Protein Expression of AEBP1, MCM4, and FABP4 Differentiate Osteogenic, Adipogenic, and Mesenchymal Stromal Stem Cells
Abstract
:1. Introduction
2. Results
2.1. Cultured Cell Populations Fulfilled Phenotypic Criteria of MSC, Adipocytic and Osteoblastic Criteria
2.2. Combined Quantitative Mass Spectrometry and Neural-Network-Based Algorithms Revealed Distinct Protein Expression Patterns of MSCs, Adipocytic and Osteoblastic Cells
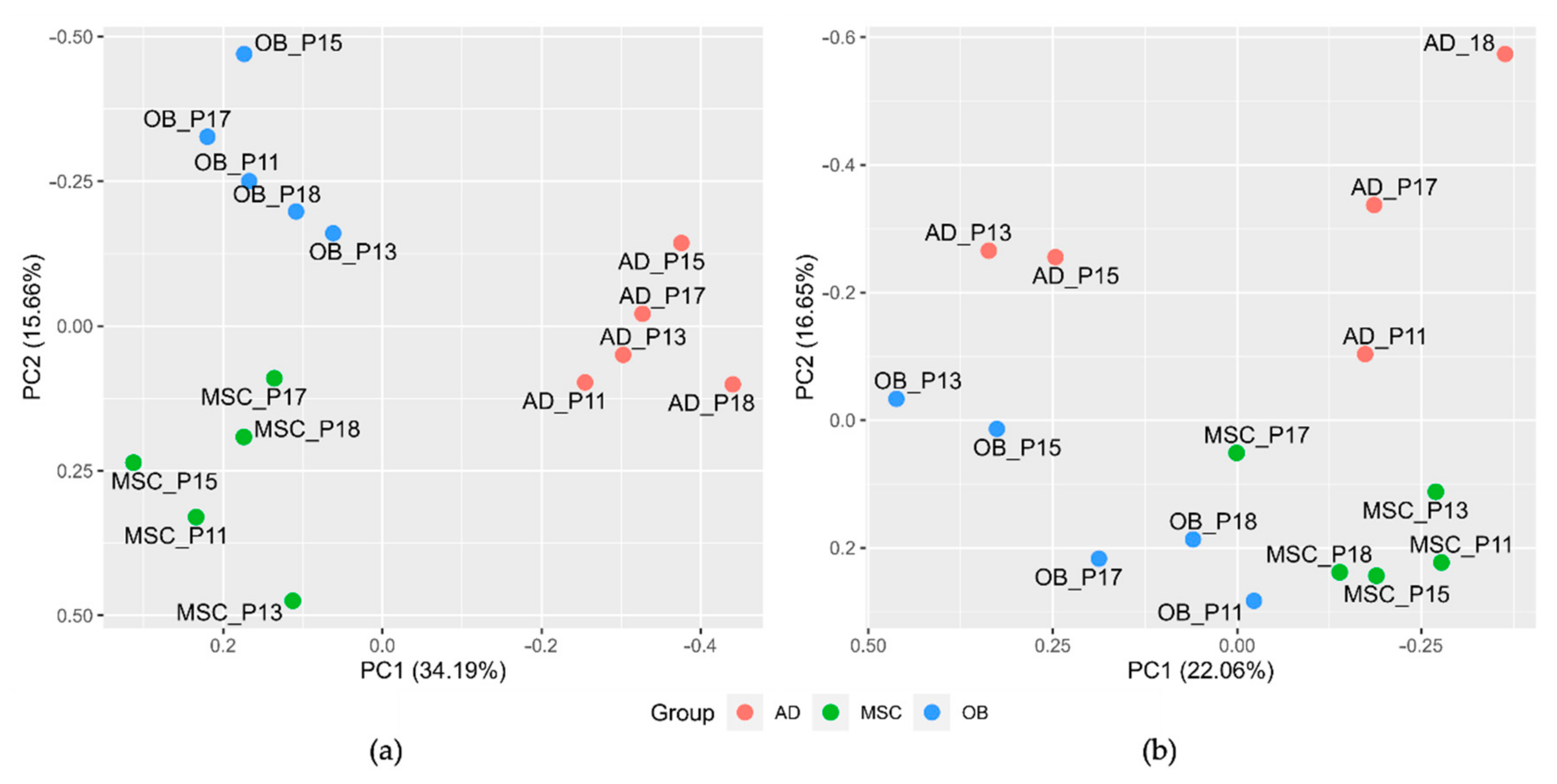
2.3. MSCs, Osteoblasts, and Adipocytes Show Distinct Clustering Behavior and Involvement of PPAR
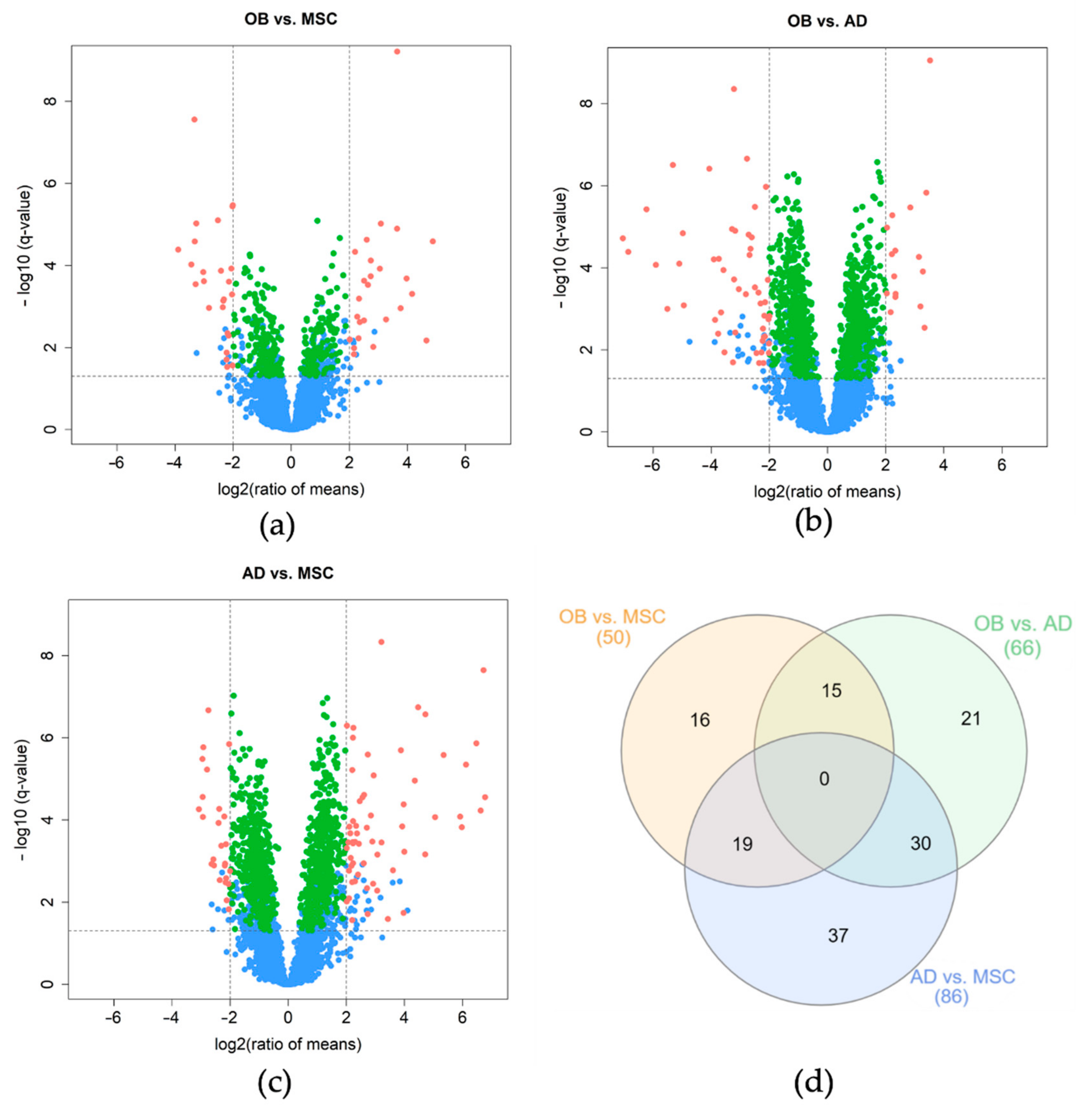
2.4. Discovery of Differentially Expressed Proteins
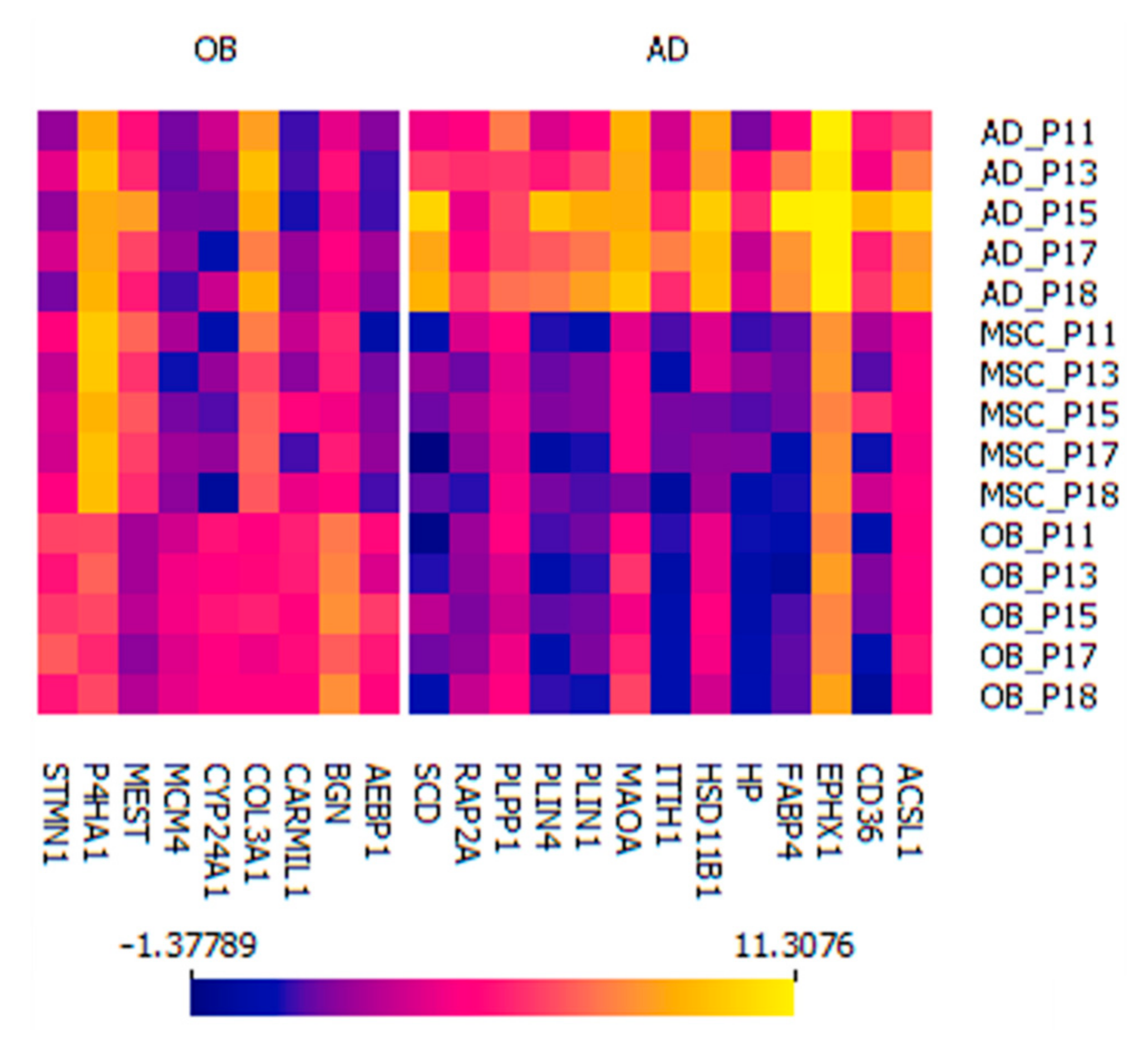
2.5. Two Distinct Protein Panels Differentiate Osteoblasts and Adipocytes from Mesenchymal Stem Cells
2.5.1. Osteoblastic Panel
2.5.2. Adipocytic Panel
2.6. MSC Marker Protein Panel Comparison
3. Discussion
3.1. Osteoblastic Panel
3.2. Adipocytic Panel
4. Materials and Methods
4.1. Donors and Materials
4.2. Cell Isolation and Culture
4.3. Colony-Forming Unit-Fibroblast (CFU-f) Assay
4.4. Cell Proliferation
4.5. In Vitro Cell Differentiation
4.5.1. Osteoblastic Differentiation
Alkaline Phosphatase (ALP) Activity
4.5.2. Adipocytic Differentiation
4.6. Flow Cytometry
4.7. Cell Isolation for Mass Spectrometry Analysis
4.8. Two-Dimensional Fluorescence Gel Electrophoresis
4.9. Sample Preparation for High-Performance Liquid Chromatography (HPLC) and Electrospray Ionization Tandem Mass Spectrometry (ESI-MS/MS)
4.10. High-Performance Liquid Chromatography (HPLC) and Electrospray Ionization Tandem Mass Spectrometry (ESI-MS/MS)
4.11. SWATH Data Processing
4.12. Quantitative Data Processing
4.13. Data Analysis
4.14. Reagents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedenstein, A.Y. Induction of bone tissue by transitional epithelium. Clin. Orthop. Relat. Res. 1968, 59, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.; Liu, D.D.; Thakor, A.S. Mesenchymal Stromal Cell Homing: Mechanisms and Strategies for Improvement. iScience 2019, 15, 421–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, N.G.; Caplan, A.I. Mesenchymal stem cells: Mechanisms of inflammation. Annu. Rev. Pathol. 2011, 6, 457–478. [Google Scholar] [CrossRef] [Green Version]
- Le Blanc, K.; Mougiakakos, D. Multipotent mesenchymal stromal cells and the innate immune system. Nat. Rev. Immunol. 2012, 12, 383–396. [Google Scholar] [CrossRef]
- Bronckaers, A.; Hilkens, P.; Martens, W.; Gervois, P.; Ratajczak, J.; Struys, T.; Lambrichts, I. Mesenchymal stem/stromal cells as a pharmacological and therapeutic approach to accelerate angiogenesis. Pharmacol. Ther. 2014, 143, 181–196. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. Enhancement of Functionality and Therapeutic Efficacy of Cell-Based Therapy Using Mesenchymal Stem Cells for Cardiovascular Disease. Int. J. Mol. Sci. 2019, 20, 982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Shapiro, L.; Flynn, A. The clinical application of mesenchymal stem cells and cardiac stem cells as a therapy for cardiovascular disease. Pharmacol. Ther. 2015, 151, 8–15. [Google Scholar] [CrossRef]
- Chung, M.-J.; Son, J.-Y.; Park, S.; Park, S.-S.; Hur, K.; Lee, S.-H.; Lee, E.-J.; Park, J.-K.; Hong, I.-H.; Kim, T.-H.; et al. Mesenchymal Stem Cell and MicroRNA Therapy of Musculoskeletal Diseases. Int. J. Stem. Cells 2021, 14, 150–167. [Google Scholar] [CrossRef]
