Deciphering the Potential Coding of Human Cytomegalovirus: New Predicted Transmembrane Proteome
Abstract
1. Introduction
2. Results
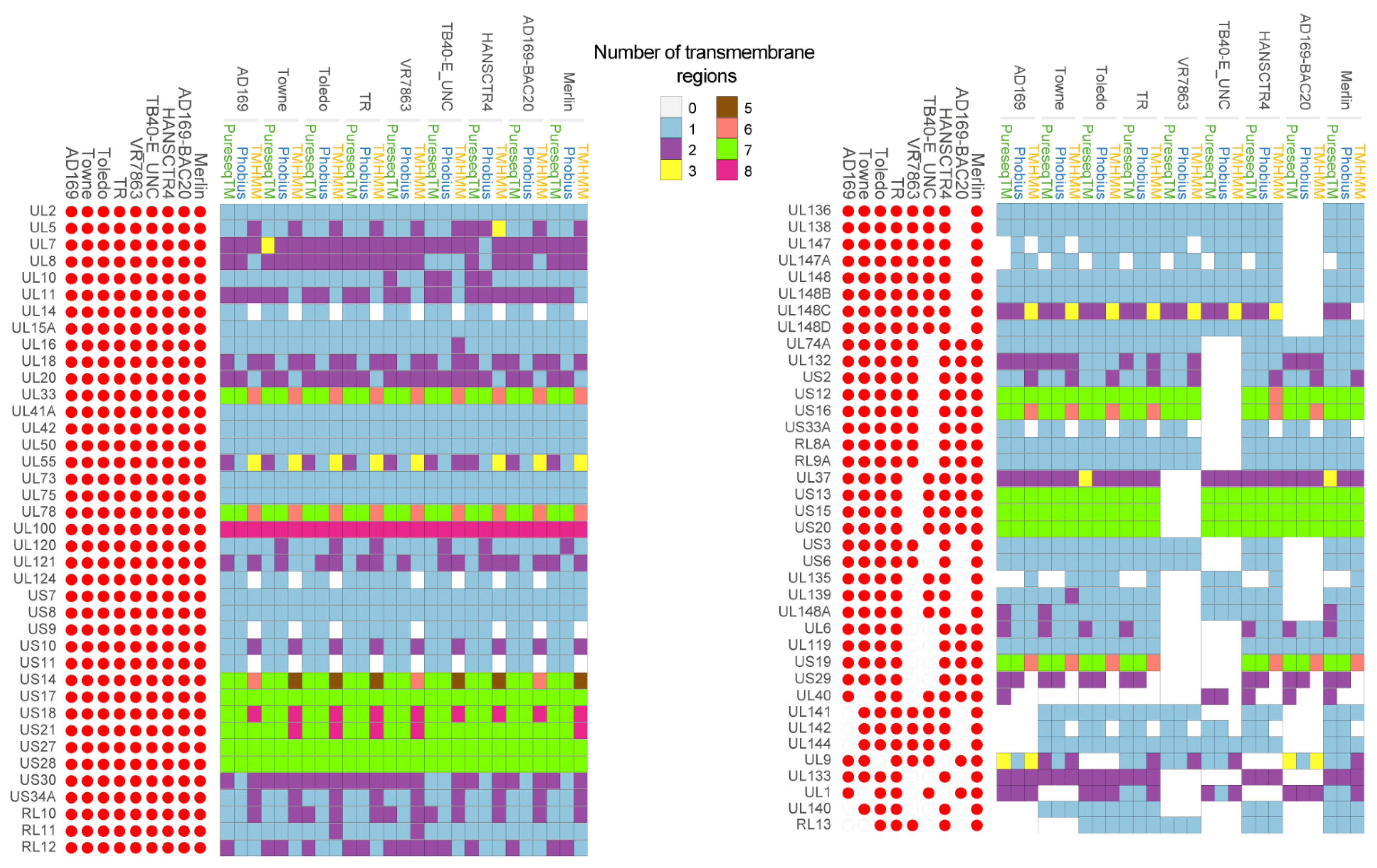
2.1. Identification of Putative Transmembrane Proteins
2.2. Homology Analysis of thePredicted Transmembrane Proteins
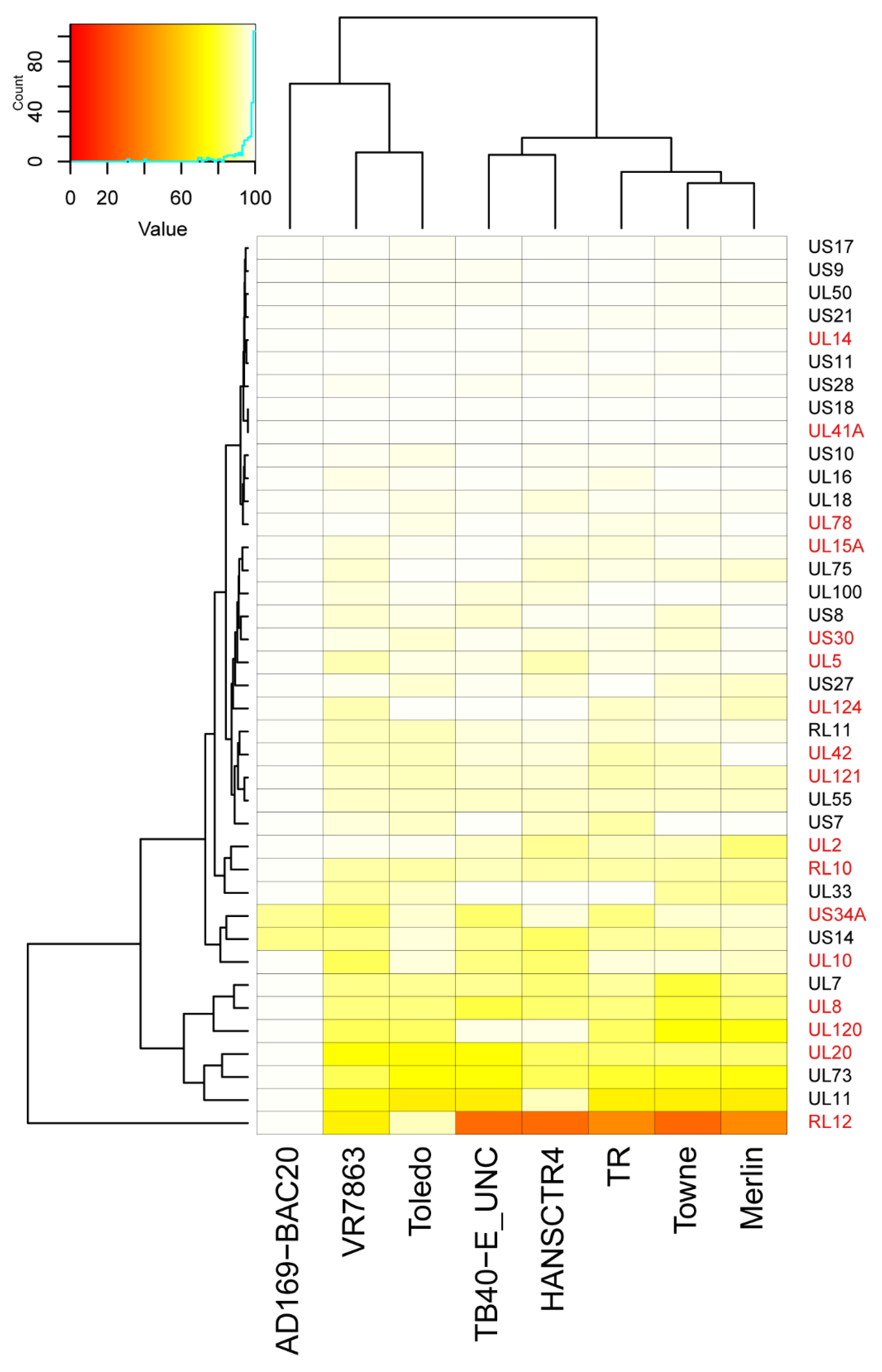
2.3. Sequence Differences among Strains
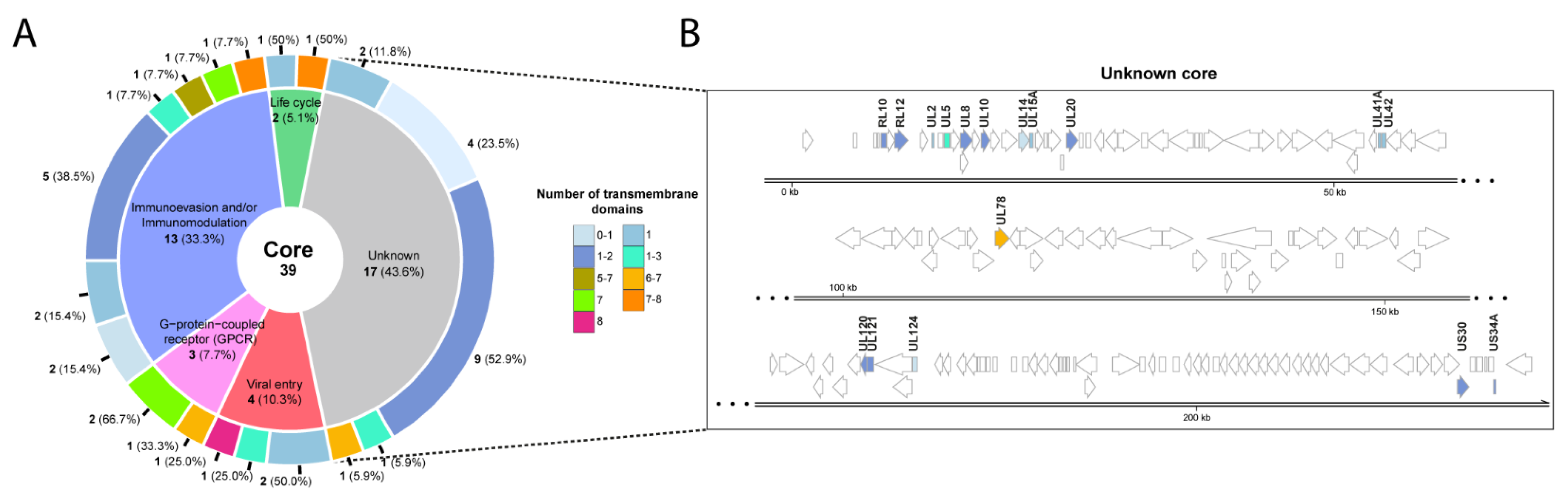
2.4. Sequence Similarities among Strains: Core Proteins
2.5. CMV Gene Families with Transmembrane Domains
3. Discussion
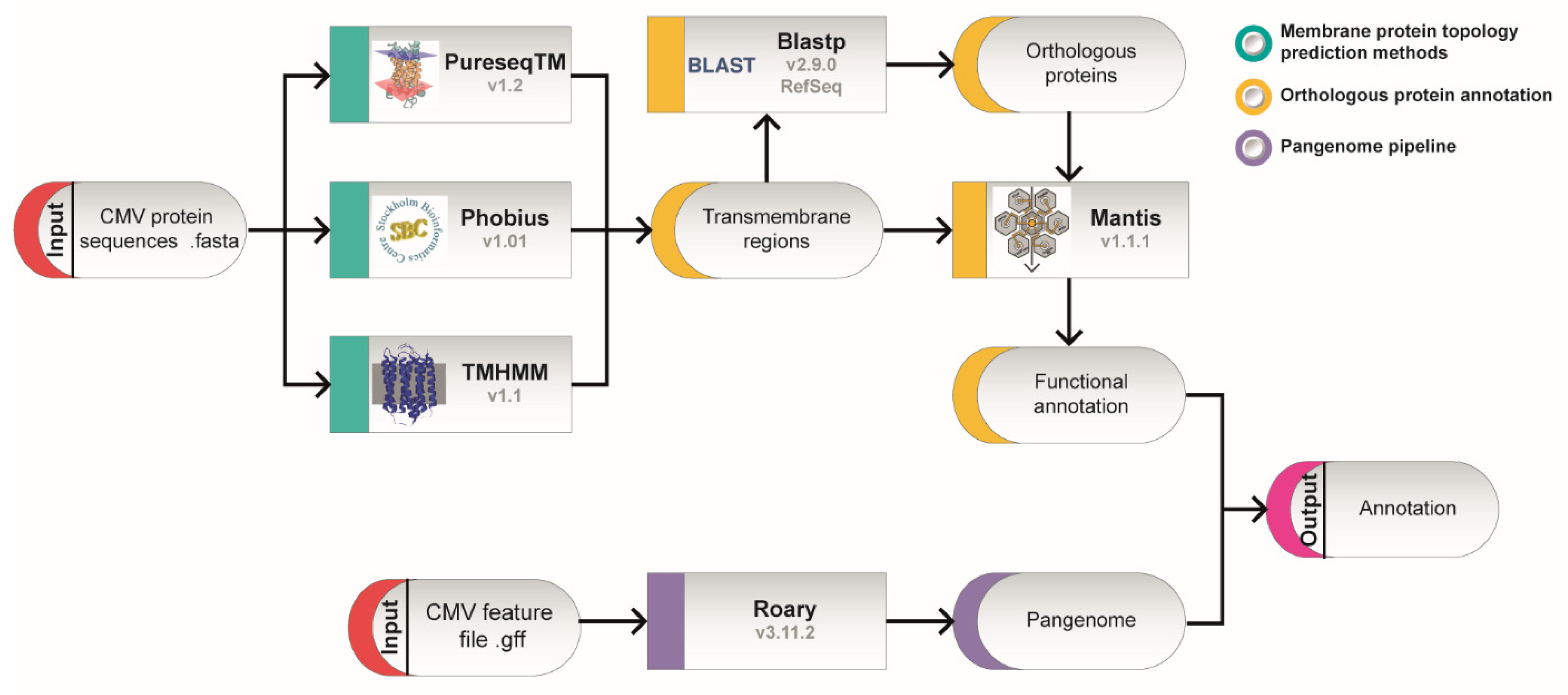
4. Materials and Methods
4.1. Transmembrane Region Analysis
4.2. Functional Annotation of the Transmembrane ORFs
4.3. Viral Pangenome Construction
4.4. Similarity Analysis
4.5. Protein Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Plotkin, S.A.; Boppana, S.B. Vaccination against the human cytomegalovirus. Vaccine 2019, 37, 7437–7442. [Google Scholar] [CrossRef] [PubMed]
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Shenk, T.E. Human Cytomegalovirus Genome; Shenk, T.E., Stinski, M.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 325, ISBN 9783540773481. [Google Scholar]
- Emery, V.C. Investigation of CMV disease in immunocompromised patients. J. Clin. Pathol. 2001, 54, 84–88. [Google Scholar] [CrossRef]
- Boeckh, M.; Geballe, A.P. Science in medicine Cytomegalovirus: Pathogen, paradigm, and puzzle. J. Clin. Investig. 2011, 121, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Seitz, R. Human Cytomegalovirus (HCMV)-Revised. Transfus. Med. Hemotherapy 2010, 37, 365–375. [Google Scholar] [CrossRef]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Stenberg, R.M.; Witte, P.R.; Stinski, M.F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 1985, 56, 665–675. [Google Scholar] [CrossRef]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar] [CrossRef]
- Van Damme, E.; Van Loock, M. Functional annotation of human cytomegalovirus gene products: An update. Front. Microbiol. 2014, 5, 218. [Google Scholar] [CrossRef]
- Balázs, Z.; Tombácz, D.; Szucs, A.; Csabai, Z.; Megyeri, K.; Petrov, A.N.; Snyder, M.; Boldogkoi, Z. Long-Read Sequencing of Human Cytomegalovirus Transcriptome Reveals RNA Isoforms Carrying Distinct Coding Potentials. Sci. Rep. 2017, 7, 15989. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Weisburd, B.; Michalski, A.; Le, V.T.K.; Hein, M.Y.; Huang, S.X.; Ma, M.; Shen, B.; Qian, S.B.; Hengel, H.; et al. Decoding human cytomegalovirus. Science 2012, 338, 1088–1093. [Google Scholar] [CrossRef]
- Galitska, G.; Coscia, A.; Forni, D.; Steinbrueck, L.; De Meo, S.; Biolatti, M.; De Andrea, M.; Cagliani, R.; Leone, A.; Bertino, E.; et al. Genetic Variability of Human Cytomegalovirus Clinical Isolates Correlates with Altered Expression of Natural Killer Cell-Activating Ligands and IFN-γ. Front. Immunol. 2021, 12, 15989. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Kamil, J.P. Viral Regulation of Cell Tropism in Human Cytomegalovirus. J. Virol. 2016, 90, 626–629. [Google Scholar] [CrossRef]
- Gerna, G.; Kabanova, A.; Lilleri, D. Human Cytomegalovirus Cell Tropism and Host Cell Receptors. Vaccines 2019, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Mocarski, E.S. Viral and host control of cytomegalovirus maturation. Trends Microbiol. 2012, 20, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ortiz, D.A.; Gurczynski, S.J.; Khan, F.; Pellett, P.E. Identification of Human Cytomegalovirus Genes Important for Biogenesis of the Cytoplasmic Virion Assembly Complex. J. Virol. 2014, 88, 9086–9099. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.I.; Munoz, F.M.; Callahan, S.T.; Rupp, R.; Wootton, S.H.; Edwards, K.M.; Turley, C.B.; Stanberry, L.R.; Patel, S.M.; Mcneal, M.M.; et al. Safety and efficacy of a cytomegalovirus glycoprotein B (gB) vaccine in adolescent girls: A randomized clinical trial. Vaccine 2016, 34, 313–319. [Google Scholar] [CrossRef]
- Pass, R.F. Development and evidence for efficacy of CMV glycoprotein B vaccine with MF59 adjuvant. J. Clin. Virol. 2009, 46, S73–S76. [Google Scholar] [CrossRef]
- Nelson, C.S.; Herold, B.C.; Permar, S.R. A new era in cytomegalovirus vaccinology: Considerations for rational design of next-generation vaccines to prevent congenital cytomegalovirus infection. NPJ Vaccines 2018, 3, 38. [Google Scholar] [CrossRef]
- Baraniak, I.; Kropff, B.; Ambrose, L.; McIntosh, M.; McLean, G.R.; Pichon, S.; Atkinson, C.; Milne, R.S.B.; Mach, M.; Griffiths, P.D.; et al. Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc. Natl. Acad. Sci. USA 2018, 115, 6273–6278. [Google Scholar] [CrossRef]
- Nelson, C.S.; Huffman, T.; Jenks, J.A.; De La Rosa, E.C.; Xie, G.; Vandergrift, N.; Pass, R.F.; Pollara, J.; Permar, S.R. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc. Natl. Acad. Sci. USA 2018, 115, 6267–6272. [Google Scholar] [CrossRef]
- Vincenti, F.; Budde, K.; Merville, P.; Shihab, F.; Ram Peddi, V.; Shah, M.; Wyburn, K.; Cassuto-Viguier, E.; Weidemann, A.; Lee, M.; et al. A randomized, phase 2 study of ASP0113, a DNA-based vaccine, for the prevention of CMV in CMV-seronegative kidney transplant recipients receiving a kidney from a CMV-seropositive donor. Am. J. Transplant. 2018, 18, 2945–2954. [Google Scholar] [CrossRef]
- Sandonís, V.; García-Ríos, E.; McConnell, M.J.; Pérez-Romero, P. Role of Neutralizing Antibodies in CMV Infection: Implications for New Therapeutic Approaches. Trends Microbiol. 2020, 28, 900–912. [Google Scholar] [CrossRef]
- Pérez-Romero, P.; Blanco, P.; Giménez, E.; Solano, C.; Navarro, D. An update on the treatment of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation. Expert Rev. Hematol. 2015, 12, 937–945. [Google Scholar] [CrossRef]
