Isolation of Platelet-Derived Exosomes from Human Platelet-Rich Plasma: Biochemical and Morphological Characterization
Abstract
:1. Introduction
2. Results
2.1. PRP Characterization
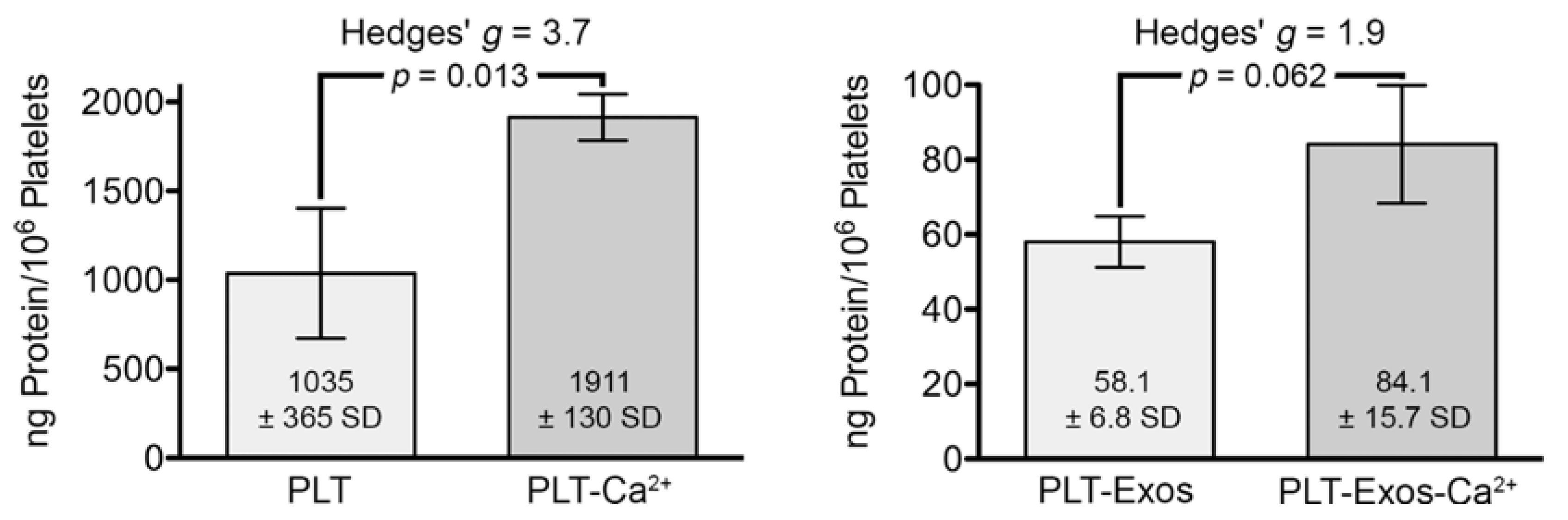
2.2. Total Protein Content Measurement in Platelets and Platelet-Derived Exosomes under Basal Conditions and Calcium-Stimulation
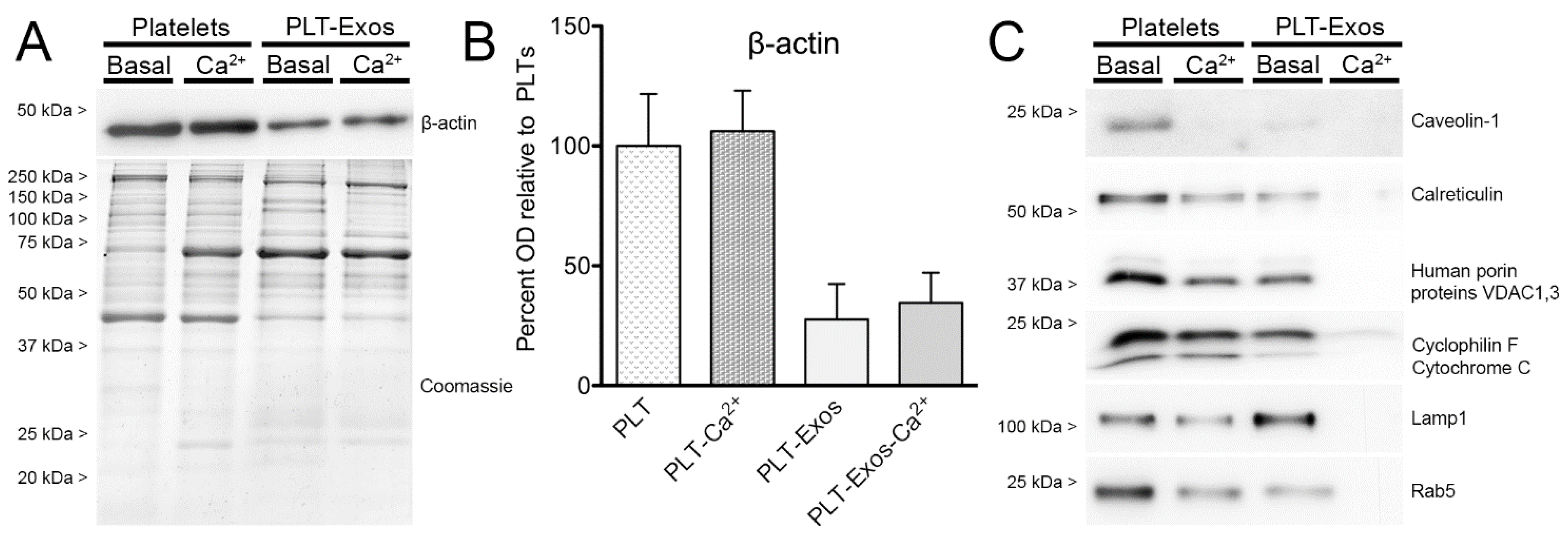
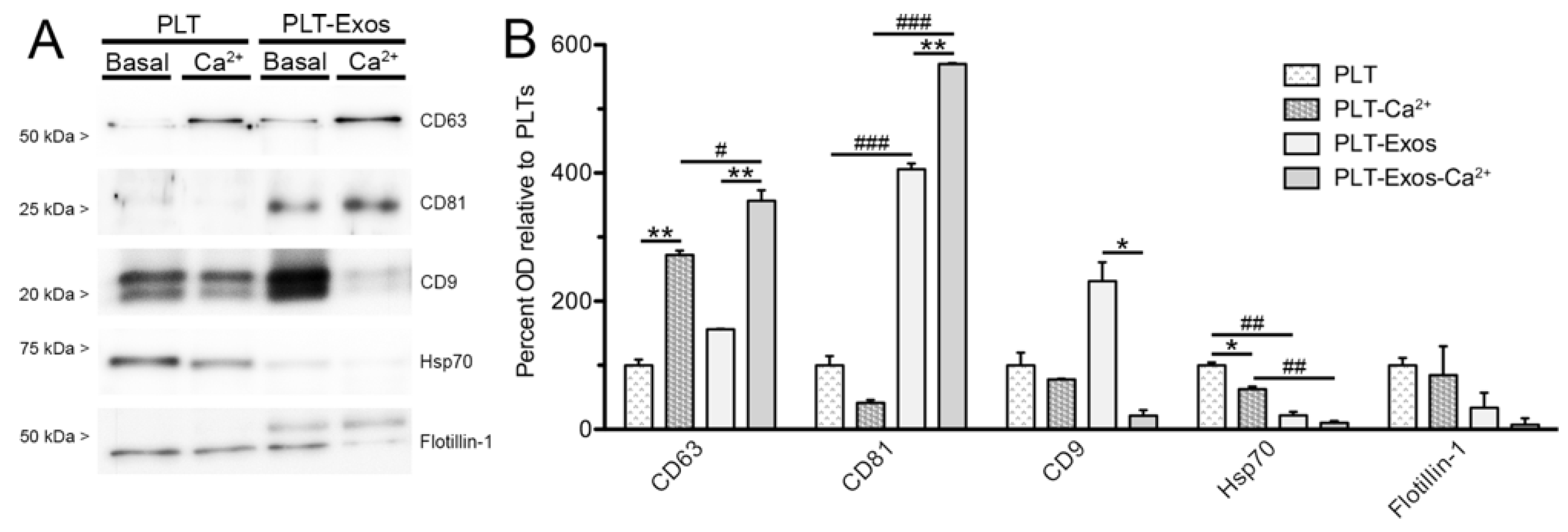
2.3. Biochemical Characterization of Human Platelet-Derived Exosomes Obtained under Basal Conditions and Calcium-Stimulation
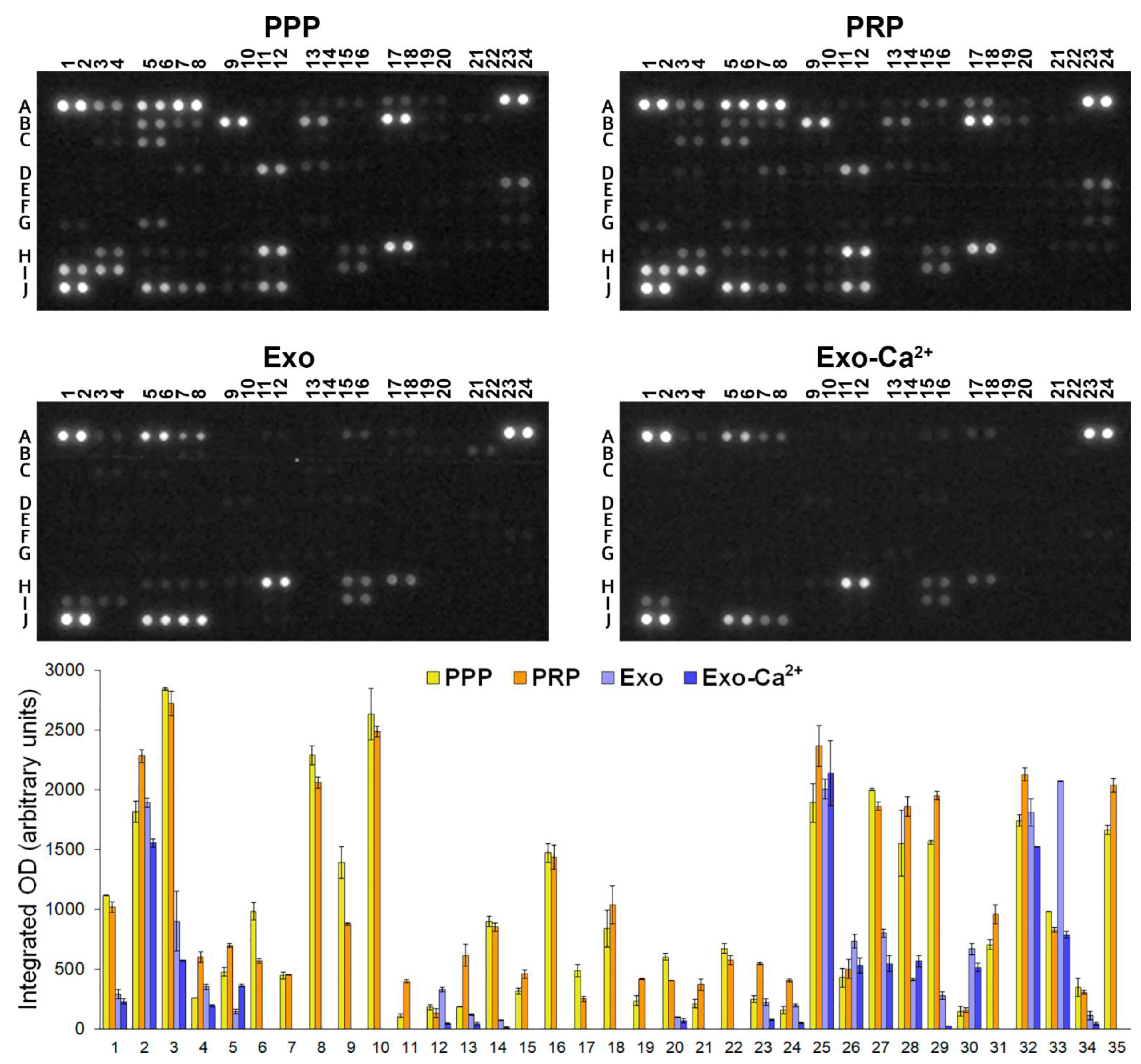
2.4. Analysis of Cytokines, Chemokines, and Growth Factors in PPP, PRP, PLT-Exos, and PLT-Exos-Ca2+
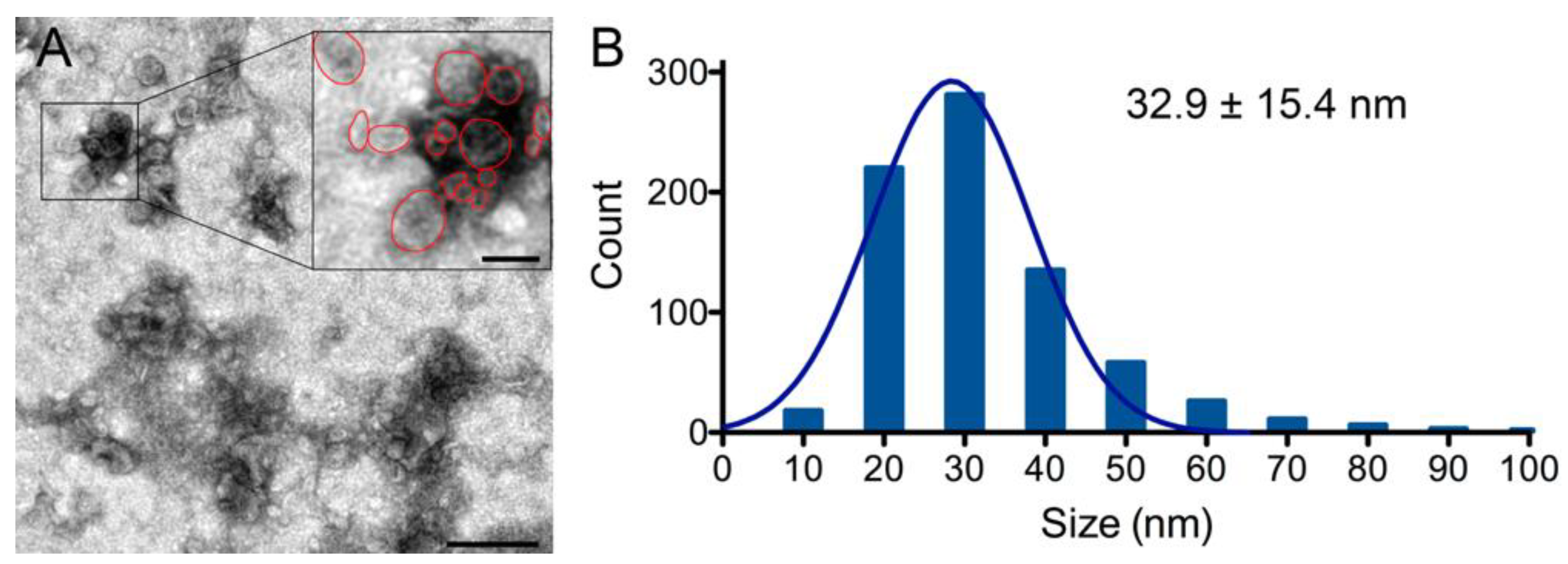
2.5. Transmission Electron Microscopy Analysis of Exosomes Isolated from Calcium-Activated Platelets
3. Discussion
4. Materials and Methods
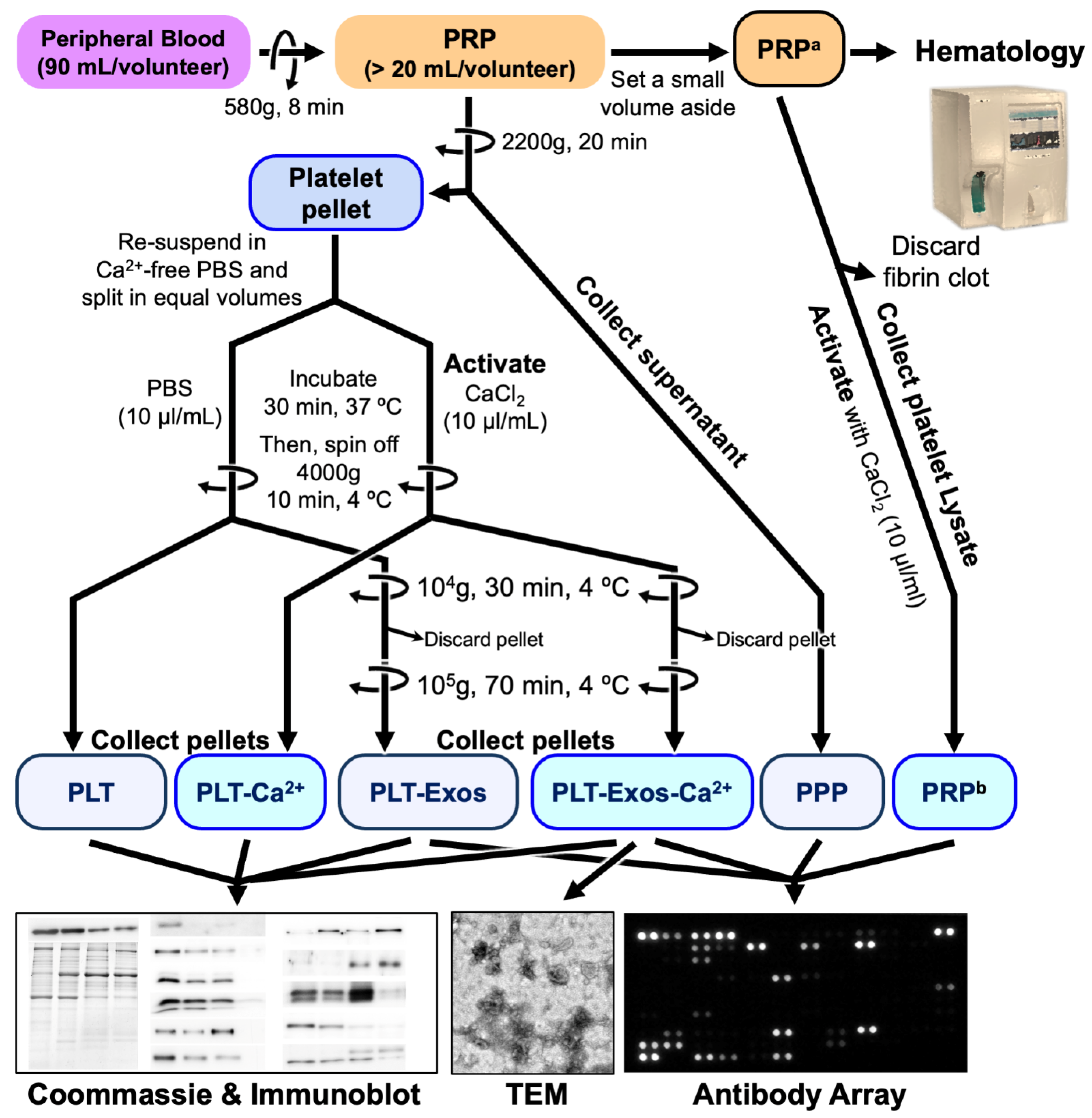
4.1. Platelet-Rich Plasma Preparation and Characterization
4.2. Isolation of Human Platelet-Derived Exosomes Obtained under Basal Conditions and CaCl2 Stimulation
4.3. Western Blot Analysis
4.4. Transmission Electron Microscopy and Analysis of Exosome Size
4.5. Analysis of Cytokines, Chemokines and Growth Factors
4.6. Data Processing, Statistical Analysis, and Figure Construction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cecerska-Heryć, E.; Goszka, M.; Serwin, N.; Roszak, M.; Grygorcewicz, B.; Heryć, R.; Dołęgowska, B. Applications of the regenerative capacity of platelets in modern medicine. Cytokine Growth Factor Rev. 2021, in press. [CrossRef] [PubMed]
- Le, A.D.K.; Enweze, L.; DeBaun, M.R.; Dragoo, J.L. Platelet-Rich Plasma. Clin. Sports Med. 2019, 38, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Anitua, E.; Delgado, D.; Sanchez, P.; Prado, R.; Orive, G.; Padilla, S. Platelet-rich plasma, a source of autologous growth factors and biomimetic scaffold for peripheral nerve regeneration. Expert Opin. Biol. Ther. 2017, 17, 197–212. [Google Scholar] [CrossRef]
- Bohren, Y.; Timbolschi, D.I.; Muller, A.; Barrot, M.; Yalcin, I.; Salvat, E. Platelet-rich plasma and cytokines in neuropathic pain: A narrative review and a clinical perspective. Eur. J. Pain 2021, 26, 43–60. [Google Scholar] [CrossRef]
- Delgado, D.; Bilbao, A.M.; Beitia, M.; Garate, A.; Sánchez, P.; González-Burguera, I.; Isasti, A.; De Jesús, M.L.; Zuazo-Ibarra, J.; Montilla, A.; et al. Effects of platelet-rich plasma on cellular populations of the central nervous system: The influence of donor age. Int. J. Mol. Sci. 2021, 22, 1725. [Google Scholar] [CrossRef]
- Delgado, D.; Garate, A.; Sánchez, P.; Bilbao, A.M.; García del Caño, G.; Salles, J.; Sánchez, M. Biological and structural effects after intraosseous infiltrations of age-dependent platelet-rich plasma: An in vivo study. J. Orthop. Res. 2020, 38, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.; Martin Cramer, E. Platelet α-granules. Blood Rev. 1993, 7, 52–62. [Google Scholar] [CrossRef]