- Kim, M.; Kim, K.H.; Song, S.U.; Yi, T.G.; Yoon, S.H.; Park, S.R.; Choi, B.H. Transplantation of human bone marrow-derived clonal mesenchymal stem cells reduces fibrotic scar formation in a rat spinal cord injury model. J. Tissue Eng. Regen. Med. 2018, 12, e1034–e1045. [Google Scholar] [CrossRef]
- Wei, X.; Yang, X.; Han, Z.P.; Qu, F.F.; Shao, L.; Shi, Y.F. Mesenchymal stem cells: A new trend for cell therapy. Acta Pharm. Sin. 2013, 34, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Yong, K.W.; Choi, J.R.; Wan Safwani, W.K. Biobanking of Human Mesenchymal Stem Cells: Future Strategy to Facilitate Clinical Applications. Adv. Exp. Med. Biol. 2016, 951, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Hong, I.-S. Double-edged sword of mesenchymal stem cells: Cancer-promoting versus therapeutic potential. Cancer Sci. 2017, 108, 1939–1946. [Google Scholar] [CrossRef]
- Knight, M.N.; Hankenson, K.D. Mesenchymal Stem Cells in Bone Regeneration. Adv. Wound Care New Rochelle 2013, 2, 306–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcacci, M.; Kon, E.; Moukhachev, V.; Lavroukov, A.; Kutepov, S.; Quarto, R.; Mastrogiacomo, M.; Cancedda, R. Stem cells associated with macroporous bioceramics for long bone repair: 6- to 7-year outcome of a pilot clinical study. Tissue Eng. 2007, 13, 947–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannotti, S.; Trombi, L.; Bottai, V.; Ghilardi, M.; D’Alessandro, D.; Danti, S.; Dell’Osso, G.; Guido, G.; Petrini, M. Use ofautologous human mesenchymal stromal cell/fibrin clot constructs in upper limb non-unions: Long-term assessment. PLoS ONE 2013, 8, e73893. [Google Scholar] [CrossRef]
- Quarto, R.; Mastrogiacomo, M.; Cancedda, R.; Kutepov, S.M.; Mukhachev, V.; Lavroukov, A.; Kon, E.; Marcacci, M. Repair of large bone defects with the use of autologous bone marrow stromal cells. N. Engl. J. Med. 2001, 344, 385–386. [Google Scholar] [CrossRef]
- Liebergall, M.; Schroeder, J.; Mosheiff, R.; Gazit, Z.; Yoram, Z.; Rasooly, L.; Daskal, A.; Khoury, A.; Weil, Y.; Beyth, S. Stem cell-based therapy for prevention of delayed fracture union: A randomized and prospective preliminary study. Mol. Ther. 2013, 21, 1631–1638. [Google Scholar] [CrossRef] [Green Version]
- Oryan, A.; Kamali, A.; Moshiri, A.; Baghaban Eslaminejad, M. Role of Mesenchymal Stem Cells in Bone Regenerative Medicine: What Is the Evidence? Cells Tissues Organs 2017, 204, 59–83. [Google Scholar] [CrossRef]
- Shi, L.; Huang, H.; Lu, X.; Yan, X.; Jiang, X.; Xu, R.; Wang, S.; Zhang, C.; Yuan, X.; Xu, Z.; et al. Effect of human umbilical cord-derived mesenchymal stem cells on lung damage in severe COVID-19 patients: A randomized, double-blind, placebo-controlled phase 2 trial. Signal. Transduct Target. Ther. 2021, 6, 58. [Google Scholar] [CrossRef]
- Ciuffreda, M.C.; Malpasso, G.; Musarò, P.; Turco, V.; Gnecchi, M. Protocols for in vitro Differentiation of Human Mesenchymal Stem Cells into Osteogenic, Chondrogenic and Adipogenic Lineages. Methods Mol. Biol. 2016, 1416, 149–158. [Google Scholar] [CrossRef]
