Characterization and Identification of a Ripening-Related Gene AaPG18 in Actinidia arguta
Abstract
:1. Introduction
2. Results
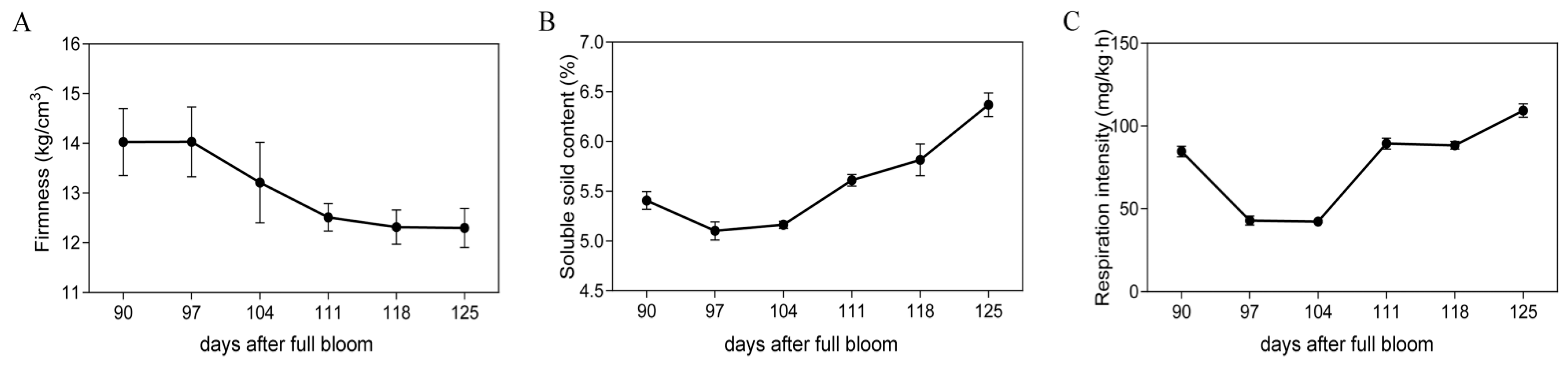
2.1. Physiological Changes in Fruit at Different Preharvest Stages
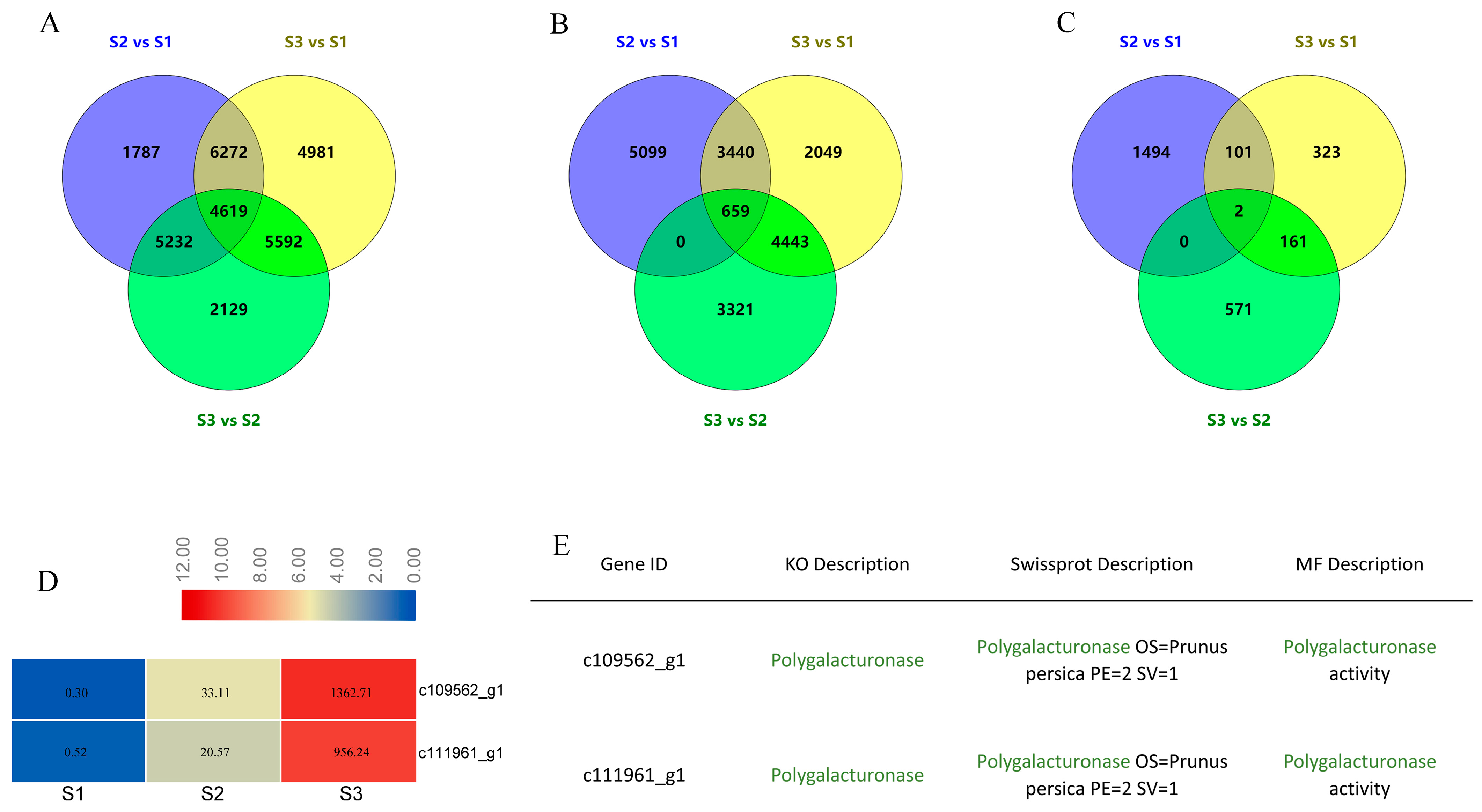
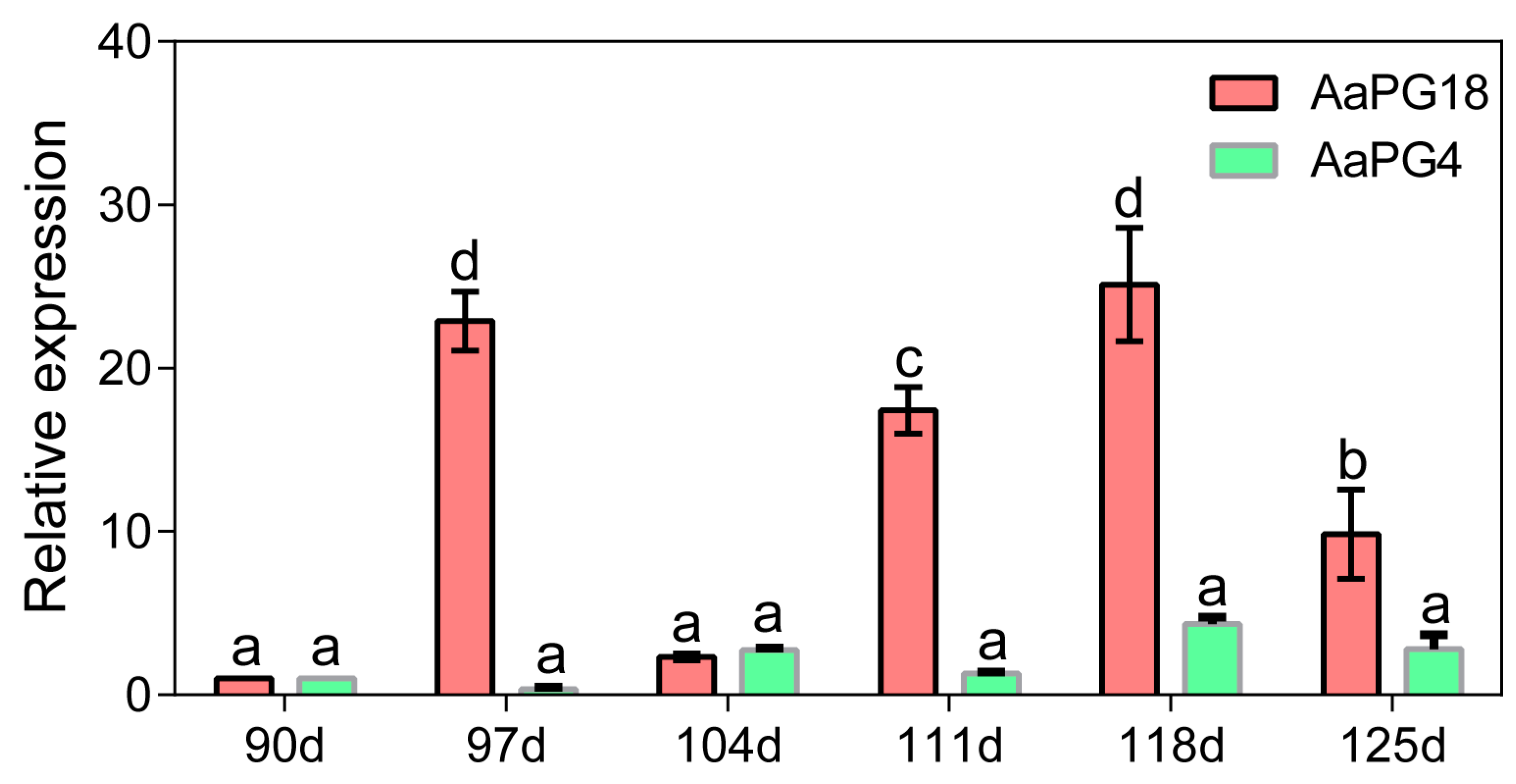
2.2. Screening of Candidate Genes Based on Transcriptome Analysis
2.3. Identification and Classification of the PG Family in Kiwifruit
2.4. A Membrane-Localizated AaPG18 Protein
2.5. Transient Overexpression of AaPG18 in Strawberry
2.6. Homologous Transient Transformation of AaPG18 in “RB-4”
3. Discussion
4. Materials and Methods
4.1. Fruits Preparation and RNA-Seq Process
4.2. Physiological Indexes Determination
4.3. Genome Availability, Pfam Search, and Chromosomal Localization
4.4. Sequence Alignment, Phylogenetic Tree Construction, and Gene Structure Model Analysis
4.5. Homologous Cloning and Vector Construction
4.6. Subcellular Localization in Nicotiana Benthamiana (N. Benthamiana)
4.7. Heterologous and Homologous Overexpression in Strawberry and Kiwifruit
4.8. qRT-PCR Analysis
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, H.W.; Ferguson, A.R. Actinidia in China: Natural diversity, phylogeographical evolution, interspecific gene flow and kiwifruit cultivar improvement. Acta Hortic. 2007, 753, 31–40. [Google Scholar] [CrossRef]
- Huang, H.W. Actinidia Genus: Classification, Resource, Domestication, Cultivation; Science Press: Beijing, China, 2013. (In Chinese) [Google Scholar]
- Barry, C.S.; Giovannoni, J.J. Ethylene and fruit ripening. Physiol. Plant. 1997, 26, 143. [Google Scholar] [CrossRef]
- Klee, H.J.; Giovannoni, J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011, 45, 41–59. [Google Scholar] [CrossRef]
- Brummell, D.A. Cell wall disassembly in ripening fruit. Funct. Plant Biol. 2006, 33, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Posé, S.; Paniagua, C.; Matas, A.J.; Gunning, A.P.; Morris, V.J.; Quesada, M.A. A nanostructural view of the cell wall disassembly process during fruit ripening and postharvest storage by atomic force microscopy. Trends Food Sci. Tech. 2009, 87, 47–58. [Google Scholar] [CrossRef]