- Sánchez-González, D.J.; Méndez-Bolaina, E.; Trejo-Bahena, N.I. Platelet-rich plasma peptides: Key for regeneration. Int. J. Pept. 2012, 2012, 532519. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Piao, Y.; Liu, Q.; Yang, X. Platelet-rich plasma-derived extracellular vesicles: A superior alternative in regenerative medicine? Cell Prolif. 2021, 54, e13123. [Google Scholar] [CrossRef]
- Torreggiani, E.; Perut, F.; Roncuzzi, L.; Zini, N.; Baglìo, S.R.; Baldini, N. Exosomes: Novel effectors of human platelet lysate activity. Eur. Cells Mater. 2014, 28, 137–151. [Google Scholar] [CrossRef]
- Tao, S.-C.; Yuan, T.; Rui, B.-Y.; Zhu, Z.-Z.; Guo, S.-C.; Zhang, C.-Q. Exosomes derived from human platelet-rich plasma prevent apoptosis induced by glucocorticoid-associated endoplasmic reticulum stress in rat osteonecrosis of the femoral head via the Akt/Bad/Bcl-2 signal pathway. Theranostics 2017, 7, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.C.; Tao, S.C.; Yin, W.J.; Qi, X.; Yuan, T.; Zhang, C.Q. Exosomes derived from platelet-rich plasma promote the re-epithelization of chronic cutaneous wounds via activation of YAP in a diabetic rat model. Theranostics 2017, 7, 81–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wang, L.; Ma, C.; Wang, G.; Zhang, Y.; Sun, S. Exosomes derived from platelet-rich plasma present a novel potential in alleviating knee osteoarthritis by promoting proliferation and inhibiting apoptosis of chondrocyte via Wnt/β-catenin signaling pathway. J. Orthop. Surg. Res. 2019, 14, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui, S.; Yuan, Y.; Du, C.; Song, P.; Chen, Y.; Wang, H.; Fan, Y.; Armstrong, D.G.; Deng, W.; Li, L. Comparison and Investigation of Exosomes Derived from Platelet-Rich Plasma Activated by Different Agonists. Cell Transplant. 2021, 30, 9636897211017832. [Google Scholar] [CrossRef] [PubMed]
- Burger, D.; Viñas, J.L.; Akbari, S.; Dehak, H.; Knoll, W.; Gutsol, A.; Carter, A.; Touyz, R.M.; Allan, D.S.; Burns, K.D. Human Endothelial Colony-Forming Cells Protect against Acute Kidney Injury Role of Exosomes. Am. J. Pathol. 2015, 185, 2309–2323. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Li, Y.; Chopp, M. Exosomes/miRNAs as mediating cell-based therapy of stroke. Front. Cell. Neurosci. 2014, 8, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunggulawa, E.J.; Wang, W.; Yin, T.; Wang, N.; Durkan, C.; Wang, Y.; Wang, G. Recent advancements in the use of exosomes as drug delivery systems 06 Biological Sciences 0601 Biochemistry and Cell Biology. J. Nanobiotechnology 2018, 16, 81. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, J.; Stewart, T.; Banks, W.A.; Zhang, J. The Transport Mechanism of Extracellular Vesicles at the Blood-Brain Barrier. Curr. Pharm. Des. 2018, 23, 6206–6214. [Google Scholar] [CrossRef]