- Krause, P.R.; Bialek, S.R.; Boppana, S.B.; Griffiths, P.D.; Laughlin, C.A.; Ljungdahl, P.O.; Mocarski, E.S.; Pass, R.F.; Read, J.S.; Schleiss, M.R.; et al. Priorities for CMV vaccine development. Vaccine 2013, 32, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Griffiths, P.D.; Reeves, M.B. The Humoral Immune Response Against the gB Vaccine: Lessons Learnt from Protection in Solid Organ Transplantation. Vaccines 2019, 7, 67. [Google Scholar] [CrossRef]
- Caposio, P.; van den Worm, S.; Crawford, L.; Perez, W.; Kreklywich, C.; Gilbride, R.M.; Hughes, C.M.; Ventura, A.B.; Ratts, R.; Marshall, E.E.; et al. Characterization of a live-attenuated HCMV-based vaccine platform. Sci. Rep. 2019, 9, 19236. [Google Scholar] [CrossRef] [PubMed]
- Schleiss, M.R. Cytomegalovirus vaccines under clinical development. J. Virus Erad. 2016, 2, 198–207. [Google Scholar] [CrossRef]
- Cui, X.; Cao, Z.; Wang, S.; Lee, R.B.; Wang, X.; Murata, H.; Adler, S.P.; McVoy, M.A.; Snapper, C.M. Novel trimeric human cytomegalovirus glycoprotein B elicits a high-titer neutralizing antibody response. Vaccine 2018, 36, 5580–5590. [Google Scholar] [CrossRef]
- Dargan, D.J.; Douglas, E.; Cunningham, C.; Jamieson, F.; Stanton, R.J.; Baluchova, K.; McSharry, B.P.; Tomasec, P.; Emery, V.C.; Percivalle, E.; et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 2010, 91, 1535–1546. [Google Scholar] [CrossRef]
- Cappadona, I.; Villinger, C.; Schutzius, G.; Mertens, T.; von Einem, J. Human Cytomegalovirus pUL47 Modulates Tegumentation and Capsid Accumulation at the Viral Assembly Complex. J. Virol. 2015, 89, 7314–7328. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Yuan, J.; Li, H.J.; Zeng, Z.F.; Luo, Z.W.; Li, S.Q.; He, C.Q.; Jia, X.F.; Zhang, X.; Zuo, H.; et al. Human cytomegalovirus UL49 encodes an early, virion-associated protein essential for virus growth in human foreskin fibroblasts. Arch. Virol. 2016, 161, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yao, Y.; Chen, J.; Wang, M. Unconserved C terminal of human cytomegalovirus tegument protein pUL76 elicits nuclear aggresome formation and induces DNA damage in transfected cells. J. Biomed. Sci. 2015, 22, 95. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Köppen-Rung, P.; Dittmer, A.; Bogner, E. Intracellular Distribution of Capsid-Associated pUL77 of Human Cytomegalovirus and Interactions with Packaging Proteins and pUL93. J. Virol. 2016, 90, 5876–5885. [Google Scholar] [CrossRef] [PubMed]
- Adair, R.; Douglas, E.R.; MacLean, J.B.; Graham, S.Y.; Aitken, J.D.; Jamieson, F.E.; Dargan, D.J. The products of human cytomegalovirus genes UL23, UL24, UL43 and US22 are tegument components. J. Gen. Virol. 2002, 83, 1315–1324. [Google Scholar] [CrossRef]
- Luo, J.; Chen, J.; Yang, E.; Shen, A.; Gong, H.; Pei, Z.; Xiao, G.; Lu, S.; Liu, F. Modulation of the Cellular Distribution of Human Cytomegalovirus Helicase by Cellular Factor Snapin. J. Virol. 2013, 87, 10628–10640. [Google Scholar] [CrossRef]
- Pizzorno, M.C.; Mullen, M.A.; Chang, Y.N.; Hayward, G.S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 1991, 65, 3839–3852. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, L.; Kankanala, J.; Wang, Z.; Geraghty, R.J. Inhibition of Human Cytomegalovirus pUL89 Terminase Subunit Blocks Virus Replication and Genome Cleavage. J. Virol. 2017, 91, e02152-16. [Google Scholar] [CrossRef]
- Connolly, S.A.; Jardetzky, T.S.; Longnecker, R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 2021, 19, 110–121. [Google Scholar] [CrossRef]
- Nguyen, C.C.; Kamil, J.P. Pathogen at the gates: Human cytomegalovirus entry and cell tropism. Viruses 2018, 10, 704. [Google Scholar] [CrossRef]
- Gao, S.; Ruan, S.; Ma, Y.; Li, M.; Wang, L.; Zheng, B.; Qi, Y.; Sun, Z.; Huang, Y.; Ruan, Q. Newly identified transcripts of UL4 and UL5 genes of human cytomegalovirus. Acta Biochim. Pol. 2015, 62, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.P.; Vesole, D.H.; Nelson, J.; Oldstone, M.B.; Stinski, M.F. Identification and expression of a human cytomegalovirus early glycoprotein. J. Virol. 1989, 63, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Vezzani, G.; Amendola, D.; Yu, D.; Chandramouli, S.; Frigimelica, E.; Maione, D.; Merola, M. The Human Cytomegalovirus UL116 Glycoprotein Is a Chaperone to Control gH-Based Complexes Levels on Virions. Front. Microbiol. 2021, 12, 630121. [Google Scholar] [CrossRef] [PubMed]
- Shikhagaie, M.; Mercé-Maldonado, E.; Isern, E.; Muntasell, A.; Albà, M.M.; López-Botet, M.; Hengel, H.; Angulo, A. The Human Cytomegalovirus-Specific UL1 Gene Encodes a Late-Phase Glycoprotein Incorporated in the Virion Envelope. J. Virol. 2012, 86, 4091–4101. [Google Scholar] [CrossRef] [PubMed]
- Anselmi, G.; Giuliani, M.; Vezzani, G.; Ferranti, R.; Gentile, M.; Cortese, M.; Amendola, D.; Pacchiani, N.; D’Aurizio, R.; Bruno, L.; et al. Characterization of pUL5, an HCMV protein interacting with the cellular protein IQGAP1. Virology 2020, 540, 57–65. [Google Scholar] [CrossRef]
- Gonzalez-Perez, A.C.; Stempel, M.; Wyler, E.; Urban, C.; Piras, A.; Hennig, T.; Ganskih, S.; Wei, Y.; Heim, A.; Landthaler, M.; et al. The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral UL4/UL5 Transcripts. MBio 2021, 12, e02683-20. [Google Scholar] [CrossRef]
- Engel, P.; Pérez-Carmona, N.; Albà, M.M.; Robertson, K.; Ghazal, P.; Angulo, A. Human cytomegalovirus UL7, a homologue of the SLAM-family receptor CD229, impairs cytokine production. Immunol. Cell Biol. 2011, 89, 753–766. [Google Scholar] [CrossRef]
