Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Preparation of Plasma
2.3. Extraction of Genomic DNA
2.4. Extraction of Circulating Cell-Free DNA
2.5. Synthetic Oligonucleotides
2.6. Sequencing Reaction
2.7. Droplet Digital™ PCR (ddPCR)
2.8. Statistical Analysis
3. Results
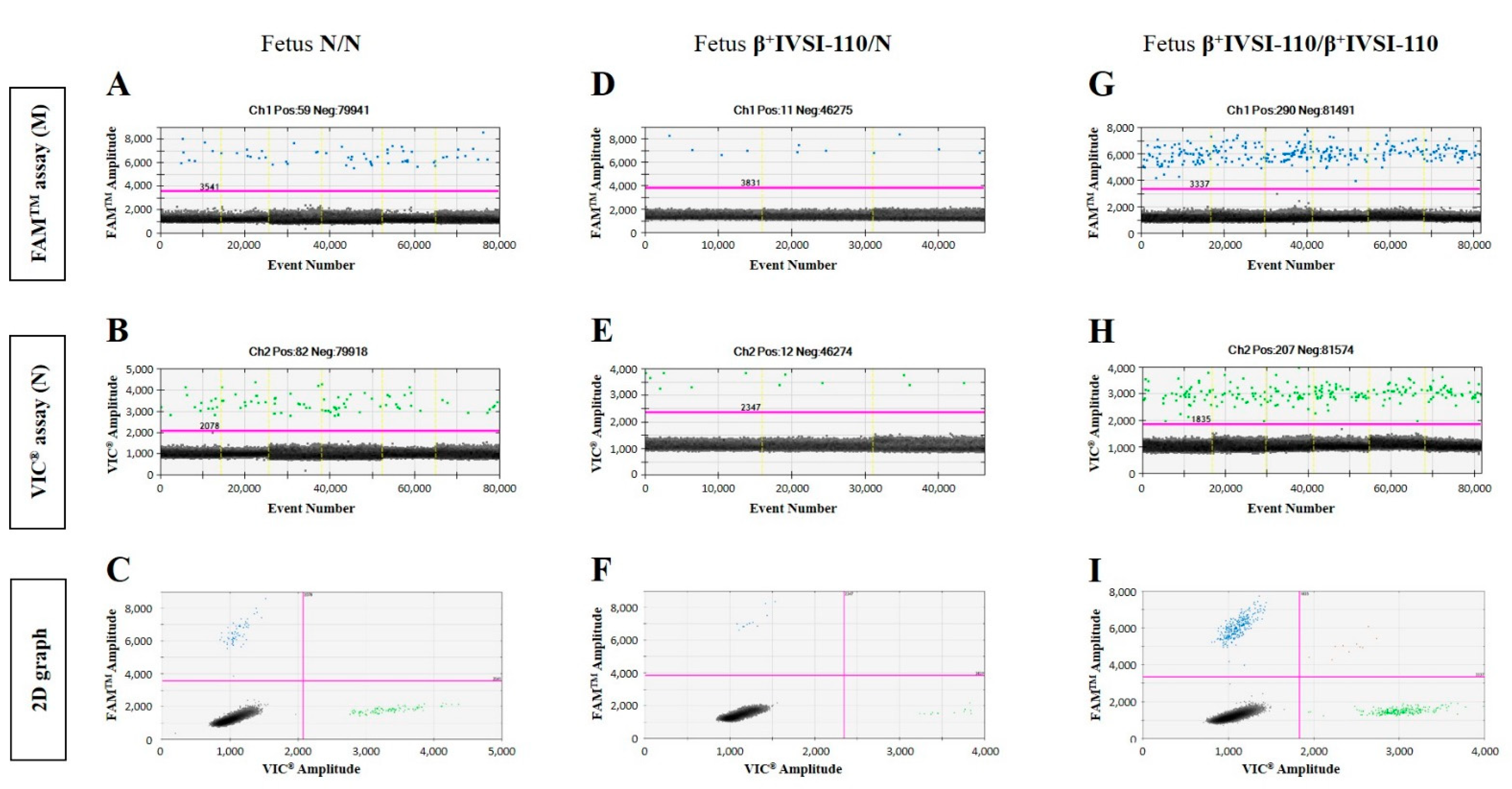
3.1. Validation of β+IVSI-110 and β039 Thalassemia ddPCR Assays by Genomic DNA Mixtures Mimicking Circulating Cell-Free Fetal DNA and Maternal DNA
3.2. NIPT of β+IVSI-110 or β039 Thalassemia Mutations Using ddPCR Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Lun, F.M.; Tsui, N.B.; Chan, K.C.; Leung, T.Y.; Lau, T.K.; Charoenkwan, P.; Chow, K.C.; Lo, W.Y.; Wanapirak, C.; Sanguansermsri, T.; et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc. Natl. Acad. Sci. USA 2008, 105, 19920–19925. [Google Scholar] [CrossRef] [PubMed]
- Illanes, S.; Denbow, M.; Kailasam, C.; Finning, K.; Soothill, P.W. Early detection of cell-free fetal DNA in maternal plasma. Early Hum. Dev. 2007, 83, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Drury, S.; Hill, M.; Chitty, L.S. Cell-Free Fetal DNA Testing for Prenatal Diagnosis. Adv. Clin. Chem. 2016, 76, 1–35. [Google Scholar] [PubMed]
- Zhou, Y.; Zhu, Z.; Gao, Y.; Yuan, Y.; Guo, Y.; Zhou, L.; Liao, K.; Wang, J.; Du, B.; Hou, Y.; et al. Effects of maternal and fetal characteristics on cell-free fetal DNA fraction in maternal plasma. Reprod. Sci. 2015, 22, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, H.J.; Park, S.Y.; Han, Y.J.; Choi, J.S.; Ryu, H.M. Early Prediction of Hypertensive Disorders of Pregnancy Using Cell-Free Fetal DNA, Cell-Free Total DNA, and Biochemical Markers. Fetal Diagn. Ther. 2016, 40, 255–262. [Google Scholar] [CrossRef]
- Vora, N.L.; Johnson, K.L.; Basu, S.; Catalano, P.M.; Hauguel-De Mouzon, S.; Bianchi, D.W. A multifactorial relationship exists between total circulating cell-free DNA levels and maternal BMI. Prenat. Diagn. 2012, 32, 912–914. [Google Scholar] [CrossRef]
- Attilakos, G.; Maddocks, D.G.; Davies, T.; Hunt, L.P.; Avent, N.D.; Soothill, P.W.; Grant, S.R. Quantification of free fetal DNA in multiple pregnancies and relationship with chorionicity. Prenat. Diagn. 2011, 31, 967–972. [Google Scholar] [CrossRef]
