Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis
Abstract
:1. Introduction
2. Results
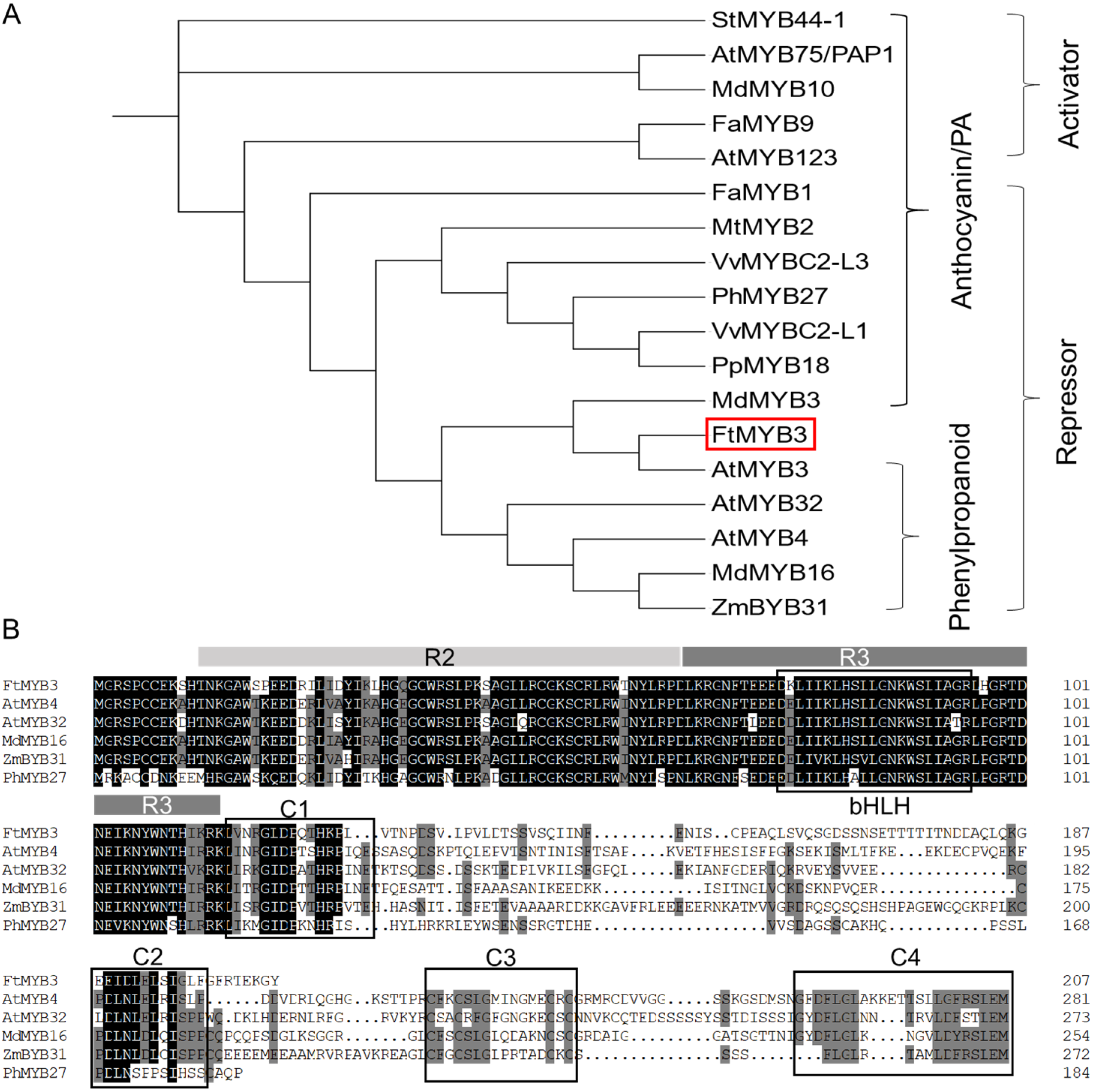
2.1. Tartary Buckwheat FtMYB3 Is an SG4 R2R3 MYB Protein
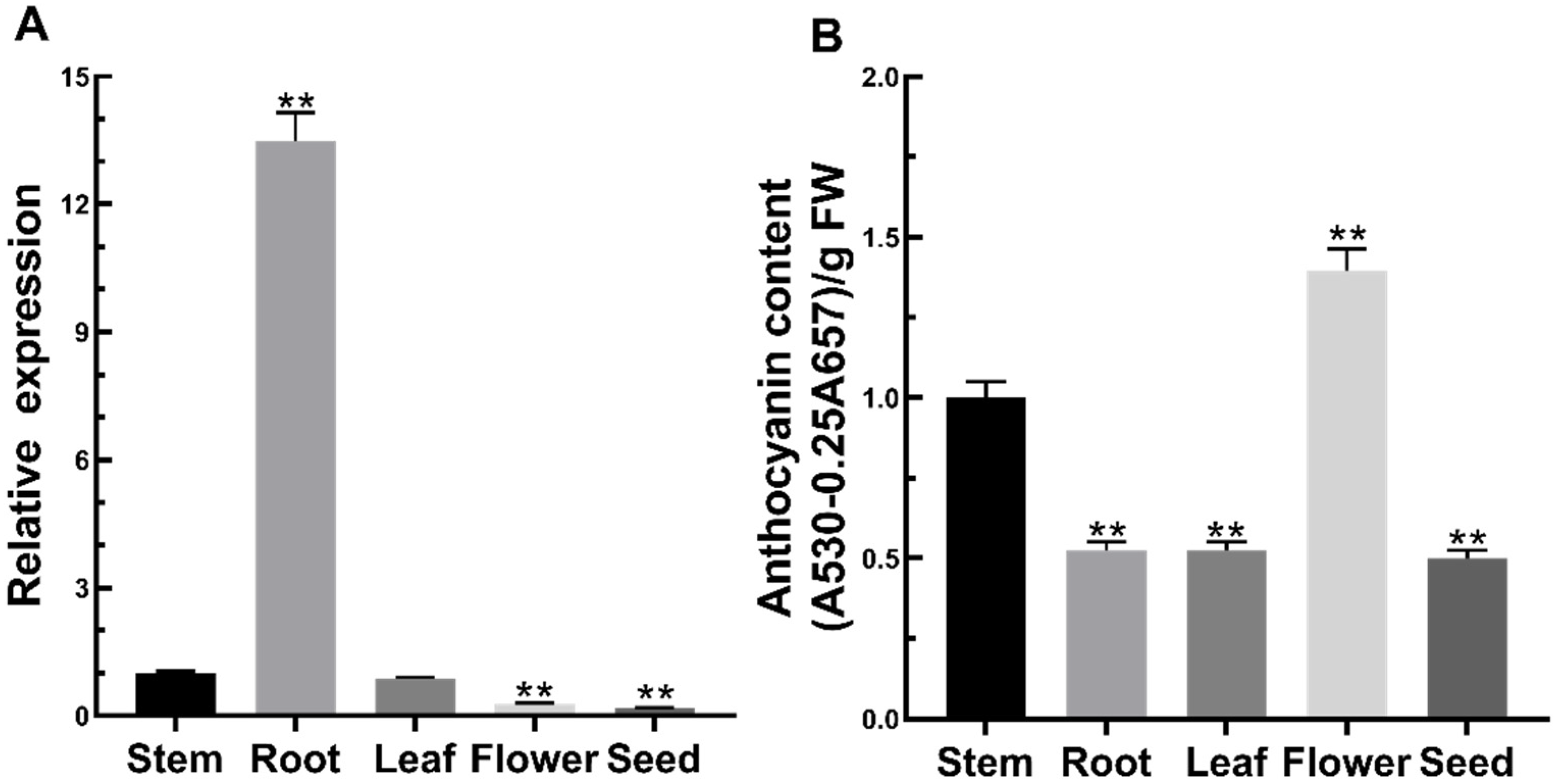
2.2. Expression Pattern of FtMYB3 and Anthocyanin Content in Tartary Buckwheat
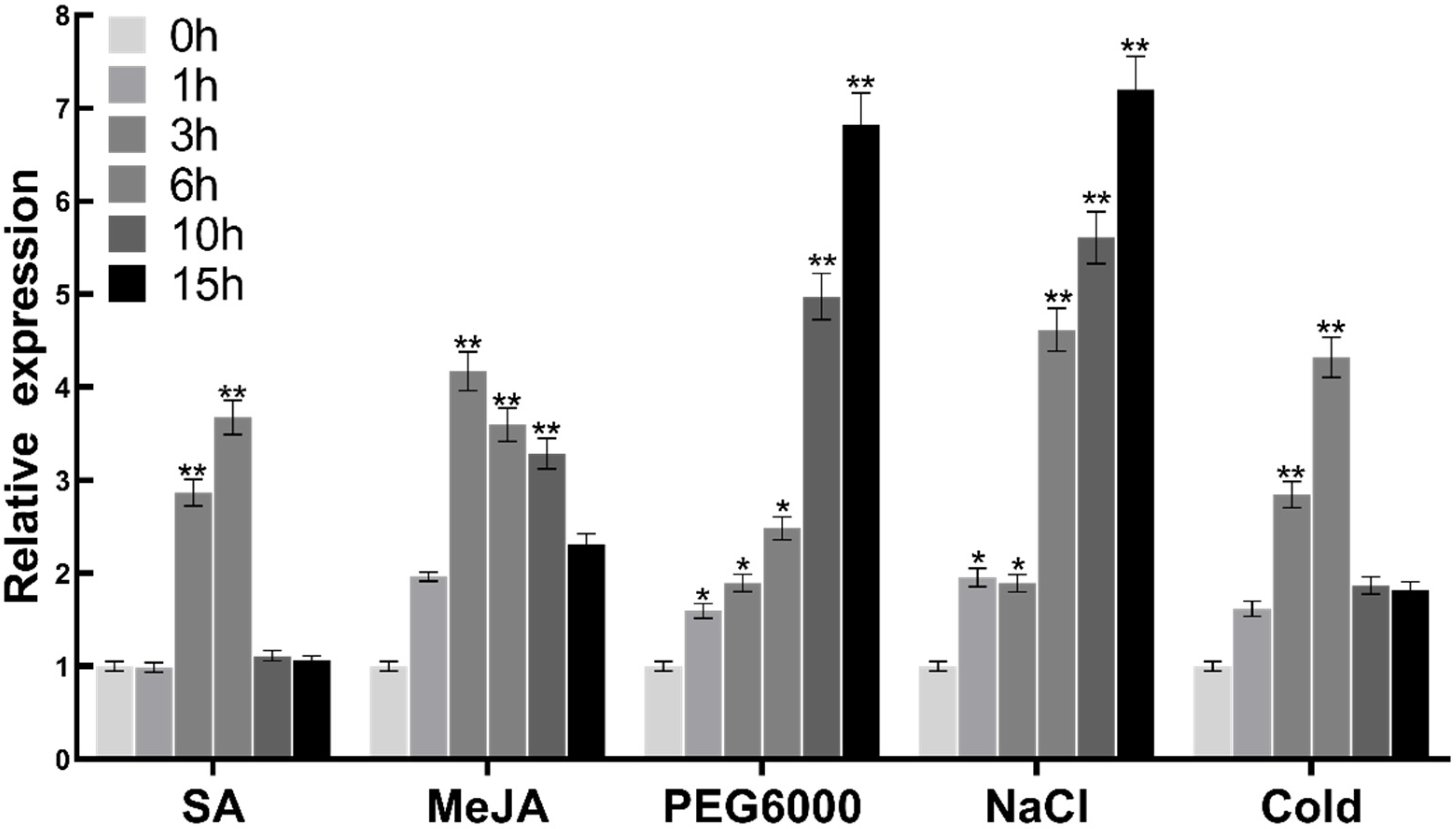
2.3. Induction of FtMYB3 by Multiple Environmental Factors and Plant Hormones
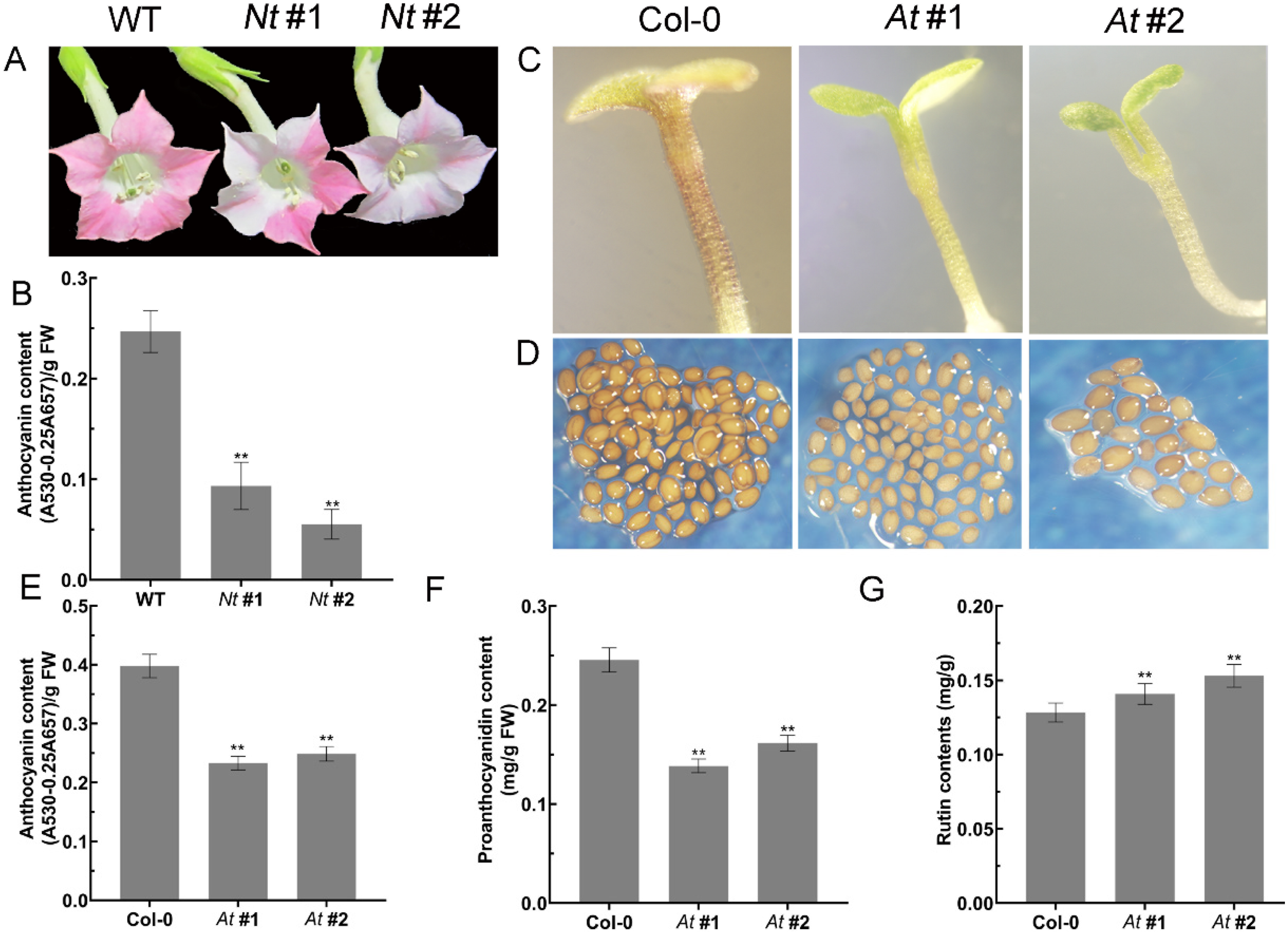
2.4. FtMYB3 Inhibits Anthocyanin/PA Accumulation
2.5. Downregulation of FtMYB3 in the Expression Levels of DFR/BAN/ANS/TT13
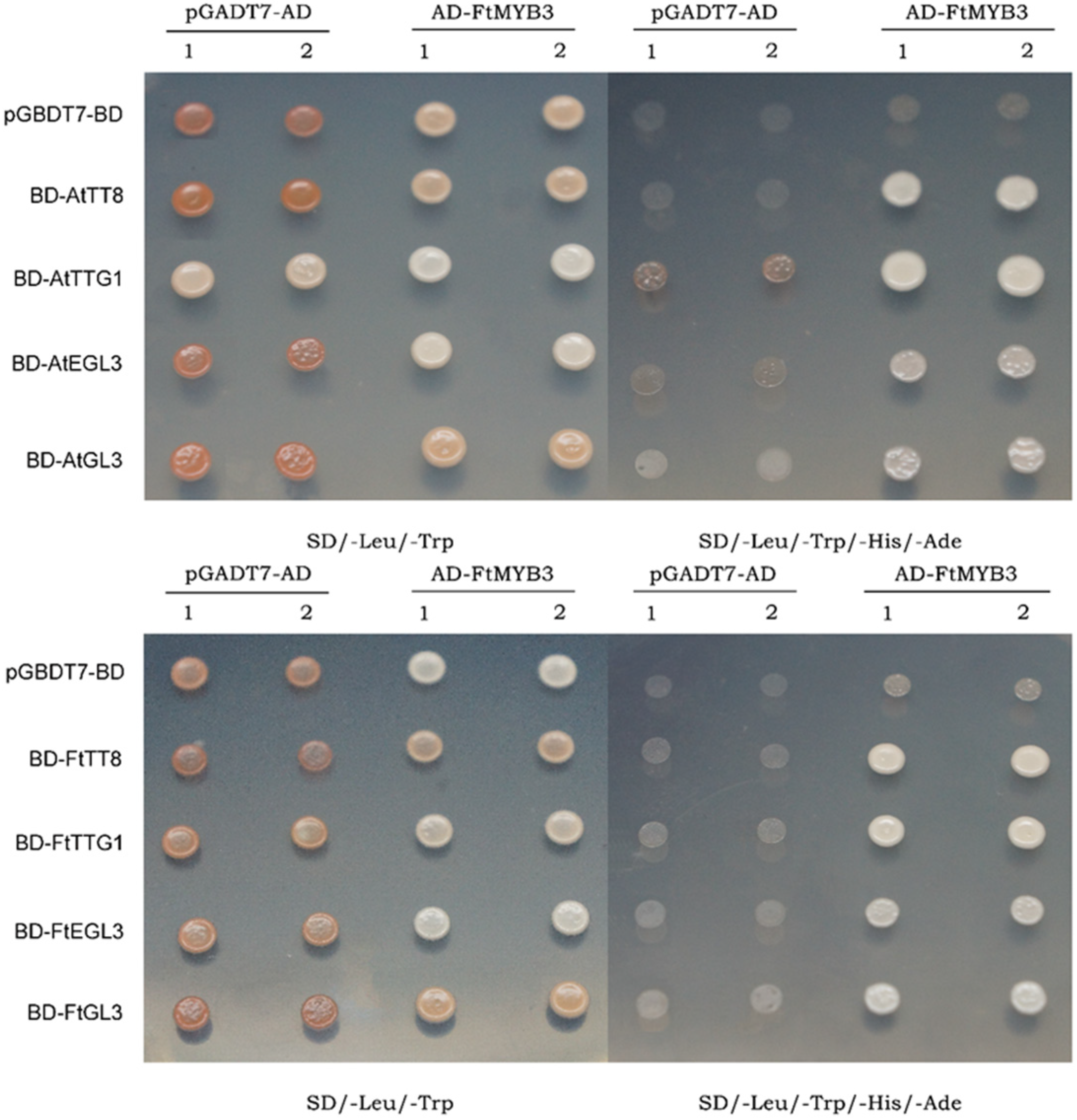
2.6. FtMYB3 Interacts with GL3/EGL3/TT8/TTG1
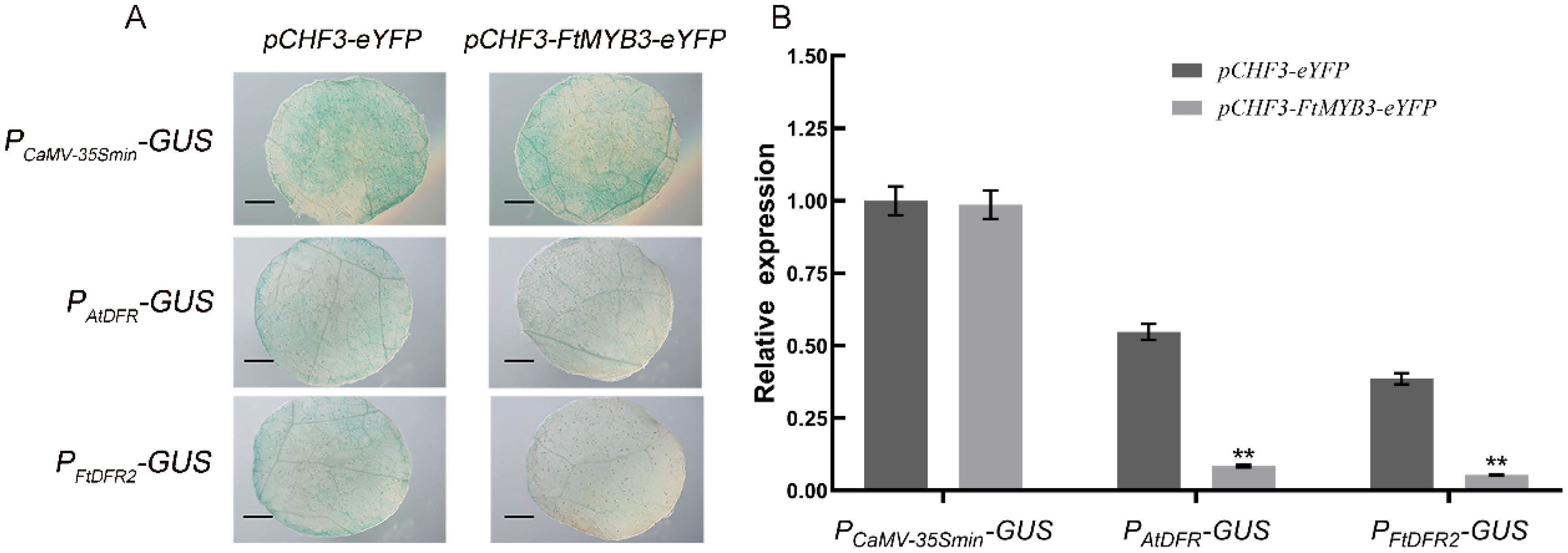
2.7. FtMYB3 Inhibits the Transcriptional Activation Activity of the DFR Promoter
3. Discussion
3.1. FtMYB3 Is a Special SG4 MYB Repressor That Reduces Anthocyanin and PA Accumulation
3.2. FtMYB3 Specifically Represses the Expression of Genes in the Late Pathway of Flavonoid Synthesis
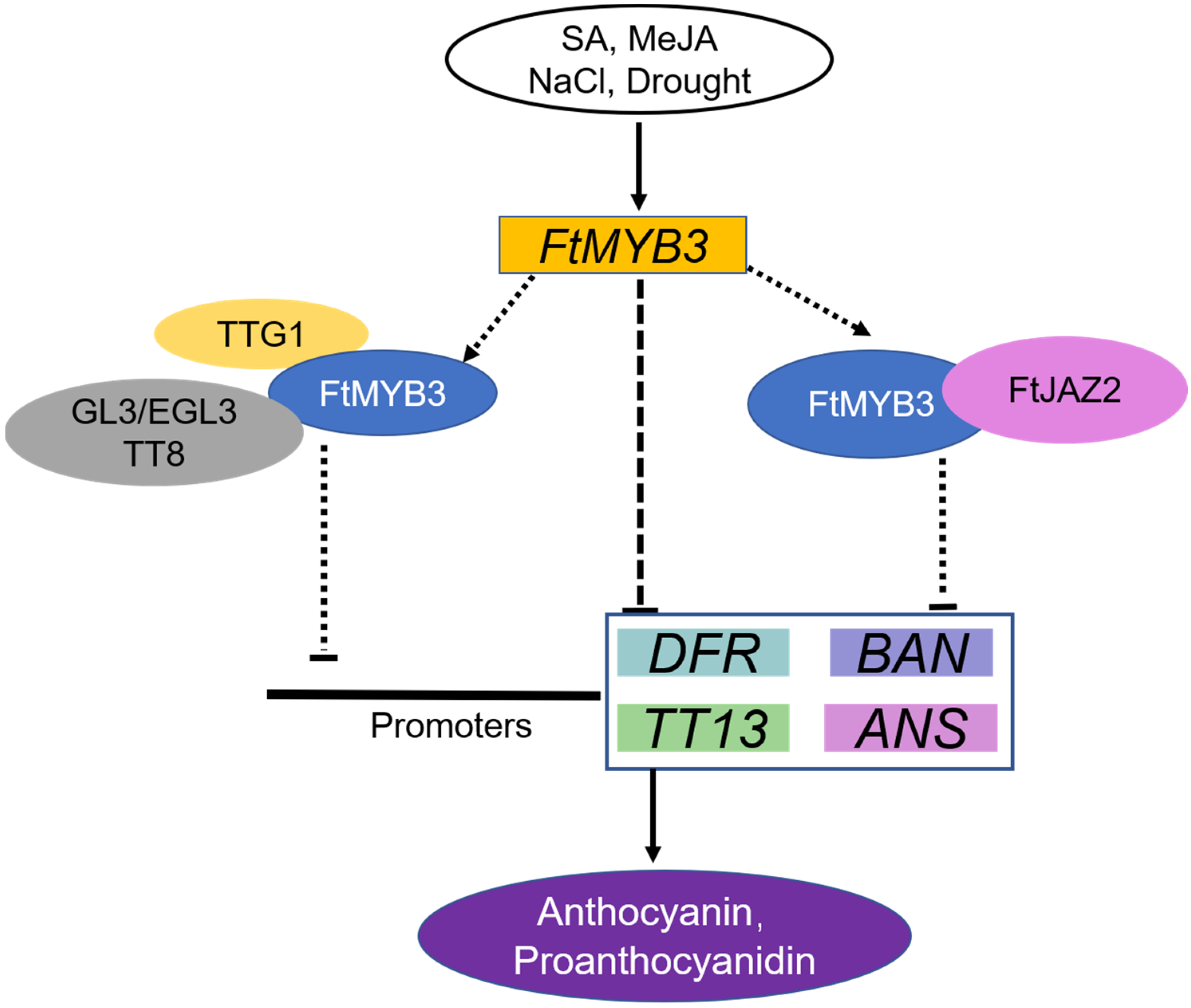
3.3. Possible Mechanism of FtMYB3 Inhibiting the Biosynthesis of Anthocyanins and PAs
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Total RNA Extraction and Gene Expression Analysis
4.3. Sequence Analysis of FtMYB3 and Its Promoter
4.4. Construction of Expression Vectors and Stable Transformation of Arabidopsis and Tobacco
4.5. Determination of Total Anthocyanin, PA and Rutin Content
4.6. Yeast Two-Hybrid Assay (Y2H)