- MacManiman, J.D.; Meuser, A.; Botto, S.; Smith, P.P.; Liu, F.; Jarvis, M.A.; Nelson, J.A.; Caposio, P. Human Cytomegalovirus-Encoded pUL7 Is a Novel CEACAM1-Like Molecule Responsible for Promotion of Angiogenesis. MBio 2014, 5, e02035. [Google Scholar] [CrossRef]
- Crawford, L.B.; Kim, J.H.; Collins-McMillen, D.; Lee, B.J.; Landais, I.; Held, C.; Nelson, J.A.; Yurochko, A.D.; Caposio, P. Human cytomegalovirus encodes a novel FLT3 receptor ligand necessary for hematopoietic cell differentiation and viral reactivation. MBio 2018, 9, e00682-18. [Google Scholar] [CrossRef]
- Pérez-Carmona, N.; Martínez-Vicente, P.; Farré, D.; Gabaev, I.; Messerle, M.; Engel, P.; Angulo, A. A Prominent Role of the Human Cytomegalovirus UL8 Glycoprotein in Restraining Proinflammatory Cytokine Production by Myeloid Cells at Late Times during Infection. J. Virol. 2018, 92, e02229-17. [Google Scholar] [CrossRef]
- Bruno, L.; Cortese, M.; Monda, G.; Gentile, M.; Caló, S.; Schiavetti, F.; Zedda, L.; Cattaneo, E.; Piccioli, D.; Schaefer, M.; et al. Human cytomegalovirus pUL10 interacts with leukocytes and impairs TCR-mediated T-cell activation. Immunol. Cell Biol. 2016, 94, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Zischke, J.; Mamareli, P.; Pokoyski, C.; Gabaev, I.; Buyny, S.; Jacobs, R.; Falk, C.S.; Lochner, M.; Sparwasser, T.; Schulz, T.F.; et al. The human cytomegalovirus glycoprotein pUL11 acts via CD45 to induce T cell IL-10 secretion. PLoS Pathog. 2017, 13, e1006454. [Google Scholar] [CrossRef] [PubMed]
- Rölle, A.; Mousavi-Jazi, M.; Eriksson, M.; Odeberg, J.; Söderberg-Nauclér, C.; Cosman, D.; Kärre, K.; Cerboni, C. Effects of Human Cytomegalovirus Infection on Ligands for the Activating NKG2D Receptor of NK Cells: Up-Regulation of UL16-Binding Protein (ULBP)1 and ULBP2 Is Counteracted by the Viral UL16 Protein. J. Immunol. 2003, 171, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, B.; Cho, S.; Shin, J.; Cho, K.; Jun, Y.; Ahn, K. Human cytomegalovirus UL18 utilizes US6 for evading the NK and T-cell responses. PLoS Pathog. 2008, 4, 1000123. [Google Scholar] [CrossRef] [PubMed]
- Jelcic, I.; Reichel, J.; Schlude, C.; Treutler, E.; Sinzger, C.; Steinle, A. The polymorphic HCMV glycoprotein UL20 is targeted for lysosomal degradation by multiple cytoplasmic dileucine motifs. Traffic 2011, 12, 1444–1456. [Google Scholar] [CrossRef] [PubMed]
- Casarosa, P.; Gruijthuijsen, Y.K.; Michel, D.; Beisser, P.S.; Holl, J.; Fitzsimons, C.P.; Verzijl, D.; Bruggeman, C.A.; Mertens, T.; Leurs, R.; et al. Constitutive Signaling of the Human Cytomegalovirus-encoded Receptor UL33 Differs from That of Its Rat Cytomegalovirus Homolog R33 by Promiscuous Activation of G Proteins of the Gq, Gi, and Gs Classes. J. Biol. Chem. 2003, 278, 50010–50023. [Google Scholar] [CrossRef]
- Van Senten, J.R.; Bebelman, M.P.; Fan, T.S.; Heukers, R.; Bergkamp, N.D.; Van Gasselt, P.; Langemeijer, E.V.; Slinger, E.; Lagerweij, T.; Rahbar, A.; et al. The human cytomegalovirus-encoded G protein- coupled receptor UL33 exhibits oncomodulatory properties. J. Biol. Chem. 2019, 294, 16297–16308. [Google Scholar] [CrossRef]
- van Senten, J.R.; Bebelman, M.P.; van Gasselt, P.; Bergkamp, N.D.; van den Bor, J.; Siderius, M.; Smit, M.J. Human cytomegalovirus-encoded G protein-coupled receptor UL33 facilitates virus dissemination via the extracellular and cell-to-cell route. Viruses 2020, 12, 594. [Google Scholar] [CrossRef]
- Xi, Y.; Harwood, S.; Wise, L.M.; Purdy, J.G. Human Cytomegalovirus pUL37x1 Is Important for Remodeling of Host Lipid Metabolism. J. Virol. 2019, 93, e00843-19. [Google Scholar] [CrossRef]
- Brune, W.; Andoniou, C.E. Die another day: Inhibition of cell death pathways by cytomegalovirus. Viruses 2017, 9, 249. [Google Scholar] [CrossRef]
- Prod’homme, V.; Tomasec, P.; Cunningham, C.; Lemberg, M.K.; Stanton, R.J.; McSharry, B.P.; Wang, E.C.Y.; Cuff, S.; Martoglio, B.; Davison, A.J.; et al. Human Cytomegalovirus UL40 Signal Peptide Regulates Cell Surface Expression of the NK Cell Ligands HLA-E and gpUL18. J. Immunol. 2012, 188, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, T.; Inoue, N. Activation of c-Jun by human cytomegalovirus UL42 through JNK activation. PLoS ONE 2020, 15, e0232635. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Z.; Guo, Y.; Zou, H.M.; Su, S.; Wang, S.Y.; Yang, Q.; Luo, M.H.; Wang, Y.Y. Human cytomegalovirus protein UL42 antagonizes cGAS/MITA-mediated innate antiviral response. PLoS Pathog. 2019, 15, e1007691. [Google Scholar] [CrossRef]
- Häge, S.; Sonntag, E.; Svrlanska, A.; Borst, E.M.; Stilp, A.C.; Horsch, D.; Müller, R.; Kropff, B.; Milbradt, J.; Stamminger, T.; et al. Phenotypical characterization of the nuclear egress of recombinant cytomegaloviruses reveals defective replication upon orf-ul50 deletion but not pul50 phosphosite mutation. Viruses 2021, 13, 165. [Google Scholar] [CrossRef]
- Weiler, N.; Paal, C.; Adams, K.; Calcaterra, C.; Fischer, D.; Stanton, R.J.; Stöhr, D.; Sampaio, K.L.; Sinzger, C. Role of envelope glycoprotein complexes in cell-associated spread of human cytomegalovirus. Viruses 2021, 13, 614. [Google Scholar] [CrossRef]
- Mach, M.; Osinski, K.; Kropff, B.; Schloetzer-Schrehardt, U.; Krzyzaniak, M.; Britt, W. The Carboxy-Terminal Domain of Glycoprotein N of Human Cytomegalovirus Is Required for Virion Morphogenesis. J. Virol. 2007, 81, 5212–5224. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Prager, A.; Boos, S.; Resch, M.; Brizic, I.; Mach, M.; Wildner, S.; Scrivano, L.; Adler, B. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog. 2017, 13, e1006281. [Google Scholar] [CrossRef]
