.; Kim, S.; Lee, Y.H. Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol. 2005, 138, 1505–1515. [Google Scholar] [CrossRef] [Green Version]
- Hansen, B.G.; Kerwin, R.E.; Ober, J.A.; Lambrix, V.M.; Mitchell-Olds, T.; Gershenzon, J.; Kliebenstein, D.J. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008, 148, 2096–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittipol, V.; He, Z.; Wang, L.; Doheny-Adams, T.; Langer, S.; Bancroft, I. Genetic architecture of glucosinolate variation in Brassica napus. J. Plant Physiol. 2019, 240, 152988. [Google Scholar] [CrossRef] [PubMed]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Sinha, A.K. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Front. Plant Sci. 2015, 6, 769. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Goto-Yamada, S.; Nakazaki, A.; Kunieda, T.; Kuwata, K.; Nagano, A.J.; Nishimura, M.; Hara-Nishimura, I. Endoplasmic reticulum-derived bodies enable a single-cell chemical defense in Brassicaceae plants. Commun. Biol. 2020, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Akinwumi, B.C.; Bordun, K.M.; Anderson, H.D. Biological Activities of Stilbenoids. Int. J. Mol. Sci. 2018, 19, 792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.I.; Zhang, X.; Pascuzzi, P.E.; Liu, C.J.; Chapple, C. Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol. 2020, 225, 154–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malka, S.K.; Cheng, Y. Possible Interactions between the Biosynthetic Pathways of Indole Glucosinolate and Auxin. Front. Plant Sci. 2017, 8, 2131. [Google Scholar] [CrossRef] [Green Version]
- Pauzaite, G.; Malakauskiene, A.; Nauciene, Z.; Zukiene, R.; Filatova, I.; Lyushkevich, V.; Mildaziene, V. Changes in Norway spruce germination and growth induced by pre-sowing seed treatment with cold plasma and electromagnetic field: Short-term versus long-term effects. Plasma Processes Polym. 2018, 15, 1700068. [Google Scholar] [CrossRef]
- Hayashi, N. Growth Enhancement Effect of Gene Expression of Plants Induced by Active Oxygen Species in Oxygen Plasma. In Agritech: Innovative Agriculture Using Microwaves and Plasmas; Horikoshi, S., Brodie, G., Takaki, K., Serpone, N., Eds.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Sirgedaitė-Šėžienė, V.; Mildažienė, V.; Žemaitis, P.; Ivankov, A.; Koga, K.; Shiratani, M.; Baliuckas, V. Long-term response of Norway spruce to seed treatment with cold plasma: Dependence of the effects on the genotype. Plasma Processes Polym. 2021, 18, 2000159. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waskow, A.; Guihur, A.; Howling, A.; Furno, I. RNA Sequencing of Arabidopsis thaliana Seedlings after Non-Thermal Plasma-Seed Treatment Reveals Upregulation in Plant Stress and Defense Pathways. Int. J. Mol. Sci. 2022, 23, 3070. https://doi.org/10.3390/ijms23063070
Waskow A, Guihur A, Howling A, Furno I. RNA Sequencing of Arabidopsis thaliana Seedlings after Non-Thermal Plasma-Seed Treatment Reveals Upregulation in Plant Stress and Defense Pathways. International Journal of Molecular Sciences. 2022; 23(6):3070. https://doi.org/10.3390/ijms23063070
Chicago/Turabian StyleWaskow, Alexandra, Anthony Guihur, Alan Howling, and Ivo Furno. 2022. "RNA Sequencing of Arabidopsis thaliana Seedlings after Non-Thermal Plasma-Seed Treatment Reveals Upregulation in Plant Stress and Defense Pathways" International Journal of Molecular Sciences 23, no. 6: 3070. https://doi.org/10.3390/ijms23063070
APA StyleWaskow, A., Guihur, A., Howling, A., & Furno, I. (2022). RNA Sequencing of Arabidopsis thaliana Seedlings after Non-Thermal Plasma-Seed Treatment Reveals Upregulation in Plant Stress and Defense Pathways. International Journal of Molecular Sciences, 23(6), 3070. https://doi.org/10.3390/ijms23063070