- Thomi, G.; Joerger-Messerli, M.; Haesler, V.; Muri, L.; Surbek, D.; Schoeberlein, A. Intranasally Administered Exosomes from Umbilical Cord Stem Cells Have Preventive Neuroprotective Effects and Contribute to Functional Recovery after Perinatal Brain Injury. Cells 2019, 8, 855. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.; Wu, Y.W.; Blyth, C.; Lichtfuss, G.; Goubran, H.; Burnouf, T. Prospective Therapeutic Applications of Platelet Extracellular Vesicles. Trends Biotechnol. 2021, 39, 598–612. [Google Scholar] [CrossRef]
- György, B.; Hung, M.E.; Breakefield, X.O.; Leonard, J.N. Therapeutic applications of extracellular vesicles: Clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 439–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of the first phase 1 clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther. 2008, 16, 782–790. [Google Scholar] [CrossRef]
- Heijnen, H.F.G.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and α-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Fiz, N.; Azofra, J.; Usabiaga, J.; Aduriz Recalde, E.; Garcia Gutierrez, A.; Albillos, J.; Gárate, R.; Aguirre, J.J.; Padilla, S.; et al. A randomized clinical trial evaluating plasma rich in growth factors (PRGF-Endoret) versus hyaluronic acid in the short-term treatment of symptomatic knee osteoarthritis. Arthrosc. J. Arthrosc. Relat. Surg. 2012, 28, 1070–1078. [Google Scholar] [CrossRef]
- Kon, E.; Di Matteo, B.; Delgado, D.; Cole, B.J.; Dorotei, A.; Dragoo, J.L.; Filardo, G.; Fortier, L.A.; Giuffrida, A.; Jo, C.H.; et al. Platelet-rich plasma for the treatment of knee osteoarthritis: An expert opinion and proposal for a novel classification and coding system. Expert Opin. Biol. Ther. 2020, 20, 1447–1460. [Google Scholar] [CrossRef]
- Everts, P.; Onishi, K.; Jayaram, P.; Lana, J.F.; Mautner, K. Platelet-rich plasma: New performance understandings and therapeutic considerations in 2020. Int. J. Mol. Sci. 2020, 21, 7794. [Google Scholar] [CrossRef]
- Padilla, S.; Sánchez, M.; Orive, G.; Anitua, E. Human-Based Biological and Biomimetic Autologous Therapies for Musculoskeletal Tissue Regeneration. Trends Biotechnol. 2017, 35, 192–202. [Google Scholar] [CrossRef]
- Oneto, P.; Zubiry, P.R.; Schattner, M.; Etulain, J. Anticoagulants Interfere With the Angiogenic and Regenerative Responses Mediated by Platelets. Front. Bioeng. Biotechnol. 2020, 8, 223. [Google Scholar] [CrossRef]
- Sánchez, M.; Anitua, E.; Delgado, D.; Sánchez, P.; Orive, G.; Padilla, S. Muscle repair: Platelet-rich plasma derivates as a bridge from spontaneity to intervention. Injury 2014, 45 (Suppl. S4), S7–S14. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef] [PubMed]
- Pienimaeki-Roemer, A.; Kuhlmann, K.; Böttcher, A.; Konovalova, T.; Black, A.; Orsõ, E.; Liebisch, G.; Ahrens, M.; Eisenacher, M.; Meyer, H.E.; et al. Lipidomic and proteomic characterization of platelet extracellular vesicle subfractions from senescent platelets. Transfusion 2015, 55, 507–521. [Google Scholar] [CrossRef]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielańczyk, Ł.; Widłak, P. Proteome profiling of exosomes purified from a small amount of human serum: The problem of co-purified serum components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, S.; Stojadinovic, O.; Golinko, M.S.; Brem, H.; Tomic-Canic, M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008, 16, 585–601. [Google Scholar] [CrossRef]