- van Senten, J.R.; Fan, T.S.; Siderius, M.; Smit, M.J. Viral G protein-coupled receptors as modulators of cancer hallmarks. Pharmacol. Res. 2020, 156, 104804. [Google Scholar] [CrossRef]
- O’Connor, C.M.; Shenk, T. Human Cytomegalovirus pUL78 G Protein-Coupled Receptor Homologue Is Required for Timely Cell Entry in Epithelial Cells but Not Fibroblasts. J. Virol. 2012, 86, 11425–11433. [Google Scholar] [CrossRef]
- Krzyzaniak, M.A.; Mach, M.; Britt, W.J. HCMV-encoded glycoprotein M (UL100) interacts with rab11 effector protein FIP4. Traffic 2009, 10, 1439–1457. [Google Scholar] [CrossRef]
- Krzyzaniak, M.; Mach, M.; Britt, W.J. The Cytoplasmic Tail of Glycoprotein M (gpUL100) Expresses Trafficking Signals Required for Human Cytomegalovirus Assembly and Replication. J. Virol. 2007, 81, 10316–10328. [Google Scholar] [CrossRef] [PubMed]
- Pandey, J.P.; Namboodiri, A.M.; Radwan, F.F.; Nietert, P.J. The decoy Fcγ receptor encoded by the cytomegalovirus UL119-UL118 gene has differential affinity to IgG proteins expressing different GM allotypes. Hum. Immunol. 2015, 76, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Landini, M.P.; Lazzarotto, T.; Xu, J.; Geballe, A.P.; Mocarski, E.S. Humoral Immune Response to Proteins of Human Cytomegalovirus Latency-Associated Transcripts. Biol. Blood Marrow Transplant. 2000, 6, 100–108. [Google Scholar] [CrossRef]
- Wu, H.; Kropff, B.; Mach, M.; Britt, W.J. Human cytomegalovirus envelope protein gpul132 regulates infectious virus production through formation of the viral assembly compartment. MBio 2020, 11, e02044-20. [Google Scholar] [CrossRef]
- Mlera, L.; Moy, M.; Maness, K.; Tran, L.N.; Goodrum, F.D. The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation. Viruses 2020, 12, 714. [Google Scholar] [CrossRef] [PubMed]
- Stanton, R.J.; Prod’Homme, V.; Purbhoo, M.A.; Moore, M.; Aicheler, R.J.; Heinzmann, M.; Bailer, S.M.; Haas, J.; Antrobus, R.; Weekes, M.P.; et al. HCMV pUL135 remodels the actin cytoskeleton to impair immune recognition of infected cells. Cell Host Microbe 2014, 16, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Bughio, F.; Umashankar, M.; Wilson, J.; Goodrum, F. Human Cytomegalovirus UL135 and UL136 Genes Are Required for Postentry Tropism in Endothelial Cells. J. Virol. 2015, 89, 6536–6550. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caviness, K.; Bughio, F.; Crawford, L.B.; Streblow, D.N.; Nelson, J.A.; Caposio, P.; Goodrum, F. Complex interplay of the UL136 isoforms balances cytomegalovirus replication and Latency. MBio 2016, 7, e01986. [Google Scholar] [CrossRef]
- Bughio, F.; Elliott, D.A.; Goodrum, F. An Endothelial Cell-Specific Requirement for the UL133-UL138 Locus of Human Cytomegalovirus for Efficient Virus Maturation. J. Virol. 2013, 87, 3062–3075. [Google Scholar] [CrossRef][Green Version]
- Han, L.; Ma, Y.; Liu, Z.; Liu, C.; Lu, Y.; Qi, Y.; Huang, Y.; Sun, Z.; Ruan, Q. Transcriptional regulation and influence on replication of the human cytomegalovirus UL1381.4 kb transcript. Mol. Med. Rep. 2017, 16, 5649–5658. [Google Scholar] [CrossRef][Green Version]
- Gelbmann, C.B.; Kalejta, R.F. The Golgi sorting motifs of human cytomegalovirus UL138 are not required for latency maintenance. Virus Res. 2019, 270, 197646. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Mao, Z.Q.; Ruan, Q.; He, R.; Ma, Y.P.; Sun, Z.R.; Ji, Y.H.; Huang, Y. Human cytomegalovirus (HCMV) UL139 open reading frame: Sequence variants are clustered into three major genotypes. J. Med. Virol. 2006, 78, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Zou, F.; Lu, Z.T.; Wang, S.; Wu, S.; Wu, Y.Y.; Sun, Z.R. Human cytomegalovirus UL141 protein interacts with CELF5 and affects viral DNA replication. Mol. Med. Rep. 2018, 17, 4657–4664. [Google Scholar] [CrossRef] [PubMed]
- Tomasec, P.; Wang, E.C.Y.; Davison, A.J.; Vojtesek, B.; Armstrong, M.; Griffin, C.; McSharry, B.P.; Morris, R.J.; Llewellyn-Lacey, S.; Rickards, C.; et al. Downregulation of natural killer cell-activating ligang CD155 by human cytomegalovirus UL141. Nat. Immunol. 2005, 6, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Nemčovičová, I.; Benedict, C.A.; Zajonc, D.M. Structure of Human Cytomegalovirus UL141 Binding to TRAIL-R2 Reveals Novel, Non-canonical Death Receptor Interactions. PLoS Pathog. 2013, 9, e1003224. [Google Scholar] [CrossRef]
- Ashiru, O.; Bennett, N.J.; Boyle, L.H.; Thomas, M.; Trowsdale, J.; Wills, M.R. NKG2D Ligand MICA Is Retained in the cis -Golgi Apparatus by Human Cytomegalovirus Protein UL142. J. Virol. 2009, 83, 12345–12354. [Google Scholar] [CrossRef]
- Bitra, A.; Nemčovičová, I.; Picarda, G.; Doukov, T.; Wang, J.; Benedict, C.A.; Zajonc, D.M. Structure of human cytomegalovirus UL144, an HVEM orthologue, bound to the B and T cell lymphocyte attenuator. J. Biol. Chem. 2019, 294, 10519–10529. [Google Scholar] [CrossRef]
- Poole, E.; Walther, A.; Raven, K.; Benedict, C.A.; Mason, G.M.; Sinclair, J. The Myeloid Transcription Factor GATA-2 Regulates the Viral UL144 Gene during Human Cytomegalovirus Latency in an Isolate-Specific Manner. J. Virol. 2013, 87, 4261–4271. [Google Scholar] [CrossRef]
- Lüttichau, H.R. The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J. Biol. Chem. 2010, 285, 9137–9146. [Google Scholar] [CrossRef]
- Seidel, E.; Dassa, L.; Schuler, C.; Oiknine-Djian, E.; Wolf, D.G.; Le-Trilling, V.T.K.; Mandelboim, O. The human cytomegalovirus protein UL147A downregulates the most prevalent MICA allele: MICA*008, to evade NK cell-mediated killing. PLoS Pathog. 2021, 17, e1008807. [Google Scholar] [CrossRef] [PubMed]