- O’Keefe, R.J.; Tuan, R.S.; Lane, N.E.; Awad, H.A.; Barry, F.; Bunnell, B.A.; Colnot, C.; Drake, M.T.; Drissi, H.; Dyment, N.A.; et al. American Society for Bone and Mineral Research-Orthopaedic Research Society Joint Task Force Report on Cell-Based Therapies. J. Bone Miner. Res. 2020, 35, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Barrena, E.; Rosset, P.; Müller, I.; Giordano, R.; Bunu, C.; Layrolle, P.; Konttinen, Y.T.; Luyten, F.P. Bone regeneration: Stem cell therapies and clinical studies in orthopaedics and traumatology. J. Cell Mol. Med. 2011, 15, 1266–1286. [Google Scholar] [CrossRef] [Green Version]
- Fakhry, M.; Hamade, E.; Badran, B.; Buchet, R.; Magne, D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J. Stem Cells 2013, 5, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Zaher, W.; Harkness, L.; Jafari, A.; Kassem, M. An update of human mesenchymal stem cell biology and their clinical uses. Arch. Toxicol. 2014, 88, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Centeno, C.J.; Al-Sayegh, H.; Freeman, M.D.; Smith, J.; Murrell, W.D.; Bubnov, R. A multi-center analysis of adverse events among two thousand, three hundred and seventy two adult patients undergoing adult autologous stem cell therapy for orthopaedic conditions. Int. Orthop. 2016, 40, 1755–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Bianco, P.; Robey, P.G.; Simmons, P.J. Mesenchymal stem cells: Revisiting history, concepts, and assays. Cell Stem Cell 2008, 2, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Post, S.; Abdallah, B.M.; Bentzon, J.F.; Kassem, M. Demonstration of the presence of independent pre-osteoblastic and pre-adipocytic cell populations in bone marrow-derived mesenchymal stem cells. Bone 2008, 43, 32–39. [Google Scholar] [CrossRef]
- Larsen, K.H.; Frederiksen, C.M.; Burns, J.S.; Abdallah, B.M.; Kassem, M. Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J. Bone Miner. Res. 2010, 25, 796–808. [Google Scholar] [CrossRef]
- McLeod, C.M.; Mauck, R.L. On the origin and impact of mesenchymal stem cell heterogeneity: New insights and emerging tools for single cell analysis. Eur. Cell Mater. 2017, 34, 217–231. [Google Scholar] [CrossRef]
- Elsafadi, M.; Manikandan, M.; Atteya, M.; Hashmi, J.A.; Iqbal, Z.; Aldahmash, A.; Alfayez, M.; Kassem, M.; Mahmood, A. Characterization of Cellular and Molecular Heterogeneity of Bone Marrow Stromal Cells. Stem Cells Int. 2016, 2016, 9378081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowal, J.M.; Schmal, H.; Halekoh, U.; Hjelmborg, J.B.; Kassem, M. Single-cell high-content imaging parameters predict functional phenotype of cultured human bone marrow stromal stem cells. Stem Cells Transl. Med. 2020, 9, 189–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keating, A. Mesenchymal stromal cells. Curr. Opin. Hematol. 2006, 13, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.J. Cryopreservation of Human Stem Cells for Clinical Application: A Review. Transfus. Med. Hemother. 2011, 38, 107–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Chen, J.; Gantz, M.; Punyanitya, M.; Heymsfield, S.B.; Gallagher, D.; Albu, J.; Engelson, E.; Kotler, D.; Pi-Sunyer, X.; et al. MRI-measured pelvic bone marrow adipose tissue is inversely related to DXA-measured bone mineral in younger and older adults. Eur. J. Clin. Nutr. 2012, 66, 983–988. [Google Scholar] [CrossRef] [Green Version]