- Wright, C.F.; Wei, Y.; Higgins, J.P.; Sagoo, G.S. Non-invasive prenatal diagnostic test accuracy for fetal sex using cell-free DNA a review and meta-analysis. BMC Res. Notes 2012, 5, 476. [Google Scholar] [CrossRef]
- Perlado-Marina, S.; Bustamante-Aragones, A.; Horcajada, L.; Trujillo-Tiebas, M.J.; Lorda-Sanchez, I.; Ruiz Ramos, M.; Plaza, J.; Rodriguez de Alba, M. Overview of Five-Years of Experience Performing Non-Invasive Fetal Sex Assessment in Maternal Blood. Diagnostics 2013, 3, 283–290. [Google Scholar] [CrossRef]
- Clausen, F.B.; Damkjær, M.B.; Dziegiel, M.H. Noninvasive fetal RhD genotyping. Transfus. Apher. Sci. 2014, 50, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Fasano, R.M. Hemolytic disease of the fetus and newborn in the molecular era. Semin. Fetal Neonatal. Med. 2016, 21, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Contro, E.; Bernabini, D.; Farina, A. Cell-Free Fetal DNA for the Prediction of Pre-Eclampsia at the First and Second Trimesters: A Systematic Review and Meta-Analysis. Mol. Diagn. Ther. 2017, 21, 125–135. [Google Scholar] [CrossRef] [PubMed]
- van Boeckel, S.R.; Davidson, D.J.; Norman, J.E.; Stock, S.J. Cell-free Fetal DNA and Spontaneous Preterm Birth. Reproduction 2017, 155, R137–R145. [Google Scholar] [CrossRef] [PubMed]
- Rolnik, D.L.; da Silva Costa, F.; Lee, T.J.; Schmid, M.; McLennan, A.C. Association between fetal fraction on cell-free DNA testing and first trimester markers for pre-eclampsia. Ultrasound Obstet. Gynecol. 2018, 52, 722–727. [Google Scholar] [CrossRef]
- Skrzypek, H.; Hui, L. Noninvasive prenatal testing for fetal aneuploidy and single gene disorders. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 42, 26–38. [Google Scholar] [CrossRef]
- Mennuti, M.T.; Chandrasekaran, S.; Khalek, N.; Dugoff, L. Cell-free DNA screening and sex chromosome aneuploidies. Prenat. Diagn. 2015, 35, 980–985. [Google Scholar] [CrossRef]
- Wright, C.F.; Burton, H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum. Reprod. Update 2009, 15, 139–151. [Google Scholar] [CrossRef]