4.7. Transient Expression Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2010, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.; Visser, R.; Arnaud, B. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Annabelle, C.; Alex, B.; Araceli, C.; Antonio, G. Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Wang, P.; Luo, Y.; Zhao, M.; Chen, F. Health benefits of anthocyanins and molecular mechanisms: Update from recent decade. Crit. Rev. Food Sci. 2017, 57, 1729–1741. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F. Proanthocyanidins in cereals and pseudocereals. Crit. Rev. Food Sci. Nutr. 2019, 59, 1521–1533. [Google Scholar] [CrossRef]
- Springob, K.; Nakajima, J.; Yamazaki, M.; Saito, K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 2003, 20, 288–303. [Google Scholar] [CrossRef]
- Li, P.; Dong, Q.; Ge, S.; He, X.; Verdier, J.; Li, D.; Zhao, J. Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanidin precursor flux in legume. Plant Biotechnol. J. 2016, 14, 1604–1618. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.; Sharma, S.; Paiva, N.; Ferreira, D.; Dixon, R. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wu, J.; Hu, K.D.; Wei, H.; Sun, H.; Hu, L.; Han, Z.; Yao, G.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Marles, M.; Ray, H.; Gruber, M. New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 2003, 64, 367–383. [Google Scholar] [CrossRef]
- Zhang, Z.; Tian, C.; Zhang, Y.; Li, C.; Li, X.; Yu, Q.; Wang, S.; Wang, X.; Chen, X.; Feng, X. Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear. BMC Plant Biol. 2020, 20, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Zhang, J.; Han, Z.; Song, T.; Li, J.; Wang, Y.; Yao, Y. McMYB12 Transcription Factors Co-regulate Proanthocyanidin and Anthocyanin Biosynthesis in Malus Crabapple. Sci. Rep. 2017, 7, 43715. [Google Scholar] [CrossRef] [Green Version]
- An, X.; Tian, Y.; Chen, K.; Liu, X.; Liu, D.; Xie, X.; Cheng, C.; Cong, P.; Hao, Y. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, M.; Yu, W.; Yao, H.; Zhou, P.; Allan, A.; Zeng, L. NtMYB3, an R2R3-MYB from Narcissus, Regulates Flavonoid Biosynthesis. Int. J. Mol. Sci. 2019, 20, 5456. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef]
- Cheng, J.; Yu, K.; Shi, Y.; Wang, J.; Duan, C. Transcription Factor VviMYB86 Oppositely Regulates Proanthocyanidin and Anthocyanin Biosynthesis in Grape Berries. Front Plant Sci. 2020, 11, 2263. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, H.; Jiang, X.; Wang, P.; Dai, X.; Chen, W.; Gao, L.; Xia, T. A WD40 Repeat Protein from Camellia sinensis Regulates Anthocyanin and Proanthocyanidin Accumulation through the Formation of MYB-bHLH-WD40 Ternary Complexes. Int. J. Mol. Sci. 2018, 19, 1686. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.; Lloyd, A. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- An, J.; Xu, R.; Liu, X.; Zhang, J.; Wang, X.; You, C.; Hao, Y. Jasmonate induces anthocyanin and proanthocyanidin biosynthesis in apple by mediating the JAZ1-TRB1-MYB9 complex. Plant J. 2021, 106, 1414–1430. [Google Scholar] [CrossRef]
