Identification and Functional Analysis of a Defensin CcDef2 from Coridius chinensis
Abstract
1. Introduction
2. Results
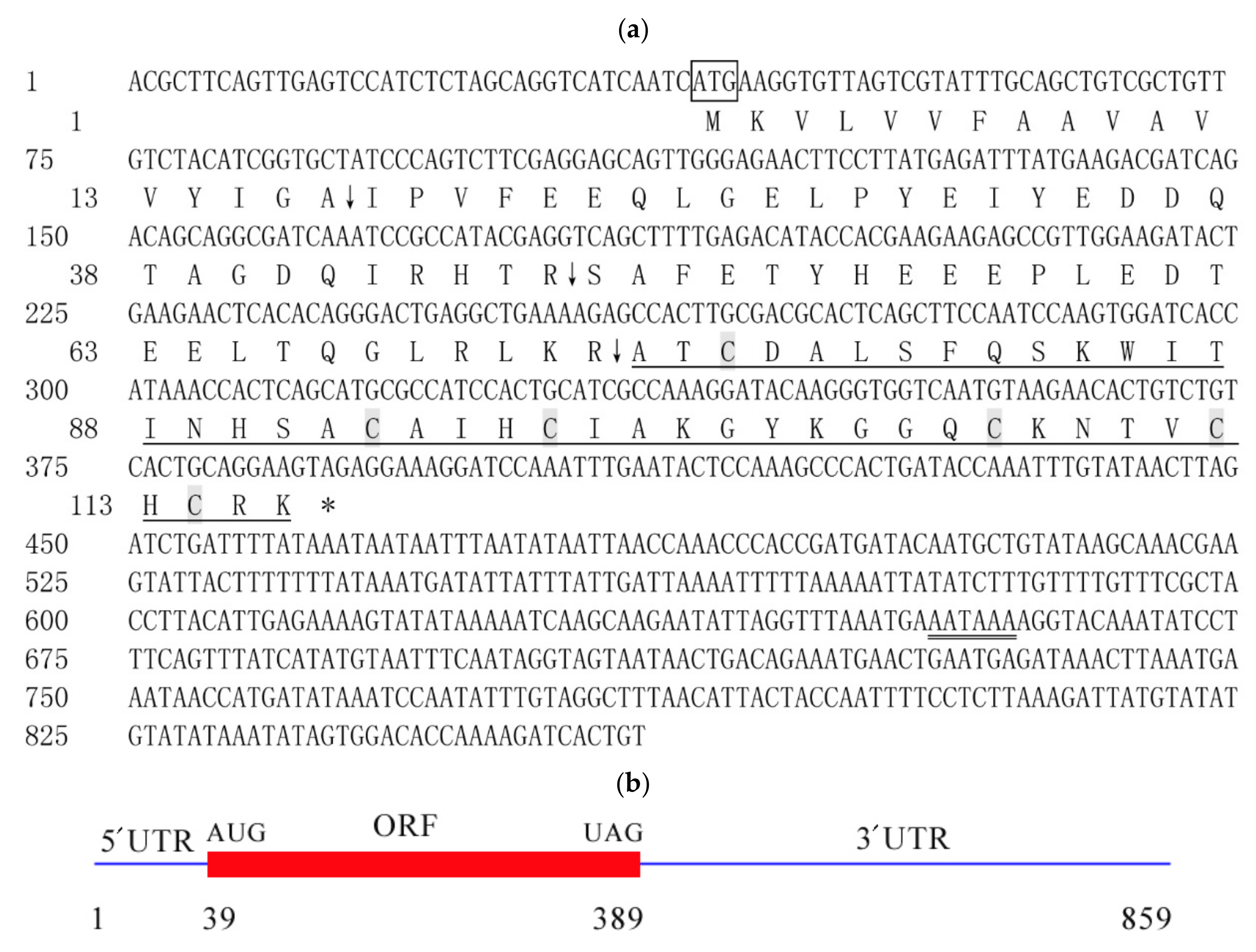
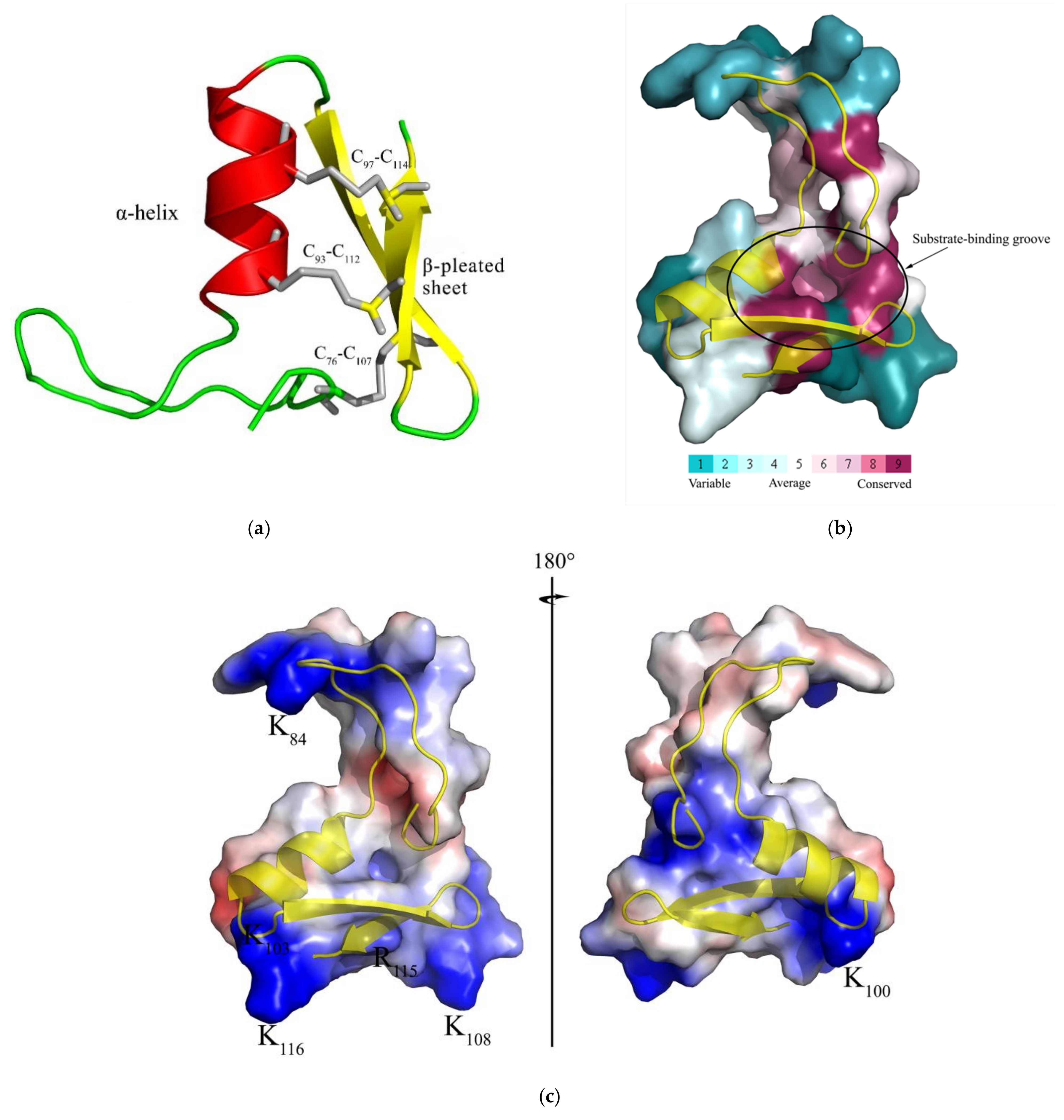
2.1. Characteristic Analysis of CcDef2
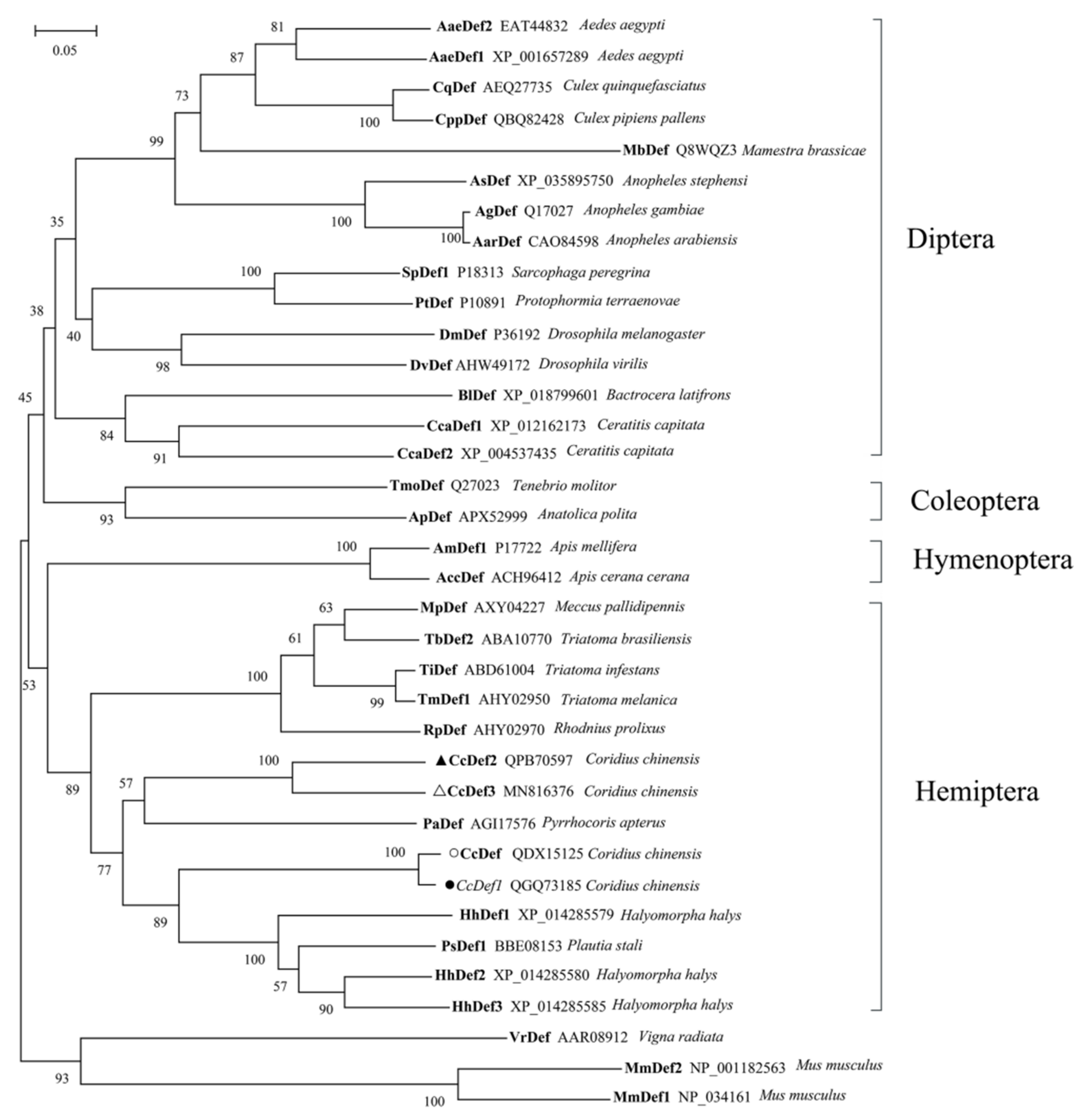
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
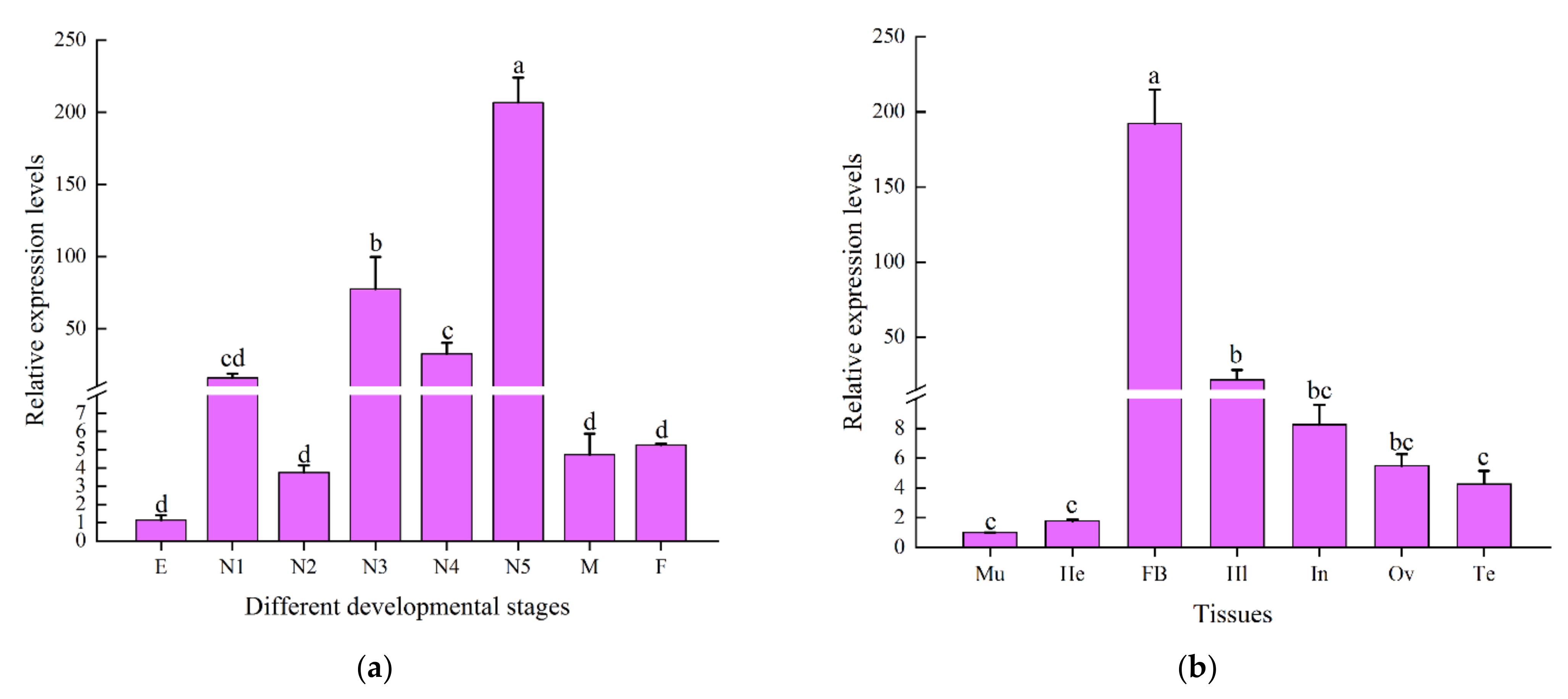
2.3. Spatiotemporal Expression Profile
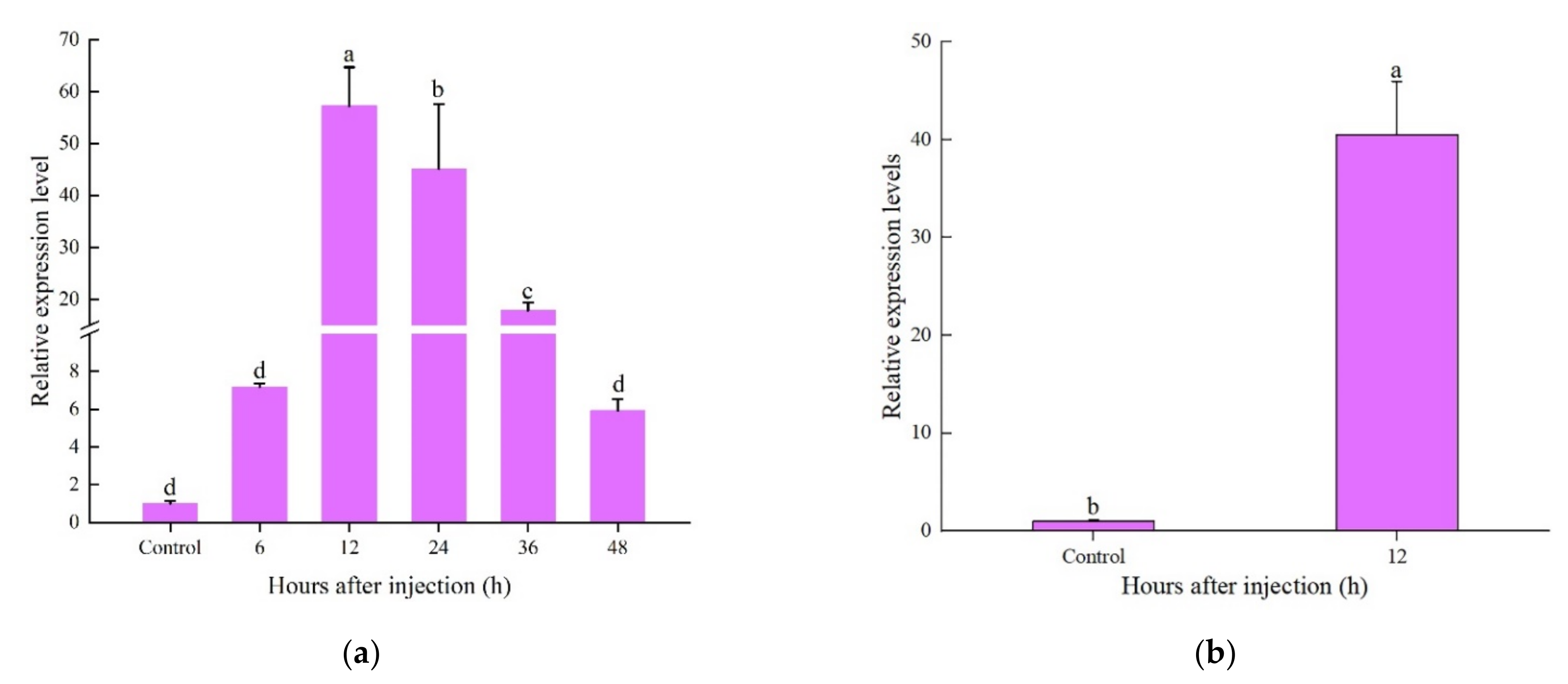
2.4. Expression Patterns of CcDef2 upon Bacterial Challenge
2.5. Expression Analysis of Recombinant Protein
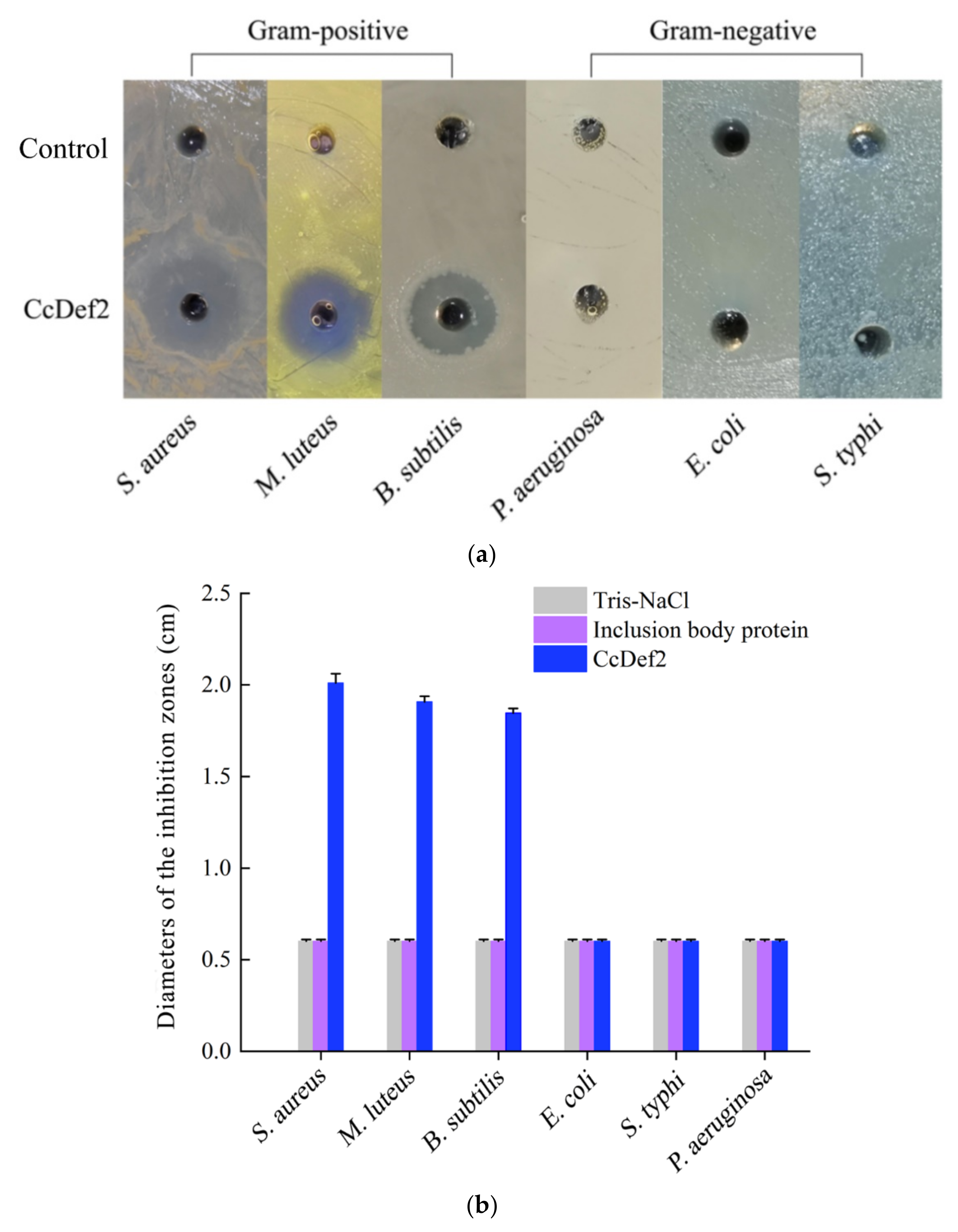
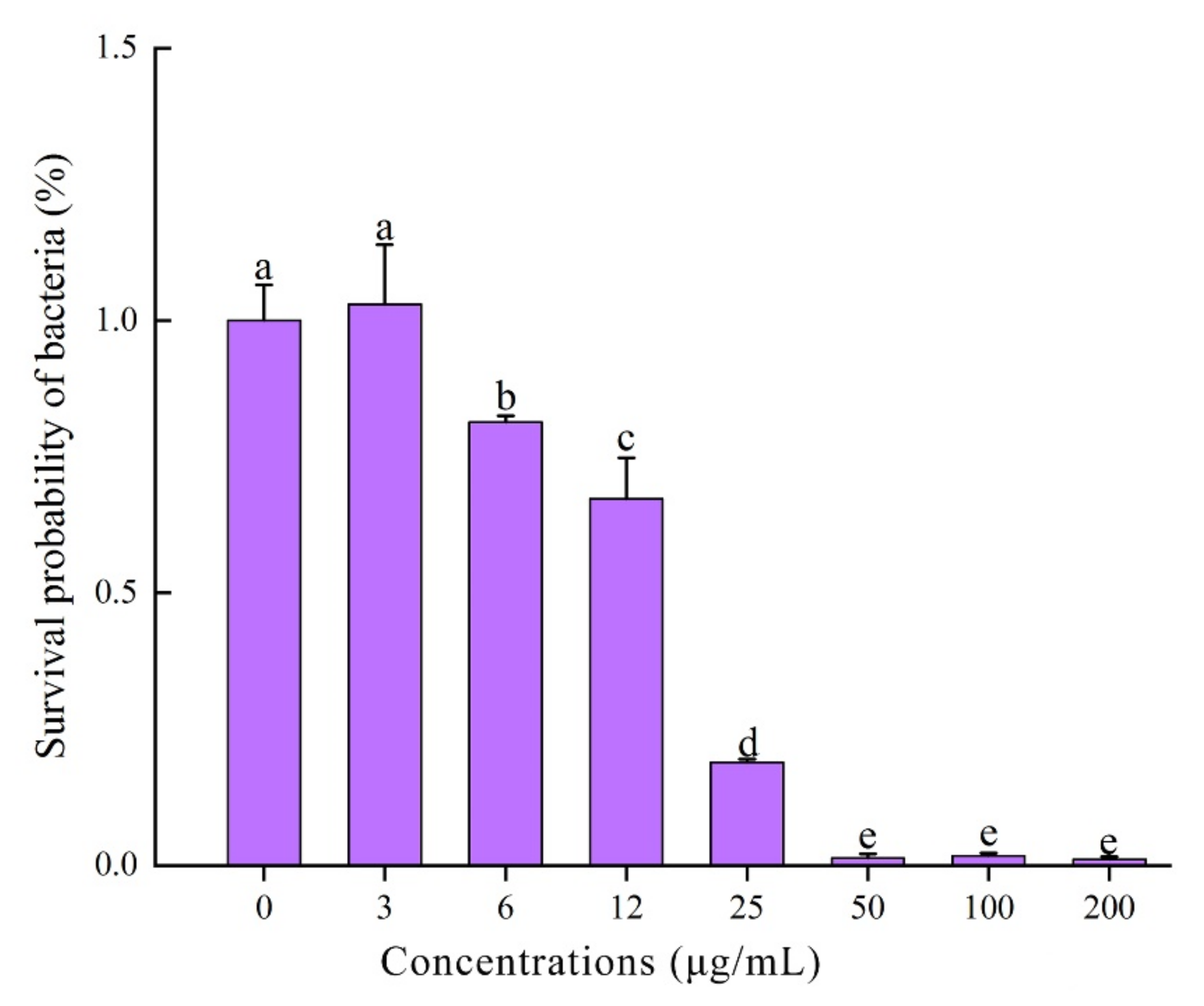
2.6. Antibacterial Spectrum and MIC
3. Discussion
4. Materials and Methods
4.1. Insects and Bacteria
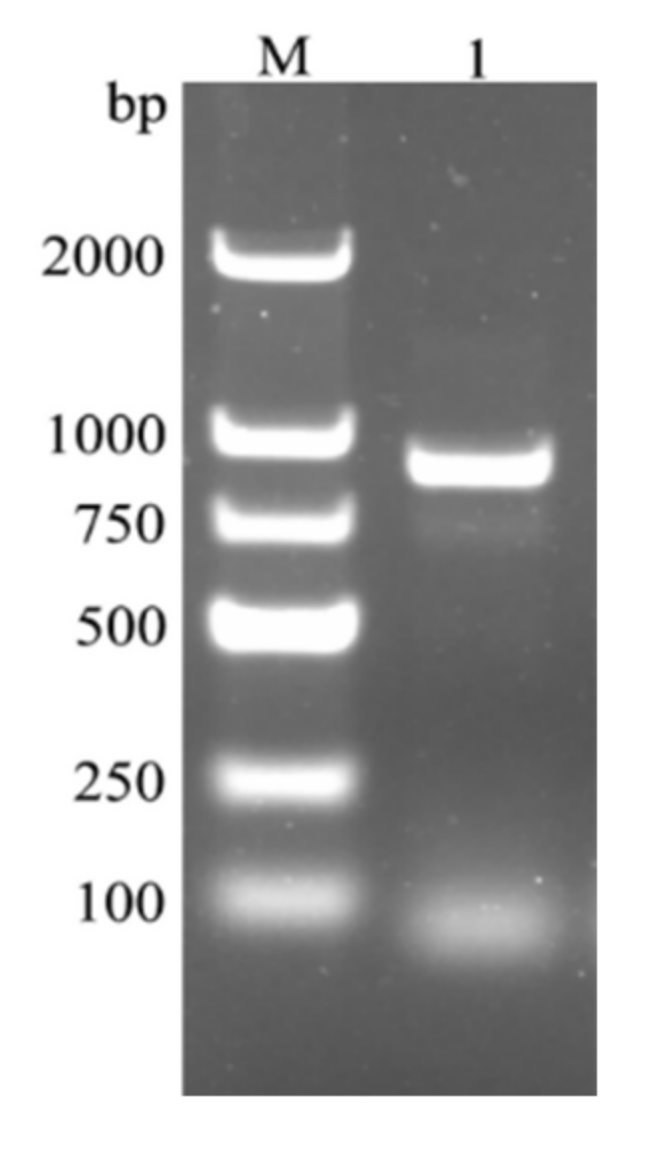
4.2. RNA Extraction and Gene Cloning
4.3. Bioinformatic Analyses
4.4. Spatiotemporal Expression Profile
4.5. Bacterial Infection Assay
4.6. Construction of Recombinant Expression Vector
4.7. Heterologous Expression, Purification and Refolding
4.8. Antimicrobial Assay
4.9. MIC Determination of CcDef2
4.10. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cytryńska, M.; Wojda, I.; Jakubowicz, T. How insects combat infections. In Lessons in Immunity; Ballarin, L., Cammarata, M., Eds.; Academic Press: Amsterdam, The Netherlands, 2016; pp. 117–128. [Google Scholar]
- Hoffmann, J.A. Innate Immunity of Insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Nakhleh, J.; El Moussawi, L.; Osta, M.A. The melanization response in insect immunity. In Advances Insect Physiology; Ligoxygakis, P., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 83–109. [Google Scholar]
- Nappi, A.J.; Christensen, B.M. Melanogenesis and associated cytotoxic reactions: Applications to insect innate immunity. Insect Biochem. Mol. Biol. 2005, 35, 443–459. [Google Scholar] [CrossRef] [PubMed]