- Li, C.J.; Cheng, P.; Liang, M.K.; Chen, Y.S.; Lu, Q.; Wang, J.Y.; Xia, Z.Y.; Zhou, H.D.; Cao, X.; Xie, H.; et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Investig. 2015, 125, 1509–1522. [Google Scholar] [CrossRef] [Green Version]
- Liao, L.; Yang, X.; Su, X.; Hu, C.; Zhu, X.; Yang, N.; Chen, X.; Shi, S.; Shi, S.; Jin, Y. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis. 2013, 4, e600. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.M.; Möller, S.; Ali, D.; Figeac, F.; Barington, T.; Schmal, H.; Kassem, M. Identification of a clinical signature predictive of differentiation fate of human bone marrow stromal cells. Stem Cell Res. 2021, 12, 265. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Liao, Y.-J.; Tang, P.-C.; Chen, Y.-H.; Chu, F.-H.; Kang, T.-C.; Chen, L.-R.; Yang, J.-R. Porcine induced pluripotent stem cell-derived osteoblast-like cells prevent glucocorticoid-induced bone loss in Lanyu pigs. PLoS ONE 2018, 13, e0202155. [Google Scholar] [CrossRef]
- Yamada, M.; Watanabe, J.; Ueno, T.; Ogawa, T.; Egusa, H. Cytoprotective Preconditioning of Osteoblast-Like Cells with N-Acetyl-L-Cysteine for Bone Regeneration in Cell Therapy. Int. J. Mol. Sci. 2019, 20, 5199. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, L.P.; Chen, L.; Nielsen, M.O.; Qanie, D.W.; Kratchmarova, I.; Kassem, M.; Andersen, J.S. Temporal profiling and pulsed SILAC labeling identify novel secreted proteins during ex vivo osteoblast differentiation of human stromal stem cells. Mol. Cell Proteom. 2012, 11, 989–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, L.J.; Zeemann, P.A.; Li, C.; Mann, M.; Jensen, O.N.; Kassem, M. Differential expression profiling of membrane proteins by quantitative proteomics in a human mesenchymal stem cell line undergoing osteoblast differentiation. Stem Cells 2005, 23, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Granéli, C.; Thorfve, A.; Ruetschi, U.; Brisby, H.; Thomsen, P.; Lindahl, A.; Karlsson, C. Novel markers of osteogenic and adipogenic differentiation of human bone marrow stromal cells identified using a quantitative proteomics approach. Stem Cell Res. 2014, 12, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aasebø, E.; Brenner, A.K.; Hernandez-Valladares, M.; Birkeland, E.; Berven, F.S.; Selheim, F.; Bruserud, Ø. Proteomic Comparison of Bone Marrow Derived Osteoblasts and Mesenchymal Stem Cells. Int. J. Mol. Sci 2021, 22, 5665. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Q.; Luo, S.; Liu, Z.; Luo, D.; Zhang, B.; Zhang, D.; Rao, P.; Xiao, J. PPARγ and Wnt Signaling in Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells. Curr Stem Cell Res. 2016, 11, 216–225. [Google Scholar] [CrossRef]
- Parisuthiman, D.; Mochida, Y.; Duarte, W.R.; Yamauchi, M. Biglycan modulates osteoblast differentiation and matrix mineralization. J. Bone Miner. Res. 2005, 20, 1878–1886. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-D.; Fisher, L.W.; Robey, P.G.; Young, M.F. The small leucine-rich proteoglycan biglycan modulates BMP-4-induced osteoblast differentiation. FASEB J. 2004, 18, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Hu, W.; Guo, F.; Zhang, W.; Wang, J.; Chen, A. Glycosaminoglycan chains of biglycan promote bone morphogenetic protein-4-induced osteoblast differentiation. Int. J. Mol. Med. 2012, 30, 1075–1080. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Pennisi, A.; Yaccoby, S. Role of decorin in the antimyeloma effects of osteoblasts. Blood 2008, 112, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Pathak, J.L.; Zhu, Y.-Q.; Bureik, M. Comparison of cytochrome P450 expression in four different human osteoblast models. Biol. Chem. 2017, 398, 1327–1334. [Google Scholar] [CrossRef]