- Mao, Z.; Jiang, H.; Wang, S.; Wang, Y.; Yu, L.; Zou, Q.; Liu, W.; Jiang, S.; Wang, N.; Zhang, Z.; et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021, 306, 110848. [Google Scholar] [CrossRef]
- Li, G.; Meng, X.; Zhu, M.; Li, Z. Research Progress of Betalain in Response to Adverse Stresses and Evolutionary Relationship Compared with Anthocyanin. Molecules 2019, 24, 3078. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Huang, T.; Tian, B.; Zhan, J. Advances in Biosynthesis and Biological Functions of Proanthocyanidins in Horticultural Plants. Foods 2020, 9, 1774. [Google Scholar] [CrossRef]
- Joshi, D.C.; Zhang, K.; Wang, C.; Chandora, R.; Khurshid, M.; Li, J.; He, M.; Georgiev, M.; Zhou, M. Strategic enhancement of genetic gain for nutraceutical development in buckwheat: A genomics-driven perspective. Biotechnol. Adv. 2020, 39, 107479. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F. Chemical composition and health effects of Tartary buckwheat. Food Chem. 2016, 203, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, V.; D’Amelia, V.; Esposito, M.; Amitrano, C.; Carillo, P.; Carputo, D.; Maggio, A. Anthocyanins are Key Regulators of Drought Stress Tolerance in Tobacco. Biology 2021, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.; Kim, C. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef]