- Qian, Y.; Han, Q.; Chen, W.; Song, J.; Zhao, X.; Ouyang, Y.; Yuan, W.; Fan, C. Platelet-rich plasma derived growth factors contribute to stem cell differentiation in musculoskeletal regeneration. Front. Chem. 2017, 5, 89. [Google Scholar] [CrossRef] [Green Version]
- Nedeau, A.E.; Bauer, R.J.; Gallagher, K.; Chen, H.; Liu, Z.J.; Velazquez, O.C. A CXCL5- and bFGF-dependent effect of PDGF-B-activated fibroblasts in promoting trafficking and differentiation of bone marrow-derived mesenchymal stem cells. Exp. Cell Res. 2008, 314, 2176–2186. [Google Scholar] [CrossRef] [Green Version]
- Chung, E.; Ahn, W.; Son, Y. CXCL5 abundant in the wound fluid at the late phase of wound healing, possibly promoting migration of mesenchymal stem cells and vascular tube formation. Tissue Eng. Regen. Med. 2014, 11, 317–322. [Google Scholar] [CrossRef]
- Liu, Y.F.; Liang, J.J.; Ng, T.K.; Hu, Z.; Xu, C.; Chen, S.; Chen, S.L.; Xu, Y.; Zhuang, X.; Huang, S.; et al. CXCL5/CXCR2 modulates inflammation-mediated neural repair after optic nerve injury. Exp. Neurol. 2021, 341, 113711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, R.; Wang, Z.; Lin, G.; Lue, T.F.; Lin, C.S. Adipose Tissue-Derived Stem Cells Secrete CXCL5 Cytokine with Neurotrophic Effects on Cavernous Nerve Regeneration. J. Sex. Med. 2011, 8, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Reid, I.R.; Comish, J. Direct Actions of Leptin on Bone Remodeling. Calcif. Tissue Int. 2004, 74, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Han, F.; Liu, L.; Yu, F.; Gui, Z.; Wang, X.; Li, B.; Fang, B.; Xia, L. The Effects of Leptin on the Proliferation and Differentiation of Primary Chondrocytes in Vitro and Cartilage Regeneration In Vivo. ACS Biomater. Sci. Eng. 2019, 5, 1907–1919. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.X.; Yu, K.; Dong, J.; Zhao, L.; Liu, Z.; Zhang, Q.; Li, S.; Du, Y.; Cheng, H. Precise prediction of calpain cleavage sites and their aberrance caused by mutations in cancer. Front. Genet. 2019, 10, 715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, K.; Hata, S.; Kawabata, Y.; Sorimachi, H. Structure, Activation, and Biology of Calpain. Diabetes 2004, 53, S12–S18. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Patron, C.; Martinez-Cuesta, M.A.; Salas, E.; Sawicki, G.; Wozniak, M.; Radomski, M.W.; Davidge, S.T. Differential regulation of platelet aggregation by matrix metalloproteinases-9 and -2. Thromb. Haemost. 1999, 82, 1730–1735. [Google Scholar] [CrossRef]
- Chen, R.; Jin, G.; McIntyre, T.M. The soluble protease ADAMDEC1 released from activated platelets hydrolyzes platelet membrane pro-epidermal growth factor (EGF) to active high-molecular-weight EGF. J. Biol. Chem. 2017, 292, 10112–10122. [Google Scholar] [CrossRef] [Green Version]