- Siddiquey, M.N.A.; Schultz, E.P.; Yu, Q.; Amendola, D.; Vezzani, G.; Yu, D.; Maione, D.; Lanchy, J.-M.; Ryckman, B.J.; Merola, M.; et al. The human cytomegalovirus protein UL116 interacts with the viral ER resident glycoprotein UL148 and promotes the incorporation of gH/gL complexes into virions. J. Virol. 2021, 95, e0220720. [Google Scholar] [CrossRef] [PubMed]
- Dassa, L.; Seidel, E.; Oiknine-Djian, E.; Yamin, R.; Wolf, D.G.; Le-Trilling, V.T.K.; Mandelboim, O. The Human Cytomegalovirus Protein UL148A Downregulates the NK Cell-Activating Ligand MICA to Avoid NK Cell Attack. J. Virol. 2018, 92, e00162-18. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.H.; Rong Sun, Z.; Ruan, Q.; Guo, J.J.; He, R.; Qi, Y.; Ma, Y.P.; Mao, Z.Q.; Huang, Y.J. Polymorphisms of human cytomegalovirus UL148A, UL148B, UL148C, UL148D genes in clinical strains. J. Clin. Virol. 2006, 37, 252–257. [Google Scholar] [CrossRef]
- Gabor, F.; Jahn, G.; Sedmak, D.D.; Sinzger, C. In vivo Downregulation of MHC Class I Molecules by HCMV Occurs during All Phases of Viral Replication but Is Not Always Complete. Front. Cell. Infect. Microbiol. 2020, 10, 283. [Google Scholar] [CrossRef]
- Park, B.; Kim, Y.; Shin, J.; Lee, S.; Cho, K.; Früh, K.; Lee, S.; Ahn, K. Human Cytomegalovirus Inhibits Tapasin-Dependent Peptide Loading and Optimization of the MHC Class I Peptide Cargo for Immune Evasion. Immunity 2004, 20, 71–85. [Google Scholar] [CrossRef]
- Dugan, G.E.; Hewitt, E.W. Structural and Functional Dissection of the Human Cytomegalovirus Immune Evasion Protein US6. J. Virol. 2008, 82, 3271–3282. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, A.; Ra, E.A.; Lee, T.A.; Choi, H.; Lee, E.; Kang, S.; Seo, J.Y.; Lee, S.; Park, B. HCMV-encoded US7 and US8 act as antagonists of innate immunity by distinctively targeting TLR-signaling pathways. Nat. Commun. 2019, 10, 4670. [Google Scholar] [CrossRef]
- Choi, H.J.; Park, A.; Kang, S.; Lee, E.; Lee, T.A.; Ra, E.A.; Lee, J.; Lee, S.; Park, B. Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type i interferon immune responses. Nat. Commun. 2018, 9, 125. [Google Scholar] [CrossRef]
- Park, B.; Spooner, E.; Houser, B.L.; Strominger, J.L.; Ploegh, H.L. The HCMV membrane glycoprotein US10 selectively targets HLA-G for degradation. J. Exp. Med. 2010, 207, 2033–2041. [Google Scholar] [CrossRef]
- Zimmermann, C.; Kowalewski, D.; Bauersfeld, L.; Hildenbrand, A.; Gerke, C.; Schwarzmüller, M.; Le-Trilling, V.T.K.; Stevanovic, S.; Hengel, H.; Momburg, F.; et al. HLA-B locus products resist degradation by the human cytomegalovirus immunoevasin US11. PLoS Pathog. 2019, 15, e1008040. [Google Scholar] [CrossRef]
- Fielding, C.A.; Weekes, M.P.; Nobre, L.V.; Ruckova, E.; Wilkie, G.S.; Paulo, J.A.; Chang, C.; Suárez, N.M.; Davies, J.A.; Antrobus, R.; et al. Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation. Elife 2017, 6, e22206. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Pellett, P.E. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology 2007, 361, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Luganini, A.; Cavaletto, N.; Raimondo, S.; Geuna, S.; Gribaudo, G. Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer. J. Virol. 2017, 91, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Gurczynski, S.J.; Das, S.; Pellett, P.E. Deletion of the Human Cytomegalovirus US17 Gene Increases the Ratio of Genomes per Infectious Unit and Alters Regulation of Immune and Endoplasmic Reticulum Stress Response Genes at Early and Late Times after Infection. J. Virol. 2014, 88, 2168–2182. [Google Scholar] [CrossRef][Green Version]
- Charpak-Amikam, Y.; Kubsch, T.; Seidel, E.; Oiknine-Djian, E.; Cavaletto, N.; Yamin, R.; Schmiedel, D.; Wolf, D.; Gribaudo, G.; Messerle, M.; et al. Human cytomegalovirus escapes immune recognition by NK cells through the downregulation of B7-H6 by the viral genes US18 and US20. Sci. Rep. 2017, 7, 8661. [Google Scholar] [CrossRef]
- Cavaletto, N.; Luganini, A.; Gribaudo, G. Inactivation of the Human Cytomegalovirus US20 Gene Hampers Productive Viral Replication in Endothelial Cells. J. Virol. 2015, 89, 11092–11106. [Google Scholar] [CrossRef]
- Luganini, A.; Di Nardo, G.; Munaron, L.; Gilardi, G.; Pla, A.F.; Gribaudo, G. Human cytomegalovirus US21 protein is a viroporin that modulates calcium homeostasis and protects cells against apoptosis. Proc. Natl. Acad. Sci. USA 2018, 115, E12370–E12377. [Google Scholar] [CrossRef]
- Boeck, J.M.; Stowell, G.A.; O’Connor, C.M.; Spencer, J.V. The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through G β γ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1. J. Virol. 2018, 92, e00644-18. [Google Scholar] [CrossRef]
- Tu, C.C.; O’Connor, C.M.; Spencer, J.V. Identification of a novel signaling complex containing host chemokine receptor CXCR4, Interleukin-10 receptor, and human cytomegalovirus US27. Virology 2020, 548, 49–58. [Google Scholar] [CrossRef]
- O’Connor, C.M.; Shenk, T. Human Cytomegalovirus pUS27 G Protein-Coupled Receptor Homologue Is Required for Efficient Spread by the Extracellular Route but Not for Direct Cell-to-Cell Spread. J. Virol. 2011, 85, 3700–3707. [Google Scholar] [CrossRef]
- Nobre, L.; Nightingale, K.; Ravenhill, B.J.; Antrobus, R.; Soday, L.; Nichols, J.; Davies, J.; Seirafian, S.; Wang, E.C.Y.; Davison, A.J.; et al. Human cytomegalovirus interactome analysis identifies degradation hubs, domain associations and viral protein functions. Elife 2019, 8, e49894. [Google Scholar] [CrossRef] [PubMed]
- Krishna, B.A.; Miller, W.E.; O’Connor, C.M. US28: HCMV’s swiss army knife. Viruses 2018, 10, 445. [Google Scholar] [CrossRef]