- Malisch, C.; Salminen, J.; Kölliker, R.; Engström, M.; Suter, D.; Studer, B.; Lüscher, A. Drought Effects on Proanthocyanidins in Sainfoin (Onobrychis viciifolia Scop.) Are Dependent on the Plant’s Ontogenetic Stage. J. Agric. Food Chem. 2016, 64, 9307–9316. [Google Scholar] [CrossRef]
- Bai, Y.; Li, C.; Zhang, J.; Li, S.; Luo, X.; Yao, H.; Chen, H.; Zhao, H.; Park, S.U.; Wu, Q. Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis. Physiol. Plant. 2014, 152, 431–440. [Google Scholar] [CrossRef]
- Luo, X.; Zhao, H.; Yao, P.; Li, Q.; Huang, Y.; Li, C.; Chen, H.; Wu, Q. An R2R3-MYB Transcription Factor FtMYB15 Involved in the Synthesis of Anthocyanin and Proanthocyanidins from Tartary Buckwheat. J. Plant Growth Regul. 2017, 37, 76–84. [Google Scholar] [CrossRef]
- Dong, Q.; Zhao, H.; Huang, Y.; Chen, Y.; Wan, M.; Zeng, Z.; Yao, P.; Li, C.; Wang, X.; Chen, H.; et al. FtMYB18 acts as a negative regulator of anthocyanin/proanthocyanidin biosynthesis in Tartary buckwheat. Plant Mol. Biol. 2020, 104, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wu, Q.; Wang, S.; Shi, J.; Dong, Q.; Yao, P.; Shi, G.; Xu, S.; Deng, R.; Li, C.; et al. FtMYB8 from Tartary buckwheat inhibits both anthocyanin/Proanthocyanidin accumulation and marginal Trichome initiation. BMC Plant Biol. 2019, 19, 263. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Li, S.; Yao, P.; Li, C.; Chen, H.; Wu, Q.; Zhao, H. The jasmonate-ZIM domain protein FtJAZ2 interacts with the R2R3-MYBtranscription factor FtMYB3 to affect anthocyanin biosynthesis in tartary buckwheat. Turk. J. Biol. 2017, 41, 526–534. [Google Scholar] [CrossRef]