- Zug, R.; Hammerstein, P. Wolbachia and the insect immune system: What reactive oxygen species can tell us about the mechanisms of Wolbachia-host interactions. Front. Microbiol. 2015, 6, 1201. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Lavine, M.D.; Strand, M.R. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002, 32, 1295–1309. [Google Scholar] [CrossRef]
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14. [Google Scholar] [CrossRef]
- Bachère, E. Anti-infectious immune effectors in marine invertebrates: Potential tools for disease control in larviculture. Aquaculture 2003, 227, 427–438. [Google Scholar] [CrossRef]
- Bulet, P.; Stöcklin, R.; Menin, L. Anti-microbial peptides: From invertebrates to vertebrates. Immunol. Rev. 2004, 198, 169–184. [Google Scholar] [CrossRef]
- Park, Y.; Hahm, K.S. Antimicrobial peptides (AMPs): Peptide structure and mode of action. J. Biochem. Mol. Biol. 2005, 38, 507–516. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engström, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Lee, J.Y.; Boman, A.; Sun, C.X.; Andersson, M.; Jörnvall, H.; Mutt, V.; Boman, H.G. Antibacterial peptides from pig intestine: Isolation of a mammalian cecropin. Proc. Natl. Acad. Sci. USA 1990, 86, 9159–9162. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Sakanaka, H.S.; Sagisaka, A.; Asaoka, A.; Taylor, D.M.; Yamakawa, M. A short peptide synthesized on the basis of coleoptericin A from Allomyrina dichotoma shows similar anti-Escherichia coli action. J. Insect Biotechnol. Sericology 2006, 75, 135–139. [Google Scholar]