- van Driel, M.; Koedam, M.; Buurman, C.J.; Hewison, M.; Chiba, H.; Uitterlinden, A.G.; Pols, H.A.; van Leeuwen, J.P. Evidence for auto/paracrine actions of vitamin D in bone: 1alpha-hydroxylase expression and activity in human bone cells. FASEB J. 2006, 20, 2417–2419. [Google Scholar] [CrossRef] [PubMed]
- Moena, D.; Merino, P.; Lian, J.B.; Stein, G.S.; Stein, J.L.; Montecino, M. Switches in histone modifications epigenetically control vitamin D3-dependent transcriptional upregulation of the CYP24A1 gene in osteoblastic cells. J. Cell Physiol. 2020, 235, 5328–5339. [Google Scholar] [CrossRef]
- Segal, E.; Friedman, N.; Koller, D.; Regev, A. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 2004, 36, 1090–1098. [Google Scholar] [CrossRef]
- Volk, S.W.; Shah, S.R.; Cohen, A.J.; Wang, Y.; Brisson, B.K.; Vogel, L.K.; Hankenson, K.D.; Adams, S.L. Type III collagen regulates osteoblastogenesis and the quantity of trabecular bone. Calcif. Tissue Int. 2014, 94, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Haugen, J.D. Human and rat osteoblast-like cells express stathmin, a growth-regulatory protein. Biochem. Biophys. Res. Commun. 1994, 201, 861–865. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, R.; Ko, S.-Y.; Oyajobi, B.O.; Papasian, C.J.; Deng, H.-W.; Zhang, S.; Zhao, M. Microtubule assembly affects bone mass by regulating both osteoblast and osteoclast functions: Stathmin deficiency produces an osteopenic phenotype in mice. J. Bone Mineral. Res. 2011, 26, 2052–2067. [Google Scholar] [CrossRef] [PubMed]
- Kuijjer, M.L.; Rydbeck, H.; Kresse, S.H.; Buddingh, E.P.; Lid, A.B.; Roelofs, H.; Bürger, H.; Myklebost, O.; Hogendoorn, P.C.W.; Meza-Zepeda, L.A.; et al. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer 2012, 51, 696–706. [Google Scholar] [CrossRef]
- Yang, C.; Pring, M.; Wear, M.A.; Huang, M.; Cooper, J.A.; Svitkina, T.M.; Zigmond, S.H. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 2005, 9, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Myllyharju, J.; Kivirikko, K.I. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004, 20, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shen, Z.; Wu, Z.; Shen, Y.; Deng, H.; Zhou, C.; Liu, H. High P4HA1 expression is an independent prognostic factor for poor overall survival and recurrent-free survival in head and neck squamous cell carcinoma. J. Clin. Lab. Anal. 2020, 34, e23107. [Google Scholar] [CrossRef]
- Eriksson, J.; Le Joncour, V.; Jahkola, T.; Juteau, S.; Laakkonen, P.; Saksela, O.; Hölttä, E. Prolyl 4-hydroxylase subunit alpha 1 (P4HA1) is a biomarker of poor prognosis in primary melanomas, and its depletion inhibits melanoma cell invasion and disrupts tumor blood vessel walls. Mol. Oncol. 2020, 14, 742–762. [Google Scholar] [CrossRef]