- Bustamante-Aragonés, A.; Rodríguez de Alba, M.; Perlado, S.; Trujillo-Tiebas, M.J.; Arranz, J.P.; Díaz-Recasens, J.; Troyano-Luque, J.; Ramos, C. Non-invasive prenatal diagnosis of single-gene disorders from maternal blood. Gene 2012, 504, 144–149. [Google Scholar] [CrossRef]
- Perlado, S.; Bustamante-Aragonés, A.; Donas, M.; Lorda-Sánchez, I.; Plaza, J.; Rodríguez de Alba, M. Fetal Genotyping in Maternal Blood by Digital PCR: Towards NIPD of Monogenic Disorders Independently of Parental Origin. PLoS ONE 2016, 11, e0153258. [Google Scholar] [CrossRef][Green Version]
- Hudecova, I. Digital PCR analysis of circulating nucleic acids. Clin. Biochem. 2015, 48, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Hui, W.W.; Jiang, P.; Tong, Y.K.; Lee, W.S.; Cheng, Y.K.; New, M.I.; Kadir, R.A.; Chan, K.C.; Leung, T.Y.; Lo, Y.M.; et al. Universal Haplotype-Based Noninvasive Prenatal Testing for Single Gene Diseases. Clin. Chem. 2017, 63, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Kagan, K.O.; Sonek, J.; Wagner, P.; Hoopmann, M. Principles of first trimester screening in the age of non-invasive prenatal diagnosis: Screening for chromosomal abnormalities. Arch. Gynecol. Obstet. 2017, 296, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Chiu, E.K.L.; Hui, W.W.I.; Chiu, R.W.K. cfDNA screening and diagnosis of monogenic disorders-where are we heading? Prenat. Diagn. 2018, 38, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Beulen, L.; Faas, B.H.W.; Feenstra, I.; van Vugt, J.M.G.; Bekker, M.N. Clinical utility of non-invasive prenatal testing in pregnancies with ultrasound anomalies. Ultrasound Obstet. Gynecol. 2017, 49, 721–728. [Google Scholar] [CrossRef]
- Hayward, J.; Chitty, L.S. Beyond screening for chromosomal abnormalities: Advances in non-invasive diagnosis of single gene disorders and fetal exome sequencing. Semin. Fetal Neonatal. Med. 2018, 23, 94–101. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, S.J.; Han, S.H.; Park, J.S.; Choi, H.J.; Ahn, J.J.; Oh, M.J.; Shim, S.H.; Cha, D.H.; Hwang, S.Y. A new approach of digital PCR system for non-invasive prenatal screening of trisomy 21. Clin. Chim. Acta 2018, 476, 75–80. [Google Scholar] [CrossRef]