- Cociancich, S.; Dupont, A.; Hegy, G.; Lanot, R.; Holder, F.; Hetru, C.; Hoffmann, J.A.; Bulet, P. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem. J. 1994, 300 Pt 2, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Casteels, P.; Ampe, C.; Riviere, L.; Damme, J.C.; Elicone, C.; Fleming, M.; Jacobs, F.; Tempst, P. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera). Eur. J. Biochem. 1990, 187, 381–386. [Google Scholar] [CrossRef]
- Casteels, P.; Tempst, P. Apidaecin-type peptide antibiotics function through a non-poreforming mechanism involving stereospecificity. Biochem. Biophys. Res. Commun. 1994, 199, 339–345. [Google Scholar] [CrossRef]
- Bulet, P.; Urge, L.; Ohresser, S.; Hetru, C.; Otvos, L. Enlarged scale chemical synthesis and range of activity of drosocin, an O-glycosylated antibacterial peptide of Drosophila. Eur. J. Biochem. 1996, 238, 64–69. [Google Scholar] [CrossRef]
- Zeya, H.I.; Spitznagel, J.K. Cationic proteins of polymorphonuclear leukocyte lysosomes. I. Resolution of antibacterial and enzymatic activities. J. Bacteriol. 1966, 91, 750–754. [Google Scholar] [CrossRef]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 1985, 76, 1427–1435. [Google Scholar] [CrossRef]
- Lambert, J.; Keppi, E.; Dimarcq, J.L.; Wicker, C.; Reichhart, J.M.; Dunbar, B.; Lepage, P.; Dorsselaer, A.; Hoffmann, J.; Fothergill, J.; et al. Insect immunity: Isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proc. Natl. Acad. Sci. USA 1989, 86, 262–266. [Google Scholar] [CrossRef]
- Matsuyama, K.J.; Natori, S.J. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J. Biol. Chem. 1988, 263, 17112–17116. [Google Scholar] [CrossRef]
- Bals, R.; Goldman, M.J.; Wilson, J.M. Mouse β-defensin 1 is a salt-sensitive antimicrobial peptide present in epithelia of the lung and urogenital tract. Infect. Immun. 1998, 66, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, Y.; Yamaguchi, Y.; Yamamoto, H.; Ishii, M.; Nagase, T.; Kurihara, H.; Akishita, M.; Ouchi, Y. In vitro and in vivo anticancer activity of human β-defensin-3 and its mouse homolog. Anticancer Res. 2016, 36, 5999–6004. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Kjos, M.; Nes, I.F.; Diep, D.B.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Cociancich, S.; Ghazi, A.; Hetru, C.; Hoffmann, J.A.; Letellier, L. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus. J. Biol. Chem. 1993, 268, 19239–19245. [Google Scholar] [CrossRef]
- Lee, E.; Kim, J.K.; Shin, S.; Jeong, K.W.; Shin, A.; Lee, J.; Lee, D.; Hwang, J.S.; Kim, Y. Insight into the antimicrobial activities of coprisin isolated from the dung beetle, Copris tripartitus, revealed by structure-activity relationships. Biochim. Biophys. Acta 2012, 1828, 271–283. [Google Scholar] [CrossRef]
- Mor, A. Peptide-based antibiotics: A potential answer to raging antimicrobial resistance. Drug Dev. Res. 2000, 50, 440–447. [Google Scholar] [CrossRef]
- Li, S.W.; Zhao, B.S.; Du, J. Isolation, purification, and detection of the antimicrobial activity of the antimicrobial peptide CcAMP1 from Coridius chinensis (Hemiptera:Dinidoridae). Acta Entomol. Sin. 2015, 58, 610–616. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.A.; Osborn, R.W. Plant defensins: Novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995, 108, 1353. [Google Scholar] [CrossRef]
- Matsuyama, K.J.; Natori, S.J. Molecular cloning of cDNA for sapecin and unique expression of the sapecin gene during the development of Sarcophaga peregrina. J. Biol. Chem. 1988, 263, 17117–17121. [Google Scholar] [CrossRef]
- Taylor, K.; Barran, P.E.; Dorin, J.R. Structure-activity relationships in beta-defensin peptides. Biopolymers 2008, 90, 1–7. [Google Scholar] [CrossRef]