- Rappaport, N.; Nativ, N.; Stelzer, G.; Twik, M.; Guan-Golan, Y.; Stein, T.I.; Bahir, I.; Belinky, F.; Morrey, C.P.; Safran, M.; et al. MalaCards: An integrated compendium for diseases and their annotation. Database 2013, 2013, bat018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, V.; Lee, B.; Marom, R. Osteogenesis imperfecta: Advancements in genetics and treatment. Curr. Opin. Pediatr. 2019, 31, 708–715. [Google Scholar] [CrossRef]
- Chen, L.; Shi, K.; Frary, C.E.; Ditzel, N.; Hu, H.; Qiu, W.; Kassem, M. Inhibiting actin depolymerization enhances osteoblast differentiation and bone formation in human stromal stem cells. Stem Cell Res. 2015, 15, 281–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prudovsky, I.; Anunciado-Koza, R.P.; Jacobs, C.G.; Kacer, D.; Siviski, M.E.; Koza, R.A. Mesoderm-specific transcript localization in the ER and ER-lipid droplet interface supports a role in adipocyte hypertrophy. J. Cell Biochem. 2018, 119, 2636–2645. [Google Scholar] [CrossRef]
- Kadota, Y.; Yanagawa, M.; Nakaya, T.; Kawakami, T.; Sato, M.; Suzuki, S. Gene expression of mesoderm-specific transcript is upregulated as preadipocytes differentiate to adipocytes in vitro. J. Physiol. Sci. 2012, 62, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Liu, W.; Kuang, S. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB J. 2013, 27, 277–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hng, C.H.; Camp, E.; Anderson, P.; Breen, J.; Zannettino, A.; Gronthos, S. HOPX regulates bone marrow-derived mesenchymal stromal cell fate determination via suppression of adipogenic gene pathways. Sci. Rep. 2020, 10, 11345. [Google Scholar] [CrossRef] [PubMed]
- Gamucci, O.; Lisi, S.; Scabia, G.; Marchi, M.; Piaggi, P.; Duranti, E.; Virdis, A.; Pinchera, A.; Santini, F.; Maffei, M. Haptoglobin deficiency determines changes in adipocyte size and adipogenesis. Adipocyte 2012, 1, 142–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christiaens, V.; Van Hul, M.; Lijnen, H.R.; Scroyen, I. CD36 promotes adipocyte differentiation and adipogenesis. Biochim. Biophys. Acta 2012, 1820, 949–956. [Google Scholar] [CrossRef]
- Do Nascimento, F.V.; Piccoli, V.; Beer, M.A.; von Frankenberg, A.D.; Crispim, D.; Gerchman, F. Association of HSD11B1 polymorphic variants and adipose tissue gene expression with metabolic syndrome, obesity and type 2 diabetes mellitus: A systematic review. Diabetol. Metab. Syndr. 2015, 7, 38. [Google Scholar] [CrossRef] [Green Version]
- Pizzinat, N.; Marti, L.; Remaury, A.; Leger, F.; Langin, D.; Lafontan, M.; Carpéné, C.; Parini, A. High expression of monoamine oxidases in human white adipose tissue: Evidence for their involvement in noradrenaline clearance. Biochem. Pharm. 1999, 58, 1735–1742. [Google Scholar] [CrossRef]
- Rodriguez-Cuenca, S.; Whyte, L.; Hagen, R.; Vidal-Puig, A.; Fuller, M. Stearoyl-CoA Desaturase 1 Is a Key Determinant of Membrane Lipid Composition in 3T3-L1 Adipocytes. PLoS ONE 2016, 11, e0162047. [Google Scholar] [CrossRef]
- Ellis, J.M.; Li, L.O.; Wu, P.-C.; Koves, T.R.; Ilkayeva, O.; Stevens, R.D.; Watkins, S.M.; Muoio, D.M.; Coleman, R.A. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 2010, 12, 53–64. [Google Scholar] [CrossRef] [Green Version]
- Himmelfarb, M.; Klopocki, E.; Grube, S.; Staub, E.; Klaman, I.; Hinzmann, B.; Kristiansen, G.; Rosenthal, A.; Dürst, M.; Dahl, E. ITIH5, a novel member of the inter-alpha-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 2004, 204, 69–77. [Google Scholar] [CrossRef]