- Jiménez-Bermúdez, S.; Redondo-Nevado, J.; Muñoz-Blanco, J.; Caballero, J.L.; López-Aranda, J.M.; Valpuesta, V. Manipulation of strawberry fruit softening by antisense expression of a pectate lyase gene. Plant Physiol. 2002, 128, 751–759. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Sutherland, P.W.; Johnston, S.L.; Gunaseelan, K.; Hallett, I.C.; Mitra, D. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus × domestica) fruit. BMC Plant Biol. 2012, 12, 129. [Google Scholar] [CrossRef] [Green Version]
- Paniagua, C.; Blanco-Portales, R.; Barceló-Muñoz, M.; García-Gago, J.A.; Waldron, K.W.; Quesada, M.A. Antisense down-regulation of the strawberry β-galactosidase gene FaβGal4 increases cell wall galactose levels and reduces fruit softening. J. Exp. Bot. 2016, 67, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Uluisik, S.; Chapman, N.H.; Smith, R.; Poole, M.; Adams, G.; Gillis, R.B. Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 2016, 34, 950–952. [Google Scholar] [CrossRef] [Green Version]
- Tacken, E.; Ireland, H.; Gunaseelan, K.; Karunairetnam, S.; Wang, D.; Schultz, K. The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol. 2010, 153, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Quesada, M.A.; Blanco-Portales, R.; Posé, S.; García-Gago, J.A.; Jiménez-Bermúdez, S.; Muñoz-Serrano, A. Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiol. 2009, 150, 1022–1032. [Google Scholar] [CrossRef] [Green Version]
- Posé, S.; Paniagua, C.; Cifuentes, M.; Blanco-Portales, R.; Quesada, M.A.; Mercado, J.A. Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 2013, 64, 3803–3815. [Google Scholar] [CrossRef]
- Wang, Z.Y.; MacRae, E.A.; Wright, M.A.; Bolitho, K.M.; Ross, G.S.; Atkinson, R.G. Polygalacturonase gene expression in kiwifruit: Relationship to fruit softening and ethylene production. Plant Mol. Biol. 2000, 42, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Mworia, E.G.; Yoshikawa, T.; Salikon, N.; Oda, C.; Asiche, W.O.; Yokotani, N. Low-temperature-modulated fruit ripening is independent of ethylene in ‘Sanuki Gold’ kiwifruit. J. Exp. Bot. 2012, 63, 963–971. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.H.; Qu, Y.; Li, T.; Yuan, H.; Wang, A.D.; Tan, D.M. Comparative transcriptome analysis of Actinidia arguta fruits reveals the involvement of various transcription factors in ripening. Hortic. Plant J. 2018, 4, 35–42. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Y.; Shen, X.; Dong, H.; Lyu, M.; Xu, L. Dissecting the complex molecular evolution and expression of polygalacturonase gene family in Brassica rapa ssp. chinensis. Plant Mol. Biol. 2015, 89, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Wang, H.; Li, Y.; Zhu, B.; Zang, Y.; He, Y. Genome-wide identification and analysis of polygalacturonase genes in Solanum lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dautt-Castro, M.; López-Virgen, A.G.; Ochoa-Leyva, A.; Contreras-Vergara, C.A.; Sortillón-Sortillón, A.P.; Martínez-Téllez, M.A. Genome-wide identification of Mango (Mangifera indica L.) polygalacturonases: Expression analysis of family members and total enzyme activity during fruit ripening. Front. Plant Sci. 2019, 10, 969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, S.; Niu, Q.F.; Qian, M.J.; Bai, S.L.; Teng, Y.W. Genome-wide identification, characterization, and expression analysis of the dehydrin gene family in Asian pear (Pyrus pyrifolia). Tree Genet. Genomes 2015, 11, 110. [Google Scholar] [CrossRef]
- Khan, N.; Fatima, F.; Haider, M.S.; Shazadee, H.; Liu, Z.; Zheng, T. Genome-wide identification and expression profiling of the polygalacturonase (PG) and pectin methylesterase (PME) genes in grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 3180. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Shao, H.; Sheng, F.; Ma, J.; Dong, Z.; Han, M. Identification and phylogenetic analysis of the polygalacturonase gene family in apple. Hortic. Plant J. 2016, 2, 241–252. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Gardner, R.C. A polygalacturonase gene from kiwifruit (Actinidia deliciosa). Plant Physiol. 1993, 103, 669–670. [Google Scholar] [CrossRef] [PubMed]
- Tavarini, S.; Degl’Innocenti, E.; Remorini, D.; Massai, R.; Guidi, L. Polygalacturonase and β-galactosidase activities in hayward kiwifruit as affected by light exposure, maturity stage and storage time. Sci. Hortic. 2009, 120, 342–347. [Google Scholar] [CrossRef]