- Li, X.; Sathasivam, R.; Park, N.I.; Wu, Q.; Park, S. Enhancement of phenylpropanoid accumulation in tartary buckwheat hairy roots by overexpression of MYB transcription factors. Ind. Crops Prod. 2020, 156, 112887. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, K.; Sun, Z.; Yan, M.; Chen, C.; Zhang, X.; Tang, Y.; Wu, Y. LNK1 and LNK2 Corepressors Interact with the MYB3 Transcription Factor in Phenylpropanoid Biosynthesis. Plant Physiol. 2017, 174, 1348–1358. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef] [Green Version]
- Grotewold, E.; Sainz, M.B.; Tagliani, L.; Hernandez, J.M.; Chandler, V.L. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 2000, 97, 13579–13584. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Appelhagen, I.; Nordholt, N.; Seidel, T.; Spelt, K.; Koes, R.; Quattrochio, F.; Sagasser, M.; Weisshaar, B. TRANSPARENT TESTA 13 is a tonoplast P3A -ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 2015, 82, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, X.; Wang, K.; Yang, M.; Chen, W.; Yang, Z.; Gao, S. MrMYB6 from Chinese Bayberry (Myrica rubra) Negatively Regulates Anthocyanin and Proanthocyanidin Accumulation. Front. Plant Sci. 2021, 12, 1218. [Google Scholar] [CrossRef]