- Gatherer, D.; Seirafian, S.; Cunningham, C.; Holton, M.; Dargan, D.J.; Baluchova, K.; Hector, R.D.; Galbraith, J.; Herzyk, P.; Wilkinson, G.W.G.; et al. High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. USA 2011, 108, 19755–19760. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.T.; Kim, Y.E.; Kim, Y.J.; Lee, M.K.; Hayward, G.S.; Ahn, J.H. Analysis of human cytomegalovirus-encoded SUMO targets and temporal regulation of SUMOylation of the immediate-early proteins IE1 and IE2 during infection. PLoS ONE 2014, 9, 103308. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lilley, B.N.; Ploegh, H.L.; Tirabassi, R.S. Human Cytomegalovirus Open Reading Frame TRL11/IRL11 Encodes an Immunoglobulin G Fc-Binding Protein. J. Virol. 2001, 75, 11218–11221. [Google Scholar] [CrossRef]
- Wilkinson, G.W.G.; Davison, A.J.; Tomasec, P.; Fielding, C.A.; Aicheler, R.; Murrell, I.; Seirafian, S.; Wang, E.C.Y.; Weekes, M.; Lehner, P.J.; et al. Human cytomegalovirus: Taking the strain. Med. Microbiol. Immunol. 2015, 204, 273–284. [Google Scholar] [CrossRef]
- Cortese, M.; Calò, S.; D’Aurizio, R.; Lilja, A.; Pacchiani, N.; Merola, M. Recombinant Human Cytomegalovirus (HCMV) RL13 Binds Human Immunoglobulin G Fc. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ren, G.; Cui, X.; Lu, Z.; Ma, Y.; Qi, Y.; Huang, Y.; Liu, Z.; Sun, Z.; Ruan, Q. Human cytomegalovirus RL13 protein interacts with host NUDT14 protein affecting viral DNA replication. Mol. Med. Rep. 2016, 13, 2167–2174. [Google Scholar] [CrossRef]
- Schultz, E.P.; Lanchy, J.-M.; Day, L.Z.; Yu, Q.; Peterson, C.; Preece, J.; Ryckman, B.J. Specialization for Cell-Free or Cell-to-Cell Spread of BAC-Cloned Human Cytomegalovirus Strains Is Determined by Factors beyond the UL128-131 and RL13 Loci. J. Virol. 2020, 94, e00034-20. [Google Scholar] [CrossRef] [PubMed]
- Lesniewski, M.; Das, S.; Skomorovska-Prokvolit, Y.; Wang, F.Z.; Pellett, P.E. Primate cytomegalovirus US12 gene family: A distinct and diverse clade of seven-transmembrane proteins. Virology 2006, 354, 286–298. [Google Scholar] [CrossRef][Green Version]
- Mozzi, A.; Biolatti, M.; Cagliani, R.; Forni, D.; Dell’Oste, V.; Pontremoli, C.; Vantaggiato, C.; Pozzoli, U.; Clerici, M.; Landolfo, S.; et al. Past and ongoing adaptation of human cytomegalovirus to its host. PLoS Pathog. 2020, 16, e1008476. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Akter, P.; Cunningham, C.; Dolan, A.; Addison, C.; Dargan, D.J.; Hassan-Walker, A.F.; Emery, V.C.; Griffiths, P.D.; Wilkinson, G.W.G. Homology between the human cytomegalovirus RL11 gene family and human adenovirus E3 genes. J. Gen. Virol. 2003, 84, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, J.A.; Paul, J.R.; Spencer, J.V. Evolution of the ability to modulate host chemokine networks via gene duplication in Human Cytomegalovirus (HCMV). Infect. Genet. Evol. 2017, 51, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Qian, Y.; Yu, W.; Guo, G.; Wang, H.; Xue, X. Functional Profile of Human Cytomegalovirus Genes and Their Associated Diseases: A Review. Front. Microbiol. 2020, 11, 2104. [Google Scholar] [CrossRef] [PubMed]
- Patro, A.R.K. Subversion of immune response by human cytomegalovirus. Front. Immunol. 2019, 10, 1155. [Google Scholar] [CrossRef]
- Weekes, M.P.; Tomasec, P.; Huttlin, E.L.; Fielding, C.A.; Nusinow, D.; Stanton, R.J.; Wang, E.C.Y.; Aicheler, R.; Murrell, I.; Wilkinson, G.W.G.; et al. Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell 2014, 157, 1460–1472. [Google Scholar] [CrossRef]
- Couté, Y.; Kraut, A.; Zimmermann, C.; Büscher, N.; Hesse, A.M.; Bruley, C.; De Andrea, M.; Wangen, C.; Hahn, F.; Marschall, M.; et al. Mass spectrometry-based characterization of the virion proteome, phosphoproteome, and associated kinase activity of human cytomegalovirus. Microorganisms 2020, 8, 820. [Google Scholar] [CrossRef] [PubMed]
- Di Bona, D.; Scafidi, V.; Plaia, A.; Colomba, C.; Nuzzo, D.; Occhino, C.; Tuttolomondo, A.; Giammanco, G.; De Grazia, S.; Montalto, G.; et al. HLA and killer cell immunoglobulin-like receptors influence the natural course of CMV infection. J. Infect. Dis. 2014, 210, 1083–1089. [Google Scholar] [CrossRef]
- Aiello, A.; Accardi, G.; Candore, G.; Caruso, C.; Colomba, C.; Di Bona, D.; Duro, G.; Gambino, C.M.; Ligotti, M.E.; Pandey, J.P. Role of immunogenetics in the outcome of HCMV infection: Implications for ageing. Int. J. Mol. Sci. 2019, 20, 685. [Google Scholar] [CrossRef]
- Di Bona, D.; Accardi, G.; Aiello, A.; Bilancia, M.; Candore, G.; Colomba, C.; Caruso, C.; Duro, G.; Gambino, C.M.; Macchia, L.; et al. Association between γ marker, human leucocyte antigens and killer immunoglobulin-like receptors and the natural course of human cytomegalovirus infection: A pilot study performed in a Sicilian population. Immunology 2018, 153, 523–531. [Google Scholar] [CrossRef]
- Pandey, J.P.; Kistner-Griffin, E.; Radwan, F.F.; Kaur, N.; Namboodiri, A.M.; Black, L.; Butler, M.A.; Carreón, T.; Ruder, A.M. Immunoglobulin genes influence the magnitude of humoral immunity to cytomegalovirus Glycoprotein B. J. Infect. Dis. 2014, 210, 1823–1826. [Google Scholar] [CrossRef] [PubMed]
- Aguttu, C.; Okech, B.A.; Mukisa, A.; Lubega, G.W. Screening and characterization of hypothetical proteins of Plasmodium falciparum as novel vaccine candidates in the fight against malaria using reverse vaccinology. J. Genet. Eng. Biotechnol. 2021, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Almén, M.S.; Nordström, K.J.V.; Fredriksson, R.; Schiöth, H.B. Mapping the human membrane proteome: A majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Orfanoudaki, G.; Economou, A. Proteome-wide subcellular topologies of E. coli polypeptides database (STEPdb). Mol. Cell. Proteom. 2014, 13, 3674–3687. [Google Scholar] [CrossRef] [PubMed]