- Taira, K.; Umikawa, M.; Takei, K.; Myagmar, B.E.; Shinzato, M.; Machida, N.; Uezato, H.; Nonaka, S.; Kariya, K. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J. Biol. Chem. 2004, 279, 49488–49496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decker, M.; Adamska, M.; Cronin, A.; Di Giallonardo, F.; Burgener, J.; Marowsky, A.; Falck, J.R.; Morisseau, C.; Hammock, B.D.; Gruzdev, A.; et al. EH3 (ABHD9): The first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J. Lipid Res. 2012, 53, 2038–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautheron, J.; Jéru, I. The Multifaceted Role of Epoxide Hydrolases in Human Health and Disease. Int. J. Mol. Sci. 2020, 22, 13. [Google Scholar] [CrossRef] [PubMed]
- Smyth, S.S.; Sciorra, V.A.; Sigal, Y.J.; Pamuklar, Z.; Wang, Z.; Xu, Y.; Prestwich, G.D.; Morris, A.J. Lipid phosphate phosphatases regulate lysophosphatidic acid production and signaling in platelets: Studies using chemical inhibitors of lipid phosphate phosphatase activity. J. Biol. Chem. 2003, 278, 43214–43223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Usatyuk, P.V.; Cummings, R.; Saatian, B.; He, D.; Watkins, T.; Morris, A.; Spannhake, E.W.; Brindley, D.N.; Natarajan, V. Lipid phosphate phosphatase-1 regulates lysophosphatidic acid-induced calcium release, NF-kappaB activation and interleukin-8 secretion in human bronchial epithelial cells. Biochem. J. 2005, 385, 493–502. [Google Scholar] [CrossRef]
- Stenderup, K.; Justesen, J.; Clausen, C.; Kassem, M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 2003, 33, 919–926. [Google Scholar] [CrossRef]
- Gemoll, T.; Epping, F.; Heinrich, L.; Fritzsche, B.; Roblick, U.J.; Szymczak, S.; Hartwig, S.; Depping, R.; Bruch, H.-P.; Thorns, C.; et al. Increased cathepsin D protein expression is a biomarker for osteosarcomas, pulmonary metastases and other bone malignancies. Oncotarget 2015, 6, 16517–16526. [Google Scholar] [CrossRef] [Green Version]
- Manza, L.L.; Stamer, S.L.; Ham, A.-J.L.; Codreanu, S.G.; Liebler, D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 2005, 5, 1742–1745. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Ovaa, H.; Huber, W.; Vermeulen, M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018, 13, 530–550. [Google Scholar] [CrossRef]
- Huber, W.; von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef]
- Lazar, C.; Gatto, L.; Ferro, M.; Bruley, C.; Burger, T. Accounting for the Multiple Natures of Missing Values in Label-Free Quantitative Proteomics Data Sets to Compare Imputation Strategies. J. Proteome Res. 2016, 15, 1116–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhmedov, M.; Martinelli, A.; Geiger, R.; Kwee, I. Omics Playground: A comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom. Bioinform. 2020, 2, lqz019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J. Stat. Softw. 2014, 61, 36. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021, 060012. [Google Scholar] [CrossRef] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2020, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]

| Gene set | Log2FC OB vs. AD | Meta-q | Avg. Expr. OB | Avg. Expr. AD |
|---|---|---|---|---|
| MYC_TARGETS_V1 | 0.575 | 0.0005 | 6.103 | 5.528 |
| E2F_TARGETS | 0.410 | 0.0005 | 7.049 | 6.638 |
| G2M_CHECKPOINT | 0.278 | 0.0005 | 5.411 | 5.133 |