- Sawakwongpra, K.; Tangmansakulchai, K.; Ngonsawan, W.; Promwan, S.; Chanchamroen, S.; Quangkananurug, W.; Sriswasdi, S.; Jantarasaengaram, S.; Ponnikorn, S. Droplet-based digital PCR for non-invasive prenatal genetic diagnosis of α and β-thalassemia. Biomed. Rep. 2021, 15, 82. [Google Scholar] [CrossRef]
- D′Aversa, E.; Breveglieri, G.; Pellegatti, P.; Guerra, G.; Gambari, R.; Borgatti, M. Non-invasive fetal sex diagnosis in plasma of early weeks pregnants using droplet digital PCR. Mol. Med. 2018, 24, 14. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Thein, S.L. The molecular basis of β-thalassemia. Cold Spring Harb. Perspect. Med. 2013, 3, 5. [Google Scholar] [CrossRef]
- Viprakasit, V.; Ekwattanakit, S. Clinical Classification, Screening and Diagnosis for Thalassemia. Hematol. Oncol. Clin. N. Am. 2018, 32, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Tan, C.; Chen, X.; Wang, F.; Wang, D.; Cao, Z.; Zhu, X.; Lu, C.; Yang, W.; Gao, N.; Gao, H.; et al. A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies. Analyst 2019, 144, 2239–2247. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.Z.; Lewis, D.E.; Simpson, J.L. Cell-free fetal DNA in maternal blood: Kinetics, source and structure. Hum. Reprod. Update 2005, 11, 59–67. [Google Scholar] [CrossRef]
- Chan, K.C.; Zhang, J.; Hui, A.B.; Wong, N.; Lau, T.K.; Leung, T.N.; Lo, K.W.; Huang, D.W.; Lo, Y.M. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004, 50, 88–92. [Google Scholar] [CrossRef]
- Li, Y.; Zimmermann, B.; Rusterholz, C.; Kang, A.; Holzgreve, W.; Hahn, S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin. Chem. 2004, 50, 1002–1011. [Google Scholar] [CrossRef]
- Liang, B.; Li, H.; He, Q.; Li, H.; Kong, L.; Xuan, L.; Xia, Y.; Shen, J.; Mao, Y.; Li, Y.; et al. Enrichment of the fetal fraction in non-invasive prenatal screening reduces maternal background interference. Sci. Rep. 2018, 8, 17675. [Google Scholar] [CrossRef]
- Breveglieri, G.; D′Aversa, E.; Finotti, A.; Borgatti, M. Non-invasive Prenatal Testing Using Fetal DNA. Mol. Diagn. Ther. 2019, 23, 291–299. [Google Scholar] [CrossRef]
