Transcriptomics of Parental Care in the Hypothalamic–Septal Region of Female Zebra Finch Brain
Abstract
1. Introduction
2. Results
2.1. RNA Sequencing
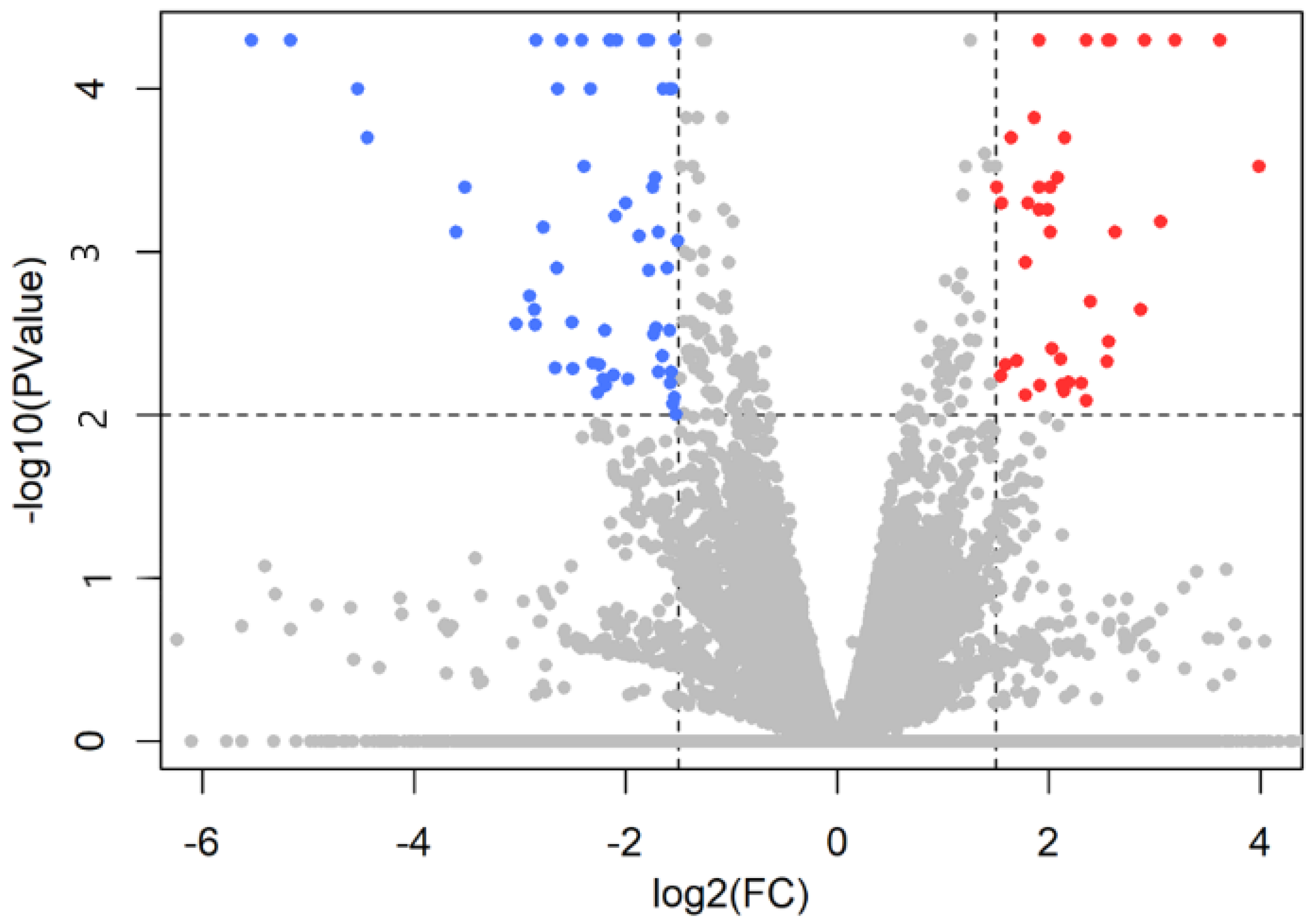
2.2. Differential Gene Expression Analysis
2.3. Validation
2.4. Parental Behaviour of Breeding Pairs
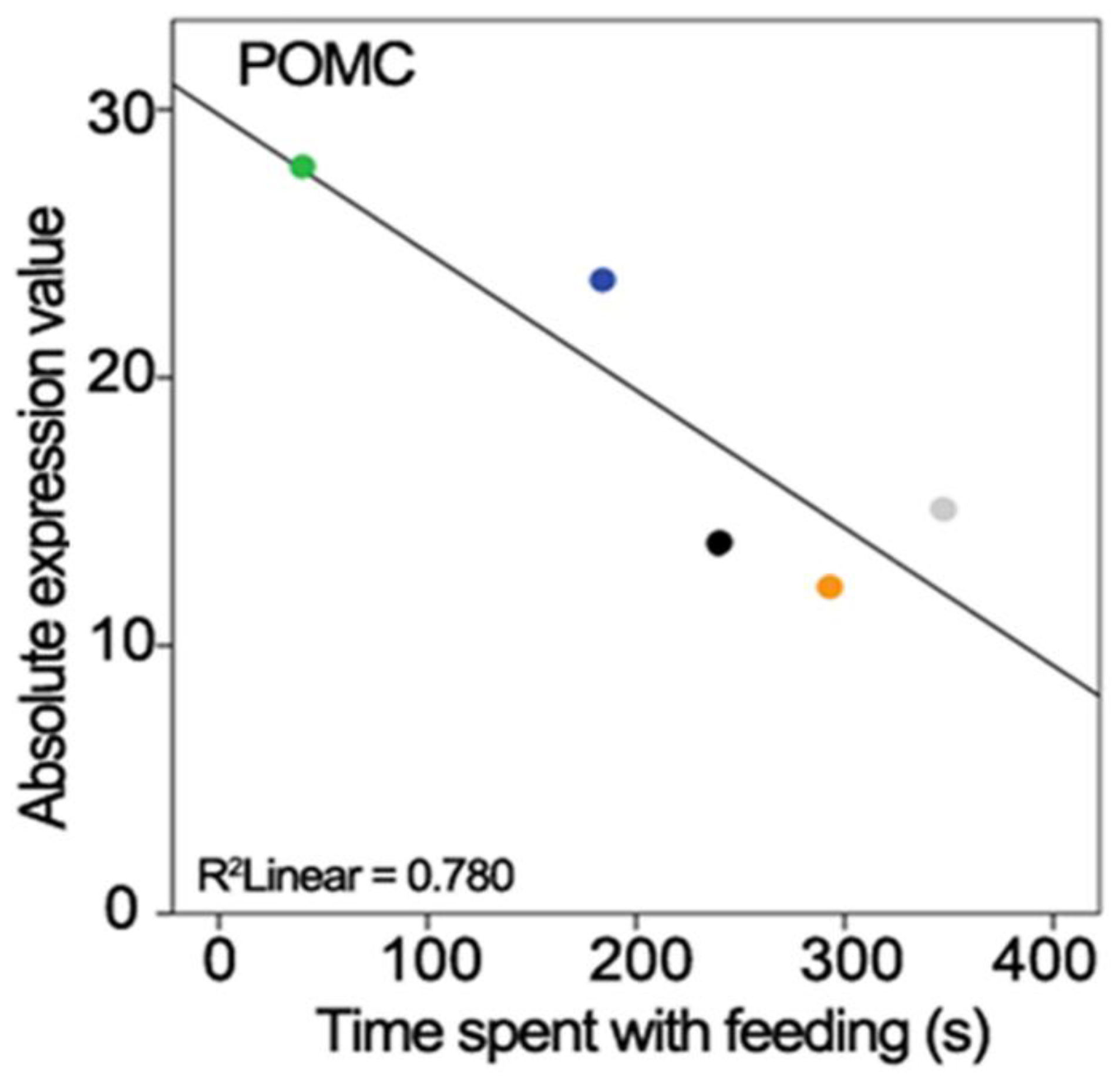
2.5. Correlation of Behaviour with Gene Expression
2.6. Relationship of DEGs
2.7. Gene Ontology Analysis
3. Discussion
3.1. Gene Expressional Studies in the Zebra Finch
3.2. Methodological Considerations
3.3. The Possible Involvement of Gene Sets and Pathways in Maternal Adaptation and Behaviour
3.3.1. Dopaminergic System
3.3.2. Serotonergic System
3.3.3. Cholinergic System