- Ceřovský, V.; Bém, R. Lucifensins, the insect defensins of biomedical importance: The story behind maggot therapy. Pharmaceuticals 2014, 7, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Lai, R. Insect antimicrobial peptides: Structures, properties and gene regulation. Zool. Res. 2010, 31, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Wanniarachchi, Y.A.; Kaczmarek, P.; Wan, A.; Nolan, E.M. Human defensin 5 disulfide array mutants: Disulfide bond deletion attenuates antibacterial activity against Staphylococcus aureus. Biochemistry 2011, 50, 8005–8017. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.; Battsetsegn, B.; Boldbaatar, D.; Miyoshi, T.; Xuan, X.; Oliver, J.M.; Fujisaki, K. Babesial vector tick defensin against Babesia sp. parasites. Infect. Immun. 2007, 75, 3633–3640. [Google Scholar] [CrossRef]
- Wilmes, M.; Cammue, B.P.; Sahl, H.G.; Thevissen, K. Antibiotic activities of host defense peptides: More to it than lipid bilayer perturbation. Nat. Prod. Rep. 2011, 28, 1350–1358. [Google Scholar] [CrossRef]
- Dimarcq, J.L.; Hoffmann, D.; Meister, M.; Bulet, P.; Lanot, R.; Reichhart, J.M.; Hoffmann, J.A. Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur. J. Biochem. 1994, 221, 201–209. [Google Scholar] [CrossRef]
- Thevissen, K.; Warnecke, D.C.; François, I.E.; Leipelt, M.; Heinz, E.; Ott, C.; Zaehringer, U.; Thomma, B.P.; Ferket, K.K.; Cammue, B.P. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 2004, 279, 3900–3905. [Google Scholar] [CrossRef]
- Dassanayake, R.S.; Silva Gunawardene, Y.I.; Tobe, S.S. Evolutionary selective trends of insect/mosquito antimicrobial defensin peptides containing cysteine-stabilized α/β motifs. Peptides 2007, 28, 62–75. [Google Scholar] [CrossRef]
- Meredith, J.M.; Hurd, H.; Lehane, M.J.; Eggleston, P. The malaria vector mosquito Anopheles gambiae expresses a suite of larval-specific defensin genes. Insect Mol. Biol. 2008, 17, 103–112. [Google Scholar] [CrossRef]