- Gruber, A.; Pacault, M.; El Khattabi, L.A.; Vaucouleur, N.; Orhant, L.; Bienvenu, T.; Girodon, E.; Vidaud, D.; Leturcq, F.; Costa, C.; et al. Non-invasive prenatal diagnosis of paternally inherited disorders from maternal plasma: Detection of NF1 and CFTR mutations using droplet digital PCR. Clin. Chem. Lab. Med. 2018, 56, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, Q.; Zhou, W.; Zhong, M.; Guo, X.; Wang, X.; Fan, X.; Yan, S.; Li, L.; Lai, Y.; et al. Cell-free DNA Barcode-Enabled Single-Molecule Test for Non invasive Prenatal Diagnosis of Monogenic Disorders: Application to β-Thalassemia. Adv. Sci. 2019, 6, 1802332. [Google Scholar] [CrossRef] [PubMed]
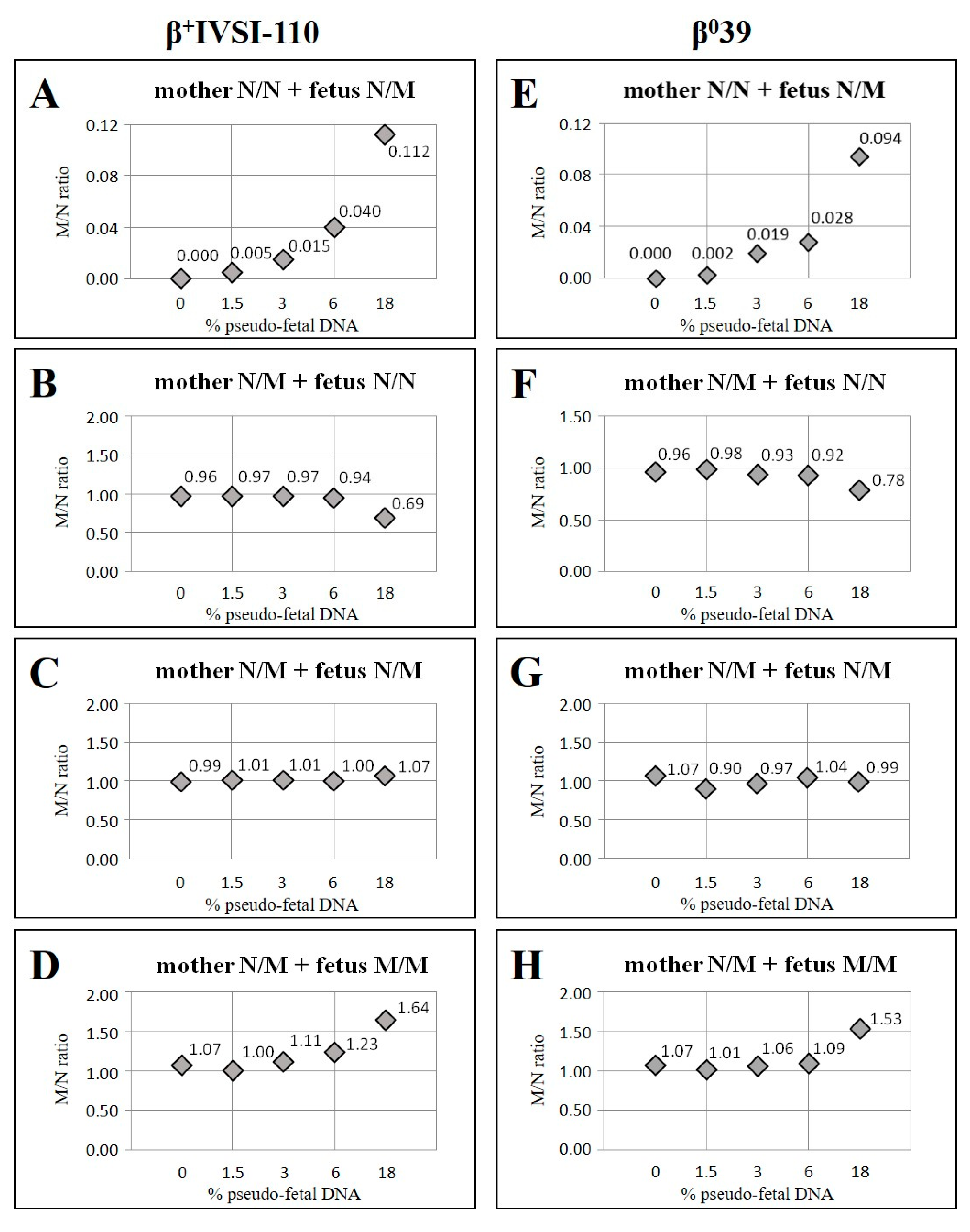


| Mother β+IVSI-110/N with Fetus N/N | Mother β039/N with Fetus N/N | ||||||
|---|---|---|---|---|---|---|---|
| Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 | Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 |
| 0 | 1.00 | 0.96 | 96 | 0 | 1.00 | 0.96 | 96 |
| 1.5 | 0.97 | 0.97 | 100 | 1.5 | 0.97 | 0.98 | 101 |
| 3 | 0.94 | 0.97 | 103 | 3 | 0.94 | 0.93 | 99 |
| 6 | 0.89 | 0.94 | 106 | 6 | 0.89 | 0.92 | 104 |
| 18 | 0.69 | 0.69 | 100 | 18 | 0.69 | 0.78 | 112 |
| Mother β+IVSI-110/N-with Fetus β+IVSI-110/β+IVSI 110 | Mother β039/N with Fetus β039/β039 | ||||||
| Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 | Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 |
| 0 | 1.00 | 1.07 | 107 | 0 | 1.00 | 1.07 | 107 |
| 1.5 | 1.03 | 1.00 | 97 | 1.5 | 1.03 | 1.01 | 98 |
| 3 | 1.06 | 1.11 | 105 | 3 | 1.06 | 1.06 | 100 |
| 6 | 1.13 | 1.23 | 109 | 6 | 1.13 | 1.09 | 97 |
| 18 | 1.44 | 1.64 | 114 | 18 | 1.44 | 1.53 | 106 |
| Mother β+IVSI-110/N with Fetus β+IVSI-110/N | Mother β039/N with Fetus β039/N | ||||||
| Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 | Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 |
| 0 | 1.00 | 0.99 | 99 | 0 | 1.00 | 1.07 | 107 |
| 1.5 | 1.00 | 1.01 | 101 | 1.5 | 1.00 | 0.90 | 90 |
| 3 | 1.00 | 1.01 | 101 | 3 | 1.00 | 0.97 | 97 |
| 6 | 1.00 | 1.00 | 100 | 6 | 1.00 | 1.04 | 104 |
| 18 | 1.00 | 1.07 | 107 | 18 | 1.00 | 0.99 | 99 |
| Pregnant Women N/N with Partner β+IVSI-110/N | Pregnant Women N/N with Partner β039/N | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Gestational Weeks | Positive Events for M Allele (no.) | Formulated Diagnosis | Fetal Genotype | Diagnosis Outcome | Sample | Gestational Weeks | Positive Events for M Allele (no.) | Formulated Diagnosis | Fetal Genotype | Diagnosis Outcome |