- Naudé, P.J.W.; Nyakas, C.; Eiden, L.E.; Ait-Ali, D.; Heide, R.; Engelborghs, S.; Luiten, P.G.M.; De Deyn, P.P.; Boer, J.A.; Eisel, U.L.M. Lipocalin 2: Novel component of proinflammatory signaling in Alzheimer’s disease. FASEB J. 2012, 26, 2811–2823. [Google Scholar] [CrossRef] [Green Version]
- Angelidis, C.; Deftereos, S.; Giannopoulos, G.; Anatoliotakis, N.; Bouras, G.; Hatzis, G.; Panagopoulou, V.; Pyrgakis, V.; Cleman, M.W. Cystatin C: An Emerging Biomarker in Cardiovascular Disease. Curr. Top. Med. Chem. 2013, 13, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, F.; Shen, J.; Zeng, H.; Li, L.; Zhao, J.; Zhao, J.; Lu, F.; Jia, W. Association between serum cystatin C and diabetic peripheral neuropathy: A cross-sectional study of a Chinese type 2 diabetic population. Eur. J. Endocrinol. 2014, 171, 641–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komabashiri, N.; Suehiro, F.; Ishii, M.; Nishimura, M. Efficacy of chitinase-3-like protein 1 as an in vivo bone formation predictable marker of maxillary/mandibular bone marrow stromal cells. Regen. Ther. 2021, 18, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yoon, S.M.; Song, S.U.; Park, S.G.; Kim, W.S.; Park, I.G.; Lee, J.; Sung, J.H. Hypoxia suppresses spontaneous mineralization and osteogenic differentiation of mesenchymal stem cells via IGFBP3 up-regulation. Int. J. Mol. Sci. 2016, 17, 1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.Q.; Lee, D.; Kim, Y.; Paek, M.; Kim, M.; Jang, K.S.; Oh, J.; Lee, Y.S.; Yeon, J.E.; Lubman, D.M.; et al. Platelet Factor 4 as a Novel Exosome Marker in MALDI-MS Analysis of Exosomes from Human Serum. Anal. Chem. 2019, 91, 13297–13305. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, H.; Kong, Y. Exosomes derived from platelet-rich plasma activate YAP and promote the fibrogenic activity of Müller cells via the PI3K/Akt pathway. Exp. Eye Res. 2020, 193, 107973. [Google Scholar] [CrossRef]
- Nilforoushzadeh, M.A.; Aghdami, N.; Taghiabadi, E. Effects of Adipose-Derived Stem Cells and Platelet-Rich Plasma Exosomes on The Inductivity of Hair Dermal Papilla Cells. Cell J. 2021, 23, 576–583. [Google Scholar] [CrossRef]
- Iyer, S.R.; Scheiber, A.L.; Yarowsky, P.; Henn, R.F., 3rd; Otsuru, S.; Lovering, R.M. Exosomes Isolated From Platelet-Rich Plasma and Mesenchymal Stem Cells Promote Recovery of Function After Muscle Injury. Am. J. Sports Med. 2020, 48, 2277–2286. [Google Scholar] [CrossRef]
- Xu, J.; Xie, G.; Yang, W.; Wang, W.; Zuo, Z.; Wang, W. Platelet-rich plasma attenuates intervertebral disc degeneration via delivering miR-141-3p-containing exosomes. Cell Cycle 2021, 20, 1487–1499. [Google Scholar] [CrossRef]
- Del Caño, G.G.; Aretxabala, X.; González-Burguera, I.; Montaña, M.; López De Jesús, M.; Barrondo, S.; Barrio, R.J.; Sampedro, C.; Goicolea, M.A.; Sallés, J. Nuclear diacylglycerol lipase-α in rat brain cortical neurons: Evidence of 2-arachidonoylglycerol production in concert with phospholipase C-β activity. J. Neurochem. 2015, 132, 489–503. [Google Scholar] [CrossRef]