- Candido-Silva, J.A.; Zanarotti, G.M.; Gallina, A.P.; Almeida, J.C. Developmental regulation of BhSGAMP-1, a gene encoding an antimicrobial peptide in the salivary glands of Bradysia hygida (Diptera, Sciaridae). Genesis 2007, 45, 630–638. [Google Scholar] [CrossRef]
- Boman, H.G. Gene-encoded peptide antibiotics and the concept of innate immunity: An update review. Scand. J. Immunol. 1998, 48, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. Chem. Immunol. Allergy 2005, 86, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R.; Jiang, H.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Kumaresan, V.; Bhatt, P.; Ganesh, M.R.; Harikrishnan, R.; Arockiaraj, J. A novel antimicrobial peptide derived from fish goose type lysozyme disrupts the membrane of Salmonella enterica. Mol. Immunol. 2015, 68, 421–433. [Google Scholar] [CrossRef]
- Baltzer, S.A.; Brown, M.H. Antimicrobial peptides: Promising alternatives to conventional antibiotics. J. Mol. Microbiol. Biotechnol. 2011, 20, 228–235. [Google Scholar] [CrossRef]
- Klüver, E.; Schulz-Maronde, S.; Scheid, S.; Meyer, B.; Forssmann, W.G.; Adermann, K. Structure-activity relation of human beta-defensin 3: Influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 2005, 44, 9804–9816. [Google Scholar] [CrossRef]
- Boulanger, N.; Lowenberger, C.; Volf, P.; Ursic, R.; Sigutova, L.; Sabatier, L.; Svobodova, M.; Beverley, S.M.; Späth, G.; Brun, R.; et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect. Immun. 2004, 72, 7140. [Google Scholar] [CrossRef]
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- Ganz, T.; Selsted, M.E.; Lehrer, R.I. Defensins. Eur. J. Haematol. 1990, 44, 1–8. [Google Scholar] [CrossRef]