- Wan, S.; Li, C.; Ma, X.; Luo, K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017, 36, 1263–1276. [Google Scholar] [CrossRef]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Yuan, Y.; Rebocho, A.B.; Sagawa, J.M.; Stanley, L.E.; Bradshaw, H.D. Competition between anthocyanin and flavonol biosynthesis produces spatial pattern variation of floral pigments between Mimulus species. Proc. Natl. Acad. Sci. USA 2016, 113, 2448–2453. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Ning, Y.; Zhang, W.; Liao, Y.; Li, L.; Cheng, H.; Cheng, S. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba. Funct. Integr. Genom. 2014, 14, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Wang, G.; Wu, J.; Saquib, W.; Andrew, A.; Zeng, L. Ectopic Overexpression of a Novel R2R3-MYB, NtMYB2 from Chinese Narcissus Represses Anthocyanin Biosynthesis in Tobacco. Molecules 2018, 23, 781. [Google Scholar]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Bioch. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-mediated Transformation of Arabidopsis Thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ning, G.; Haiyan, L.; Liao, L.; Bao, M. Single MYB-type transcription factor AtCAPRICE: A new efficient tool to engineer the production of anthocyanin in tobacco. Biochem. Biophys. Res. Commun. 2009, 388, 742–747. [Google Scholar] [CrossRef] [PubMed]
- Peel, G.J.; Dixon, R. A Detection and Quantification of Engineered Proanthocyanidins in Transgenic Plants. Nat. Prod. Commun. 2007, 2, 1009–1014. [Google Scholar] [CrossRef]