- Saumell-Esnaola, M.; Barrondo, S.; García Del Caño, G.; Goicolea, M.A.; Sallés, J.; Lutz, B.; Monory, K. Subsynaptic distribution, lipid raft targeting and g protein-dependent signalling of the type 1 cannabinoid receptor in synaptosomes from the mouse hippocampus and frontal cortex. Molecules 2021, 26, 6897. [Google Scholar] [CrossRef] [PubMed]
- Garro, M.A.; De Jesús, M.L.; De Azúa, I.R.; Callado, L.F.; Meana, J.J.; Sallés, J. Regulation of phospholipase Cβ activity by muscarinic acetylcholine and 5-HT2 receptors in crude and synaptosomal membranes from human cerebral cortex. Neuropharmacology 2001, 40, 686–695. [Google Scholar] [CrossRef]
- Hedges, L.V. Distribution Theory for Glass’s Estimator of Effect size and Related Estimators. J. Educ. Stat. 1981, 6, 107–128. [Google Scholar] [CrossRef]
| 1. PRP Preparation | |
|---|---|
| Initial blood volume | 90 mL per subject (8 mL per tube) |
| Anticoagulant | Sodium citrate 3.8% (w/v) |
| System | Close |
| Centrifugation | Yes |
| Number | 1 |
| Seed | 580 g–8 min |
| Final PRP volume | >20 mL per subject |
| 2. PRP Characteristics | |
| PRP Type | 13-00-11 |
| MPV | 8.8 ± 0.6 fL |
| Red Blood Cells | <0.01 × 106/µL |
| White Blood Cells | <0.05 × 106/µL |
| Neutrophils | - |
| Lymphocytes | - |
| Monocytes | - |
| Eosinophils | - |
| Basophils | - |
| Activation | 10 µL CaCl2 (10% w/v) per mL PRP |
| 3. Other Remarkable PRP and Study features | |
| The product used for the analysis of cytokine and growth factors was the platelet lysate obtained after activation of PRP with CaCl2. | |
| N. | Coordinates | Analyte | Integrated OD ± SD | Integrated OD ± SD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PPP | PRP | p Value | PRP/PPP Ratio | Exo | Exo-Ca2+ | p Value | Exo-Ca2+/Exo Ratio | |||
| 1 | A3, A4 | Adiponectin | 1114 ± 5 | 1017 ± 45 | 0.094 | 0.91 | 289 ± 41 | 227 ± 22 | 0.200 | 0.79 |
| 2 | A5, A6 | Apo A-I | 1817 ± 88 | 2280 ± 53 | 0.024 * | 1.26 | 1892 ± 38 | 1554 ± 35 | 0.011 * | 0.82 |
| 3 | A7, A8 | Angiogenin | 2840 ± 12 | 2720 ± 102 | 0.242 | 0.96 | 898 ± 250 | 571 ± 5 | 0.206 | 0.66 |
| 4 | A15, A16 | BDNF | 256 ± 1 | 597 ± 45 | 0.009 ** | 2.33 | 350 ± 22 | 193 ± 10 | 0.012 * | 0.55 |
| 5 | A17, A18 | Complement C5/C5a | 474 ± 35 | 695 ± 16 | 0.015 * | 1.47 | 143 ± 17 | 359 ± 9 | 0.004 ** | 2.52 |
| 6 | B5, B6 | Chitinase 3-like 1 | 981 ± 71 | 567 ± 18 | 0.015 * | 0.58 | ||||
| 7 | B7, B8 | Complement Factor D | 444 ± 26 | 452 ± 4 | 0.720 | 1.02 | ||||
| 8 | B9, B10 | C-Reactive Protein | 2287 ± 80 | 2059 ± 48 | 0.074 | 0.90 | ||||
| 9 | B13, B14 | Cystatin C | 1388 ± 134 | 876 ± 11 | 0.033 * | 0.63 | ||||
| 10 | B17, B18 | Dipeptidyl peptidase-4 (DPP4) | 2632 ± 216 | 2486 ± 45 | 0.449 | 0.95 | ||||
| 11 | B19, B20 | EGF | 108 ± 16 | 395 ± 11 | 0.002 ** | 3.70 | ||||