| 1 | 37 | 0 | N/N | N/N | √ | 25 | 39 | 196 | β039/N | β039/N | √ |
| 2 | 18 | 0 | N/N | N/N | √ | 26 | 35 | 123 | β039/N | β039/N | √ |
| 3 | 16 | 91 | β+IVSI-110/N | β+IVSI-110/N | √ | 27 | 33 | 39 | β039/N | β039/N | √ |
| 4 | 15 | 43 | β+IVSI-110/N | β+IVSI-110/N | √ | 28 | 29 | 19 | β039/N | β039/N | √ |
| 5 | 12 | 1 | N/N | β0IVSII-1/N | √ | 29 | 28 | 78 | β039/N | β039/N | √ |
| 6 | 12 | 12 | β+IVSI-110/N | β+IVSI-110/β039 | √ | 30 | 26 | 33 | β039/N | β039/N | √ |
| 7A | 10 | 178 | β+IVSI-110/N | β+IVSI-110/N | √ | 31 | 25 | 21 | β039/N | β039/N | √ |
| 8 | 9 | 62 | β+IVSI-110/N | β+IVSI-110/N | √ | 32 | 24 | 2 | N/N | N/N | √ |
| 7B | 5 | 3 | β+IVSI-110/N | β+IVSI-110/N | √ | 33 | 24 | 8 | β039/N | β039/N | √ |
| 34 | 21 | 39 | β039/N | β039/N | √ | ||||||
| 35 | 14 | 50 | β039/N | β039/N | √ | ||||||
| 36 | 13 | 4 | β039/N | β039/N | √ | ||||||
| 37 | 7 | 0 | N/N | N/N | √ | ||||||
| 38 | 5 | 26 | β039/N | β039/N | √ | ||||||
| Pregnant Women β+IVSI-110/N with Partner N/N | Pregnant Women β039/N with Partner N/N | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) | # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) |
| 9 | 38 | 0.61 | −3.46 | N/N | N/N (√) | 39 | 39 | 1.09 | 0.52 | β039/N | β039/N (√) |
| 10 | 33 | 0.63 | −3.37 | N/N | N/N(√) | 40 | 36 | 0.89 | −1.19 | β039/N | β039/N (√) |
| 11 | 30 | 0.86 | −1.41 | β+IVSI-110/N | β+IVSI-110/N (√) | 41 | 29 | 0.71 | −2.66 | N/N | N/N (√) |
| 12 | 30 | 0.83 | −1.64 | β+IVSI-110/N | β+IVSI-110/N (√) | 42A | 26 | 1.00 | −0.25 | β039/N | β039/N (√) |
| 13 | 14 | 0.91 | −1.02 | β+IVSI-110/N | β+IVSI-110/N (√) | 43 | 26 | 0.51 | −4.36 | N/N | N/N (√) |
| 14 | 12 | 1.03 | 0.00 | β+IVSI-110/N | β+IVSI-110/N (√) | 42B | 22 | 1.09 | 0.49 | β039/N | β039/N (√) |
| 15 | 12 | 0.95 | −0.67 | β+IVSI-110/N | β+IVSI-110/N (√) | 44 | 20 | 0.68 | −2.93 | N/N | N/N (√) |
| 16 | 12 | 0.91 | −1.00 | β+IVSI-110/N | β+IVSI-110/N (√) | 45 | 18 | 0.78 | −2.06 | β039/N | N/N |
| 17 | 11 | 0.91 | −1.00 | β+IVSI-110/N | β+IVSI-110/N (√) | 46 | 14 | 0.63 | −3.31 | N/N | N/N (√) |