- Dal Monte, P.; Pignatelli, S.; Rossini, G.; Landini, M.P. Genomic variants among human cytomegalovirus (HCMV) clinical isolates: The glycoprotein n (gN) paradigm. Hum. Immunol. 2004, 65, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Mattick, C.; Dewin, D.; Polley, S.; Sevilla-Reyes, E.; Pignatelli, S.; Rawlinson, W.; Wilkinson, G.; Dal Monte, P.; Gompels, U.A. Linkage of human cytomegalovirus glycoprotein gO variant groups identified from worldwide clinical isolates with gN genotypes, implications for disease associations and evidence for N-terminal sites of positive selection. Virology 2004, 318, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Whisstock, J.C.; Lesk, A.M. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003, 36, 307–340. [Google Scholar] [CrossRef]
- Gilbert, C.; Feschotte, C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010, 8, e1000495. [Google Scholar] [CrossRef]
- Tchoupa, A.K.; Schuhmacher, T.; Hauck, C.R. Signaling by epithelial members of the CEACAM family—Mucosal docking sites for pathogenic bacteria. Cell Commun. Signal. 2014, 12, 27. [Google Scholar] [CrossRef]
- Chan, C.-M.; Chu, H.; Wang, Y.; Wong, B.H.-Y.; Zhao, X.; Zhou, J.; Yang, D.; Leung, S.P.; Chan, J.F.-W.; Yeung, M.-L.; et al. Carcinoembryonic Antigen-Related Cell Adhesion Molecule 5 Is an Important Surface Attachment Factor That Facilitates Entry of Middle East Respiratory Syndrome Coronavirus. J. Virol. 2016, 90, 9114–9127. [Google Scholar] [CrossRef]
- Terakita, A. The opsins. Genome Biol. 2005, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Arav-Boger, R.; Foster, C.B.; Zong, J.C.; Pass, R.F. Human cytomegalovirus-encoded α-chemokines exhibit high sequence variability in congenitally infected newborns. J. Infect. Dis. 2006, 193, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.A.; Hynes, R.O. Distribution and Evolution of von Willebrand/Integrin A Domains: Widely Dispersed Domains with Roles in Cell Adhesion and Elsewhere. Mol. Biol. Cell 2002, 13, 4100–4109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ni, C.; Li, Z.; Li, X.; Han, R.; Zhao, F.; Xu, J.; Gao, X.; Wang, S. PureseqTM: Efficient and accurate prediction of transmembrane topology from amino acid sequence only. bioRxiv 2019. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Queirós, P.; Delogu, F.; Hickl, O.; May, P.; Wilmes, P. Mantis: Flexible and consensus-driven genome. Giga Sci. 2021, 10, giab042. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Selengut, J.D.; Richter, R.A.; Harkins, D.; Basu, M.K.; Beck, E. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013, 41, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. In Ggplot2; Springer: New York, NY, USA, 2009; pp. 1–7. ISBN 9780387981413. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
| CMV Strains | Isolation Source | Number of Culture Passages | Accession Number |
|---|---|---|---|
| AD169 | Adenoids of a 7-year-old girl | Many times in human fibroblasts | FJ527563.1 |
| Towne | Urine of a 2-month-old infant with microcephaly and hepatosplenomegaly | Many times in human fibroblasts | FJ616285.1 |
| Toledo | Urine from a congenitally infected infant | Several times in human fibroblasts | GU937742.2 |
| TR | Vitreous humor from eye of HIV-positive male | Several times in human fibroblasts | KF021605.1 |
| VR7863 | Urine samples of a congenitally infected neonate and cultured in endothelial and epithelial cells | Cultured in endothelial and epithelial cells | KX544838.1 |
| TB40-E_UNC | Throat swab of a bone marrow transplant patient | Cultured adapted | KX544839.1 |
| HANSCTR4 | Blood from stem cell transplant recipient (D-R+) | Sequenced directly from clinical material via target enrichment | KY123653.1 |
| AD169-BAC20 | - | - | MN920393.1 |
| Merlin | Urine from a congenitally infected child | 3 times in human fibroblasts | NC006273.2 |
| Gene | Localization | Function | Number of TM Domains | References |
|---|---|---|---|---|
| UL1 * | VM | Unknown. pUL1 could modulate CMV host cell tropism. | 1–2 | [45] |
| UL2 * | HM | Unknown. | 1 | - |
| UL5 * | V | Unknown. It is suggested to be involved in efficient viral assembly, propagation and replication. | 1–3 | [46,47] |
| UL6 * | HM | Unknown. | 1–2 | - |
| UL7 | UL7 is involved in immunomodulation. | 2–3 | [48,49,50] | |
| UL8 * | HM | UL8 decreases the release of a large number of pro-inflammatory factors later after infection of THP-1 myeloid cells. UL8 may exert an immunosuppressive role key for CMV survival in the host. | 1–2 | [51] |
| UL9 * | HM | Unknown function. Its deletion mutation cause enhanced growth in HFFs cells. | 1–3 | [9] |
| UL10 * | M | Unknown. Potential role in immunomodulation. | 1–2 | [52] |
| UL11 | HM, ERM | pUL11 interacts with CD45 phosphatase on T cells, inducing the IL-10 secretion. | 1–2 | [53] |
| UL14 * | HM | Unknown. | 0–1 | - |
| UL15A * | HM | Unknown. | 1 | - |
| UL16 | HM | Immunoevasion and inhibition of the activation of NK cells. | 1 | [54] |
| UL18 | HM | Immunomodulation and immunoevasion. | 1–2 | [55] |
| UL20 * | ERM | Unknown. UL20 could be destined to sequester cellular proteinases not known to date for degradation in lysosomes. | 1–2 | [56] |
| UL33 | HM | UL33 has homology with GPCR which activates different ligand-independent signalling pathways and also involved in virus dissemination. | 6–7 | [57,58,59] |
| UL37 | ERM, GM, MM | Viral replication. | 2–3 | [60,61] |
| UL40 | HM | Immunomodulation. | 0–2 | [62] |