- Lamberty, M.; Ades, S.; Uttenweiler-Joseph, S.; Brookhart, G.; Bushey, D.; Hoffmann, J.A.; Bulet, P. Insect immunity. Isolation from the lepidopteran Heliothis virescens of a novel insect defensin with potent antifungal activity. J. Biol. Chem. 1999, 274, 9320–9326. [Google Scholar] [CrossRef]
- Lowenberger, C.; Bulet, P.; Charlet, M.; Hetru, C.; Hodgeman, B.M.; Christensen, B.; Hoffmann, J.A. Insect immunity: Isolation of three novel inducible antibacterial defensins from the vector mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 1995, 25, 867–873. [Google Scholar] [CrossRef]
- Kaushal, A.; Gupta, K.; van Hoek, M.L. Characterization of Cimex lectularius (bedbug) defensin peptide and its antimicrobial activity against human skin microflora. Biochem. Biophys. Res. Commun. 2016, 470, 955–960. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, L.; Mu, L.X.; Wang, Y.P.; Bian, H.; Li, J.; Lu, Y.L.; Han, Y.; Liu, T.; Lv, J.; Feng, C.P.; et al. Purification and characterization of a novel defensin from the salivary glands of the black fly, Simulium bannaense. Parasit. Vectors 2015, 8, 71. [Google Scholar] [CrossRef]
- Wilde, C.G.; Griffith, J.E.; Marra, M.N.; Snable, J.L.; Scott, R.W. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J. Biol. Chem. 1989, 264, 11200–11203. [Google Scholar] [CrossRef]
- Valore, E.V.; Park, C.H.; Quayle, A.J.; Wiles, K.R.; McCray, P.B.; Ganz, T. Human β-defensin-1: An antimicrobial peptide of urogenital tissues. J. Clin. Investig. 1998, 101, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Bartels, J.; Christophers, E.; Schröder, J.M. A peptide antibiotic from human skin. Nature 1997, 387, 861. [Google Scholar] [CrossRef]
- Starner, T.D.; Agerberth, B.; Gudmundsson, G.H.; McCray, P.B. Expression and activity of beta-defensins and LL-37 in the developing human lung. J. Immunol. 2005, 174, 1608. [Google Scholar] [CrossRef]