| PEROXISOME | −0.223 | 0.0366 | 6.925 | 7.148 |
| BILE_ACID_METABOLISM | −0.317 | 0.0006 | 6.762 | 7.079 |
| FATTY_ACID_METABOLISM | −0.350 | 0.0006 | 6.535 | 6.885 |
| CHOLESTEROL_HOMEOSTASIS | −0.377 | 0.0315 | 6.318 | 6.695 |
| OXIDATIVE_PHOSPHORYLATION | −0.438 | 0.0006 | 5.028 | 5.466 |
| ADIPOGENESIS | −0.491 | 0.0006 | 5.782 | 6.273 |
| Gene set | Log2FC OB vs. MSC | Meta-q | Avg. Expr. OB | Avg. Expr. MSC |
| MYC_TARGETS_V2 | 0.279 | 0.0435 | 5.745 | 5.466 |
| E2F_TARGETS | 0.274 | 0.0008 | 7.034 | 6.76 |
| MYC_TARGETS_V1 | 0.241 | 0.0098 | 6.076 | 5.835 |
| Gene set | Log2FC AD vs. MSC | Meta-q | Avg. Expr. AD | Avg. Expr. MSC |
| OXIDATIVE_PHOSPHORYLATION | 0.659 | 0.0006 | 5.590 | 4.931 |
| CHOLESTEROL_HOMEOSTASIS | 0.533 | 0.0006 | 6.654 | 6.121 |
| ADIPOGENESIS | 0.529 | 0.0006 | 6.206 | 5.677 |
| FATTY_ACID_METABOLISM | 0.478 | 0.0006 | 6.909 | 6.431 |
| BILE_ACID_METABOLISM | 0.411 | 0.0006 | 7.096 | 6.685 |
| PEROXISOME | 0.337 | 0.0028 | 7.282 | 6.945 |
| XENOBIOTIC_METABOLISM | 0.271 | 0.0011 | 5.857 | 5.585 |
| PI3K_AKT_MTOR_SIGNALING | −0.203 | 0.0359 | 7.127 | 7.331 |
| MYC_TARGETS_V1 | −0.334 | 0.0105 | 5.605 | 5.939 |
| MITOTIC_SPINDLE | −0.335 | 0.0006 | 6.635 | 6.97 |
| Protein | Gene Name | CV | q-Value | Log2FC AD vs. MSC | Log2FC OB vs. MSC | Log2FC OB vs. AD |
|---|---|---|---|---|---|---|
| CD105+ | ENG | 0.104 | 0.25 | 0.121 | −0.884 | −0.763 |
| CD73+ | NT5E | 0.064 | 0.43 | 0.426 | 0.157 | 0.583 |
| CD90+ | THY1 | 0.121 | 0.41 | 0.355 | −1.046 | −0.691 |
| CD45- | PTPRC | Not Identified | ||||
| CD34- | CD34 | Not Identified | ||||
| CD14- | CD14 | Not Identified | ||||
| CD11b- | ITGAM | Not Identified | ||||
| CD79a- | CD79a | Not Identified | ||||
| CD19- | CD19 | Not Identified | ||||
| HLA-DR- | HLA-DR | Not Identified | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sauer, T.; Facchinetti, G.; Kohl, M.; Kowal, J.M.; Rozanova, S.; Horn, J.; Schmal, H.; Kwee, I.; Schulz, A.-P.; Hartwig, S.; et al. Protein Expression of AEBP1, MCM4, and FABP4 Differentiate Osteogenic, Adipogenic, and Mesenchymal Stromal Stem Cells. Int. J. Mol. Sci. 2022, 23, 2568. https://doi.org/10.3390/ijms23052568
Sauer T, Facchinetti G, Kohl M, Kowal JM, Rozanova S, Horn J, Schmal H, Kwee I, Schulz A-P, Hartwig S, et al. Protein Expression of AEBP1, MCM4, and FABP4 Differentiate Osteogenic, Adipogenic, and Mesenchymal Stromal Stem Cells. International Journal of Molecular Sciences. 2022; 23(5):2568. https://doi.org/10.3390/ijms23052568
Chicago/Turabian StyleSauer, Thorben, Giulia Facchinetti, Michael Kohl, Justyna M. Kowal, Svitlana Rozanova, Julia Horn, Hagen Schmal, Ivo Kwee, Arndt-Peter Schulz, Sonja Hartwig, and et al. 2022. "Protein Expression of AEBP1, MCM4, and FABP4 Differentiate Osteogenic, Adipogenic, and Mesenchymal Stromal Stem Cells" International Journal of Molecular Sciences 23, no. 5: 2568. https://doi.org/10.3390/ijms23052568
APA StyleSauer, T., Facchinetti, G., Kohl, M., Kowal, J. M., Rozanova, S., Horn, J., Schmal, H., Kwee, I., Schulz, A.-P., Hartwig, S., Kassem, M., Habermann, J. K., & Gemoll, T. (2022). Protein Expression of AEBP1, MCM4, and FABP4 Differentiate Osteogenic, Adipogenic, and Mesenchymal Stromal Stem Cells. International Journal of Molecular Sciences, 23(5), 2568. https://doi.org/10.3390/ijms23052568