| UL41A * | VM | Unknown. UL41A not to code for proteins. | 1 | [10] |
| UL42 * | HM, C | Unknown. Potential role in immunoevasion. | 1 | [63,64] |
| UL50 | HNM | Assembly, maturation and egress of virions. | 1 | [65] |
| UL55 | VM, HM, GM | Glycoprotein B participates in viral entry. | 1–3 | [66] |
| UL73 | VM, HM, GM | Glycoprotein N is involved in the binding of the virus to the host cell, viral spread and virion morphogenesis. | 1 | [67] |
| UL74A * | VM | Unknown | 1 | - |
| UL75 | HM, VM | Glycoprotein H participates in viral entry. It is part of the trimeric and pentameric complexes. | 1 | [68] |
| UL78 * | HM, ERM | Unknown. UL78 is a G protein-coupled receptor. | 6–7 | [69,70] |
| UL100 | HM, VM | Envelope glycoprotein M participates in viral entry. | 8 | [71,72] |
| UL119 | VM | Immunoevasion. | 1 | [73] |
| UL120 * | HM | Unknown. | 1–2 | - |
| UL121 * | HM | Unknown. | 1–2 | - |
| UL124 * | HM | Potential role in latency. | 0–1 | [74] |
| UL132 | VM | Essential for CMV assembly compartment formation and the efficient production of infectious particles. | 1–2 | [75] |
| UL133 | GM | UL133 forms a complex with UL138 and UL136. It is involved in the establishment of CMV latency. | 2 | [76] |
| UL135 | HM, GM | Immunomodulation. Post entry Tropism in Endothelial Cells. | 0–1 | [77,78] |
| UL136 | HM | Replication, latency, and dissemination. Post entry Tropism in Endothelial Cells. | 1 | [76,78,79,80] |
| UL138 | GM | Latency and DNA replication. | 1 | [81,82] |
| UL139 * | HM | Unknown. Potential role in immunomodulation. | 1–2 | [83] |
| UL140 * | HM | Unknown. | 1 | - |
| UL141 | ERM | Immunomodulation and DNA replication. | 1 | [84,85,86] |
| UL142 | ERM | Immunomodulation. | 0–1 | [87] |
| UL144 | HM | Inhibition of T-cell activation and latency. | 1 | [88,89] |
| UL147 * | EXR | Unknown. Potential role in immunomodulation. | 0–1 | [90] |
| UL147A | HM | Immunomodulation. | 0–1 | [91] |
| UL148 | ERM | Viral ER-resident glycoprotein that interacts with UL116 promoting the incorporation of gH/gL complexes into virions. | 1 | [92] |
| UL148A | HM | Immunoevasion of NK cells. | 1–2 | [93] |
| UL148B * | HM | Unknown. | 1 | [94] |
| UL148C * | HM | Unknown. | 0–3 | [94] |
| UL148D * | HM | Unknown. | 1 | [94] |
| US2 | ERM | Immunomodulation. | 1–2 | [95] |
| US3 | ERM | Immunoevasion. | 1 | [96] |
| US6 | ERM | Immunomodulation. | 1 | [97] |
| US7 | ERM | Immunoevasion. | 1 | [98] |
| US8 | ERM, GM | Immunomodulation. | 1 | [98] |
| US9 | ERM, GM, CK | Glycoprotein US9 is an antagonist of IFN signalling to persistently evade host innate antiviral responses. | 0–1 | [99] |
| US10 | ERM | Inhibition of the host immune response. | 1–2 | [100] |
| US11 | ERM | Inhibition of the host immune response. | 0–1 | [101] |
| US12 | HM | Inmunomodulation of NK cells activation. | 6–7 | [102] |
| US13 * | HM | Unknown. | 7 | - |
| US14 | HM | Inmunomodulation of NK cells activation. Potential role in virions maturation and egress. | 5–7 | [102,103] |
| US15 * | HM | Unknown. | 7 | - |
| US16 | HM, C | Tropism in endothelial and epithelial cells. | 6–7 | [104] |
| US17 | HM | Immunomodulation. | 7 | [105] |
| US18 | HM. | Immunoevasion of NK cell. | 7–8 | [106] |
| US19 * | HM | Unknown. Its delection affect NK cell activation. | 6–7 | [102] |
| US20 | M | Inhibition NK cell activation. Also participates in the viral replication process in endothelial cells. | 7 | [106,107] |
| US21 | HM | Viroporin that modulates calcium homeostasis and protects cells against apoptosis. | 7–8 | [108] |
| US27 | V, HM | Immunomodulation. Also is required for efficient viral spread by the extracellular route. | 7 | [109,110,111] |
| US28 | HM | Immunomodulation. Lytic and latent CMV infection. Possible role in regulation of the actin cytoskeleton or cytoskeletal remodelling. | 7 | [112,113] |
| US29 * | HM | Unknown. | 0–2 | - |
| US30 * | HM | Unknown. | 1–2 | - |
| US33A * | - | Unknown. | 0–1 | [114] |
| US34A * | HM. | Unknown. Potential target of SUMO complex. | 1–2 | [115] |
| RL8A * | HM | Unknown. | 1 | - |
| RL9A * | HM | Unknown. | 1 | - |
| RL10 * | VM | Unknown. | 1–2 | - |
| RL11 | HM | Immunomodulation. RL11 is a type I transmembrane glycoproteins which bind immunoglobulin G Fc. I | 1–2 | [116] |
| RL12 * | VM | Unknown. RL12 is a Fc binding protein. | 1–2 | [117] |
| RL13 * | VM | Unknown. Potential role in replication, immunoevasión and viral spread by cell-free or cell-to-cell mechanisms. | 1 | [118,119,120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancebo, F.J.; Parras-Moltó, M.; García-Ríos, E.; Pérez-Romero, P. Deciphering the Potential Coding of Human Cytomegalovirus: New Predicted Transmembrane Proteome. Int. J. Mol. Sci. 2022, 23, 2768. https://doi.org/10.3390/ijms23052768
Mancebo FJ, Parras-Moltó M, García-Ríos E, Pérez-Romero P. Deciphering the Potential Coding of Human Cytomegalovirus: New Predicted Transmembrane Proteome. International Journal of Molecular Sciences. 2022; 23(5):2768. https://doi.org/10.3390/ijms23052768
Chicago/Turabian StyleMancebo, Francisco J., Marcos Parras-Moltó, Estéfani García-Ríos, and Pilar Pérez-Romero. 2022. "Deciphering the Potential Coding of Human Cytomegalovirus: New Predicted Transmembrane Proteome" International Journal of Molecular Sciences 23, no. 5: 2768. https://doi.org/10.3390/ijms23052768
APA StyleMancebo, F. J., Parras-Moltó, M., García-Ríos, E., & Pérez-Romero, P. (2022). Deciphering the Potential Coding of Human Cytomegalovirus: New Predicted Transmembrane Proteome. International Journal of Molecular Sciences, 23(5), 2768. https://doi.org/10.3390/ijms23052768