- Su, T.; Mulla, M.S. Quantitative determination of free amino acids in the hemolymph of autogenous and anautogenous strains of Culex tarsalis (Diptera: Culicidae). J. Med. Entomol. 1997, 34, 729–734. [Google Scholar] [CrossRef]


| Bacterial Strain | MIC (μM) |
|---|---|
| Gram-positive bacteria | |
| S. aureus ATCC 25923 | 0.92 |
| M. luteus CMCC 28001 | 1.24 |
| B. subtilis CMCC 63501 | 1.56 |
| Gram-negative bacteria | |
| E. coli ATCC 25922 | ND |
| P. aeruginosa CMCC 10104 | ND |
| S. typhi CMCC 50071 | ND |
| Primer Name | Sequence | Primer Usage |
|---|---|---|
| Def2-F | 5′-ACGCTTCAGTTGAGTCCATCT-3′ | Cloning |
| Def2-R | 5′-ACAGTGATCTTTTGGTGTCCACT-3′ | |
| Def2-qF | 5′-GCTGTCGCTGTTGTCTACATCGGT-3′ | RT-qPCR |
| Def2-qR | 5′-CGGCTCTTCTTCGTGGTATGTCTC-3′ | |
| Actin-F | 5′-ACCGCTGAGAGGGAAATCG-3′ | |
| Actin-R | 5′-CAAGAAGGAAGGCTGGAAGAG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, T.; Du, J.; Li, S.-W.; Huang, H.; Qi, X.-L. Identification and Functional Analysis of a Defensin CcDef2 from Coridius chinensis. Int. J. Mol. Sci. 2022, 23, 2789. https://doi.org/10.3390/ijms23052789
Gong T, Du J, Li S-W, Huang H, Qi X-L. Identification and Functional Analysis of a Defensin CcDef2 from Coridius chinensis. International Journal of Molecular Sciences. 2022; 23(5):2789. https://doi.org/10.3390/ijms23052789
Chicago/Turabian StyleGong, Tao, Juan Du, Shang-Wei Li, Hai Huang, and Xiao-Lang Qi. 2022. "Identification and Functional Analysis of a Defensin CcDef2 from Coridius chinensis" International Journal of Molecular Sciences 23, no. 5: 2789. https://doi.org/10.3390/ijms23052789
APA StyleGong, T., Du, J., Li, S.-W., Huang, H., & Qi, X.-L. (2022). Identification and Functional Analysis of a Defensin CcDef2 from Coridius chinensis. International Journal of Molecular Sciences, 23(5), 2789. https://doi.org/10.3390/ijms23052789