- Minas, I.S.; Vicente, A.R.; Dhanapal, A.P.; Manganaris, G.A.; Goulas, V.; Vasilakakis, M. Ozone-induced kiwifruit ripening delay is mediated by ethylene biosynthesis inhibition and cell wall dismantling regulation. Plant Sci. 2014, 229, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Prakash, R.; Hallett, I.C.; Wong, S.F.; Johnston, S.L.; O’Donoghue, E.M.; McAtee, P.A. Cell separation in kiwifruit without development of a specialised detachment zone. BMC Plant Biol. 2017, 17, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitalo, O.W.; Tokiwa, S.; Kondo, Y.; Otsuki, T.; Galis, I.; Suezawa, K. Low temperature storage stimulates fruit softening and sugar accumulation without ethylene and aroma volatile production in kiwifruit. Front. Plant Sci. 2019, 10, 888. [Google Scholar] [CrossRef]
- Huang, W.J.; Chen, M.Y.; Zhao, T.T.; Han, F.; Zhang, Q.; Liu, X.L.; Jiang, C.Y.; Zhong, C.H. Genome-wide identification and expression analysis of polygalacturonase gene family in kiwifruit (Actinidia chinensis) during fruit softening. Plants 2020, 9, 327. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.T.; Lai, R.L.; Cheng, C.Z.; Feng, X.; Qu, M.M.; Liu, P.Y.; Chen, W.G.; Wu, R.J. Cloning and expression of polygalacturonase gene (AcPG) during fruit softening of kiwifruit (Actinidia chinensis). J. Agric. Biotechnol. 2017, 25, 205–213. (In Chinese) [Google Scholar] [CrossRef]
- Tisza, V.; Kovács, L.; Balogh, A.; Heszky, L.; Kiss, E. Characterization of FaSPT, a SPATULA gene encoding a bHLH transcriptional factor from the non-climacteric strawberry fruit. Plant Physiol. Biochem. 2010, 48, 822–826. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Merchante, C.; Sánchez-Sevilla, J.F.; de Luis Balaguer, M.A.; Pott, D.M. Characterizing the involvement of FaMADS9 in the regulation of strawberry fruit receptacle development. Plant Biotechnol. J. 2019, 18, 929–943. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Liu, C.; Han, L.; Wang, S.; Xue, Z. NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genom. 2016, 291, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, Y.; Zhao, L.; Li, C.; Yu, J.; Li, T. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020, 40, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.D.; Wang, W.Q.; Tong, Y.; Li, M.J.; Grierson, D.; Ferguson, I. Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 2018, 178, 850–863. [Google Scholar] [CrossRef] [Green Version]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.Q.; Wang, J.; Wu, Y.Y.; Li, D.W.; Allan, A.C.; Yin, X.R. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020, 225, 1618–1634. [Google Scholar] [CrossRef]