- Yao, P.; Huang, Y.; Dong, Q.; Wan, M.; Wang, A.; Chen, Y.; Li, C.; Wu, Q.; Chen, H.; Zhao, H. FtMYB6, a Light-Induced SG7 R2R3-MYB Transcription Factor, Promotes Flavonol Biosynthesis in Tartary Buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 2020, 68, 13685–13696. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Halladay, J.; Craig, E.A. Genomic Libraries and a Host Strain Designed for Highly Efficient Two-Hybrid Selection in Yeast. Genetics 1996, 144, 1425–1436. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Deng, R.; Bai, Y.; Wu, H.; Li, C.; Wu, Q.; Zhao, H. Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis. Int. J. Mol. Sci. 2022, 23, 2775. https://doi.org/10.3390/ijms23052775
Wang L, Deng R, Bai Y, Wu H, Li C, Wu Q, Zhao H. Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis. International Journal of Molecular Sciences. 2022; 23(5):2775. https://doi.org/10.3390/ijms23052775
Chicago/Turabian StyleWang, Lei, Renyu Deng, Yuechen Bai, Huala Wu, Chenglei Li, Qi Wu, and Haixia Zhao. 2022. "Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis" International Journal of Molecular Sciences 23, no. 5: 2775. https://doi.org/10.3390/ijms23052775
APA StyleWang, L., Deng, R., Bai, Y., Wu, H., Li, C., Wu, Q., & Zhao, H. (2022). Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis. International Journal of Molecular Sciences, 23(5), 2775. https://doi.org/10.3390/ijms23052775