| 12 | C3, C4 | Emmprin | 180 ± 23 | 131 ± 36 | 0.246 | 0.74 | 328 ± 19 | 42 ± 7 | 0.003 ** | 0.13 |
| 13 | C3, C4 | CXCL5 (ENA-78) | 183 ± 4 | 613 ± 91 | 0.022 * | 3.35 | 116 ± 7 | 39 ± 13 | 0.018 * | 0.34 |
| 14 | C5, C6 | Endoglin | 895 ± 46 | 851 ± 35 | 0.394 | 0.95 | 70 ± 5 | 9 ± 7 | 0.010 * | 0.13 |
| 15 | D7, D8 | ICAM-1 | 314 ± 23 | 457 ± 38 | 0.044 * | 1.46 | ||||
| 16 | D11, D12 | IGFBP-2 | 1470 ± 80 | 1434 ± 104 | 0.735 | 0.98 | ||||
| 17 | D13, D14 | IGFBP-3 | 485 ± 48 | 250 ± 22 | 0.025 * | 0.52 | ||||
| 18 | E23, E24 | IL18-Binding Protein | 839 ± 155 | 1034 ± 157 | 0.338 | 1.25 | ||||
| 19 | G1, G2 | Leptin | 234 ± 40 | 415 ± 5 | 0.024 * | 1.80 | ||||
| 20 | G5, G6 | Lipocalin-2 | 601 ± 30 | 403 ± 3 | 0.011 * | 0.67 | 96 ± 4 | 70 ± 4 | 0.193 | 0.73 |
| 21 | G23, G24 | MMP-9 | 209 ± 32 | 369 ± 47 | 0.057 | 1.78 | ||||
| 22 | H3, H4 | Osteopontin | 670 ± 40 | 573 ± 39 | 0.135 | 0.86 | ||||
| 23 | H5, H6 | PDGF-AA | 247 ± 27 | 544 ± 11 | 0.005 ** | 2.22 | 220 ± 28 | 73 ± 7 | 0.019 * | 0.34 |
| 24 | H7, H8 | PDGF-AB/BB | 156 ± 32 | 404 ± 12 | 0.010 ** | 2.65 | 194 ± 14 | 47 ± 7 | 0.006 ** | 0.24 |
| 25 | H11, H12 | Platelet factor 4 (PF4) | 1889 ± 162 | 2367 ± 172 | 0.103 | 1.26 | 2004 ± 84 | 2135 ± 272 | 0.642 | 1.07 |
| 26 | H15, H16 | CCL5 | 427 ± 78 | 500 ± 81 | 0.455 | 1.19 | 731 ± 57 | 531 ± 64 | 0.081 | 0.73 |
| 27 | H17, H18 | RBP-4 | 2000 ± 7 | 1861 ± 32 | 0.028 * | 0.93 | 799 ± 34 | 544 ± 68 | 0.042 * | 0.68 |
| 28 | I1, I2 | PAI-1 | 1548 ± 275 | 1860 ± 84 | 0.265 | 1.22 | 411 ± 9 | 564 ± 50 | 0.051 | 1.37 |
| 29 | I3, I4 | SHBG | 1560 ± 15 | 1949 ± 35 | 0.005 ** | 1.25 | 279 ± 32 | 23 ± 1 | 0.008 ** | 0.08 |
| 30 | I9, I10 | Trefoil Factor 3 (TFF3) | 146 ± 39 | 156 ± 17 | 0.780 | 1.11 | 667 ± 48 | 513 ± 37 | 0.069 | 0.77 |
| 31 | I15, I16 | Thrombospondin-1 | 703 ± 39 | 957 ± 77 | 0.053 | 1.36 | ||||
| 32 | J5, J6 | Vitamin D BP (VDB) | 1740 ± 47 | 2126 ± 55 | 0.017 * | 1.22 | 1809 ± 115 | 1520 ± 2 | 0.070 | 0.84 |
| 33 | J7, J8 | PECAM-1 (CD31) | 977 ± 1 | 828 ± 20 | 0.009 ** | 0.85 | 2070 ± 2 | 784 ± 26 | 0.0002 *** | 0.38 |
| 34 | J9, J10 | TIM-3 | 345 ± 75 | 304 ± 18 | 0.529 | 0.90 | 109 ± 40 | 40 ± 13 | 0.124 | 0.38 |
| 35 | J11, J12 | VCAM-1 | 1662 ± 38 | 2035 ± 59 | 0.017 * | 1.22 | ||||
| Target | Dilution | Clonality (Clone) | Species (Isotype) | Immunizing Antigen | Use | Source, Catalog N. |
|---|---|---|---|---|---|---|
| β-actin | 1:5000 | Monoclonal (AC-15) | Mouse (IgG1) | Epitope mapping at the N-terminal end of β-actin | Loading control | Sigma, A5441, |
| Caveolin-1 | 1:750 | Polyclonal | Rabbit (IgG) | Synthetic peptide mapping at the N-terminus of caveolin-1 | Components of cell surface machinery for endosome formation | Santa Cruz Biotech., sc-894 |
| Human Porin proteins VDAC1,3 | 1:1000 | Monoclonal (20B12AF2) | Mouse (IgG2b) | Full length protein | Mitochondrial marker | Abcam, ab14734 |
| Cyclophilin F | 1:1000 | Monoclonal (E11AE12BD4) | Mouse (IgG1) | Full length Rat Cyclophilin F | Mitochondrial marker | Abcam, ab110324 |