- Fu, B.L.; Wang, W.Q.; Liu, X.F.; Duan, X.W.; Allan, A.C.; Grierson, D. An ethylene-hypersensitive methionine sulfoxide reductase regulated by NAC transcription factors increases methionine pool size and ethylene production during kiwifruit ripening. New Phytol. 2021, 232, 237–251. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G. Method of determination of respiration strength for vegetables and fruits after reaping. Phys. Test. Chem. Anal. Part B Chem. Analgsis 2005, 8, 596–597. (In Chinese) [Google Scholar]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, J.T.; Kong, Y.Z.; Wang, Q.; Sun, Y.H.; Gong, D.P.; Lv, J. Mapgene2chrom, a tool to draw gene physical map based on perl and svg languages. Hereditas 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ampomah-Dwamena, C.; McGhie, T.; Reginald, W.; Montefiori, M.; Hellens, R.P.; Allan, A.C. The kiwifruit lycopene beta-cyclase plays a significant role in carotenoid accumulation in fruit. J. Exp. Bot. 2009, 60, 3765–3779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; Preter, D.P.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Huang, H.; Abid, M.; Gu, H.; Fang, J.; Cheng, Z.; Qi, X. Characterization and Identification of a Ripening-Related Gene AaPG18 in Actinidia arguta. Int. J. Mol. Sci. 2022, 23, 2597. https://doi.org/10.3390/ijms23052597
Li Y, Huang H, Abid M, Gu H, Fang J, Cheng Z, Qi X. Characterization and Identification of a Ripening-Related Gene AaPG18 in Actinidia arguta. International Journal of Molecular Sciences. 2022; 23(5):2597. https://doi.org/10.3390/ijms23052597
Chicago/Turabian StyleLi, Yukuo, Hailei Huang, Muhammad Abid, Hong Gu, Jinbao Fang, Zhongping Cheng, and Xiujuan Qi. 2022. "Characterization and Identification of a Ripening-Related Gene AaPG18 in Actinidia arguta" International Journal of Molecular Sciences 23, no. 5: 2597. https://doi.org/10.3390/ijms23052597
APA StyleLi, Y., Huang, H., Abid, M., Gu, H., Fang, J., Cheng, Z., & Qi, X. (2022). Characterization and Identification of a Ripening-Related Gene AaPG18 in Actinidia arguta. International Journal of Molecular Sciences, 23(5), 2597. https://doi.org/10.3390/ijms23052597