| 18 | 10 | 1.04 | 0.09 | β+IVSI-110/N | β+IVSI-110/N (√) | 42C | 13 | 1.12 | 0.75 | β039/N | β039/N (√) |
| 19 | 9 | 0.72 | −2.59 | N/N | N/N (√) | 6 * | 12 | 1.07 | 0.33 | β039/N | β039/β+IVSI-110 (√) |
| Pregnant Women β+IVSI-110/N with Partner β+IVSI-110/N | 47 | 9 | 0.88 | −1.28 | β039/N | β039/N (√) | |||||
| # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) | 48 | 7 | 1.10 | 0.58 | β039/N | β039/N (√) |
| Pregnant Women β039/N with Partner β039/N | |||||||||||
| # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) | ||||||
| 20 | 12 | 0.94 | −0.73 | β+IVSI-110/N | β+IVSI-110/N (√) | ||||||
| 21 | 10 | 0.70 | −2.71 | N/N | N/N (√) | ||||||
| 22 | 8 | 1.04 | 0.09 | β+IVSI-110/N | β+IVSI-110/N (√) | 49 | 32 | 0.69 | −2.82 | N/N | N/N (√) |
| 23 | 7 | 1.40 | 3.09 | β+IVSI-110/β+IVSI-110 | β+IVSI-110/β+IVSI-110 (√) | ||||||
| 24 | 7 | 0.90 | −0.94 | β+IVSI-110/N | β+IVSI-110/N (√) | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aversa, E.; Breveglieri, G.; Boutou, E.; Balassopoulou, A.; Voskaridou, E.; Pellegatti, P.; Guerra, G.; Scapoli, C.; Gambari, R.; Borgatti, M. Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma. Int. J. Mol. Sci. 2022, 23, 2819. https://doi.org/10.3390/ijms23052819
D’Aversa E, Breveglieri G, Boutou E, Balassopoulou A, Voskaridou E, Pellegatti P, Guerra G, Scapoli C, Gambari R, Borgatti M. Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma. International Journal of Molecular Sciences. 2022; 23(5):2819. https://doi.org/10.3390/ijms23052819
Chicago/Turabian StyleD’Aversa, Elisabetta, Giulia Breveglieri, Effrossyni Boutou, Angeliki Balassopoulou, Ersi Voskaridou, Patrizia Pellegatti, Giovanni Guerra, Chiara Scapoli, Roberto Gambari, and Monica Borgatti. 2022. "Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma" International Journal of Molecular Sciences 23, no. 5: 2819. https://doi.org/10.3390/ijms23052819
APA StyleD’Aversa, E., Breveglieri, G., Boutou, E., Balassopoulou, A., Voskaridou, E., Pellegatti, P., Guerra, G., Scapoli, C., Gambari, R., & Borgatti, M. (2022). Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma. International Journal of Molecular Sciences, 23(5), 2819. https://doi.org/10.3390/ijms23052819