| Cytochrome C | 1:1000 | Monoclonal (37BA11) | Mouse (IgG2a) | Bovine heart Cytochrome C | Mitochondrial marker | Abcam, ab110325 |
| Calreticulin | 1:1000 | Monoclonal (FMC 75) | Mouse (IgG) | Fusion protein comprising calreticulin | Endoplasmic reticulum marker | Abcam, ab22683 |
| Lamp1 | 1:500 | Monoclonal (H4A3) | Mouse (IgG1) | Adherent spleen cells | Lysosome marker | Santa Cruz Biotech., sc-20011 |
| Rab5 | 1:1000 | Monoclonal (D-11) | Mouse (IgG2b) | Full-length Rab5A | Early endosome marker | Santa Cruz Biotech., sc-46692 |
| Flotillin-1 | 1:1000 | Polyclonal | Rabbit (IgG) | Residues 1-100 of human flotillin-1 conjugated to KLH | Lipid raft marker, often present in exosome membranes | Abcam, ab41927 |
| Hsp70 | 1:1000 | Polyclonal | Rabbit (ns) | License protected | Platelet cytosolic protein identified as a cargo protein in some exosomes | System Biosciences, EXOAB-Hsp70A-1 |
| CD63 | 1:3000 | Polyclonal | Rabbit (ns) | License protected | Widely used as exosome surface marker | System Biosciences, EXOAB-CD63A-1 |
| CD81 | 1:1000 | Polyclonal | Rabbit (ns) | License protected | Widely used as exosome surface marker | System Biosciences, EXOAB-CD81A-1 |
| CD9 | 1:1000 | Polyclonal | Rabbit (ns) | License protected | Widely used as exosome surface marker | System Biosciences, EXOAB-CD9A-1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saumell-Esnaola, M.; Delgado, D.; García del Caño, G.; Beitia, M.; Sallés, J.; González-Burguera, I.; Sánchez, P.; López de Jesús, M.; Barrondo, S.; Sánchez, M. Isolation of Platelet-Derived Exosomes from Human Platelet-Rich Plasma: Biochemical and Morphological Characterization. Int. J. Mol. Sci. 2022, 23, 2861. https://doi.org/10.3390/ijms23052861
Saumell-Esnaola M, Delgado D, García del Caño G, Beitia M, Sallés J, González-Burguera I, Sánchez P, López de Jesús M, Barrondo S, Sánchez M. Isolation of Platelet-Derived Exosomes from Human Platelet-Rich Plasma: Biochemical and Morphological Characterization. International Journal of Molecular Sciences. 2022; 23(5):2861. https://doi.org/10.3390/ijms23052861
Chicago/Turabian StyleSaumell-Esnaola, Miquel, Diego Delgado, Gontzal García del Caño, Maider Beitia, Joan Sallés, Imanol González-Burguera, Pello Sánchez, Maider López de Jesús, Sergio Barrondo, and Mikel Sánchez. 2022. "Isolation of Platelet-Derived Exosomes from Human Platelet-Rich Plasma: Biochemical and Morphological Characterization" International Journal of Molecular Sciences 23, no. 5: 2861. https://doi.org/10.3390/ijms23052861
APA StyleSaumell-Esnaola, M., Delgado, D., García del Caño, G., Beitia, M., Sallés, J., González-Burguera, I., Sánchez, P., López de Jesús, M., Barrondo, S., & Sánchez, M. (2022). Isolation of Platelet-Derived Exosomes from Human Platelet-Rich Plasma: Biochemical and Morphological Characterization. International Journal of Molecular Sciences, 23(5), 2861. https://doi.org/10.3390/ijms23052861