3.3.4. Brain Function of Thyroid Hormones
3.3.5. Pro-Opiomelanocortin
3.3.6. Neuro-Immunological Alterations
4. Materials and Methods
4.1. Animals
4.2. Experimental Design
4.3. Video Recording of Parental Behaviour
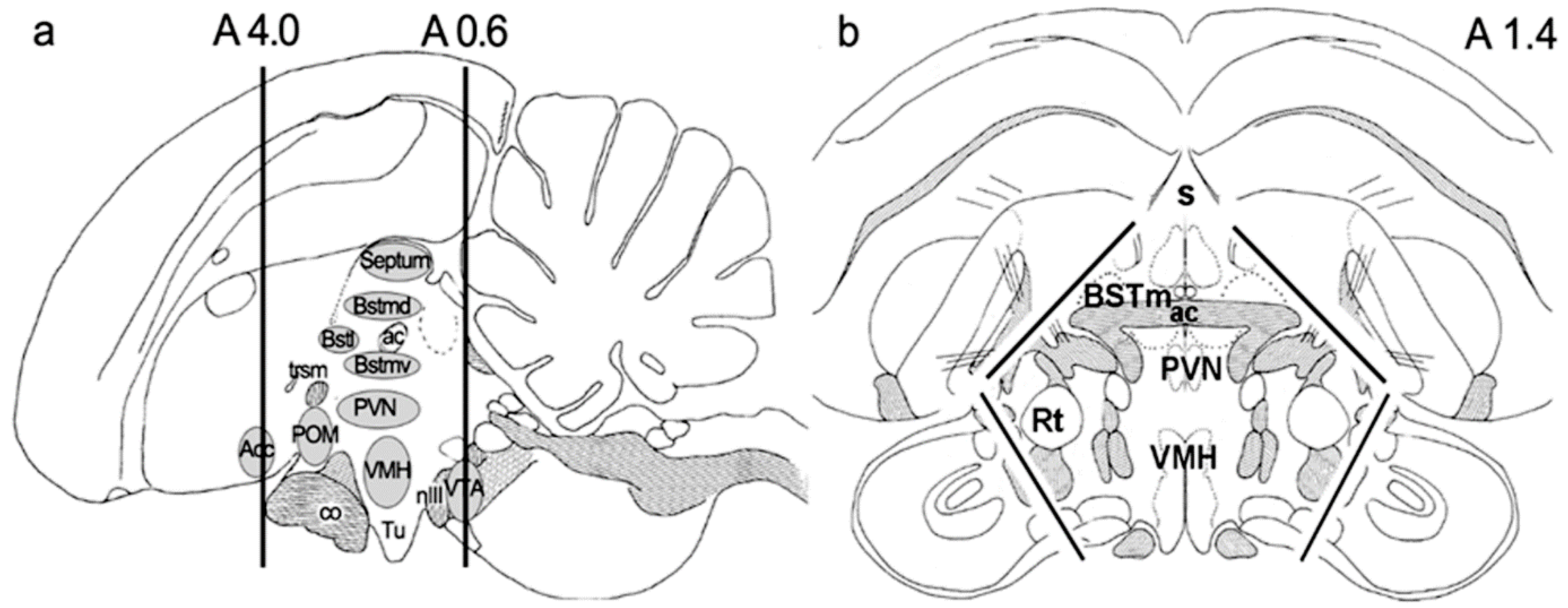
4.4. Microdissection of Brain Samples
4.5. RNA Sequencing
4.5.1. Library Construction
4.5.2. Handling of Sequencing Data
4.5.3. Data Pre-Processing and Alignment
4.5.4. Differential Gene Expression Analysis
4.5.5. Gene Set Enrichment Analysis
4.6. Real-Time PCR Validation
4.7. Statistical Analysis
4.8. Protein–Protein Interaction Analysis of Differentially Expressed Genes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dulac, C.; O’connell, L.A.; Wu, Z. Neural control of maternal and paternal behaviors. Science 2014, 345, 765–770. [Google Scholar] [CrossRef]
- Reynolds, J.D.; Goodwin, N.B.; Freckleton, R.P. Evolutionary transitions in parental care and live bearing in vertebrates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002, 357, 269–281. [Google Scholar] [CrossRef]
- Robinson, G.E. Genomics. Beyond nature and nurture. Science 2004, 304, 397–399. [Google Scholar] [CrossRef]
- Driessen, T.M.; Eisinger, B.E.; Zhao, C.; Stevenson, S.A.; Saul, M.C.; Gammie, S.C. Genes showing altered expression in the medial preoptic area in the highly social maternal phenotype are related to autism and other disorders with social deficits. BMC Neurosci. 2014, 15, 11. [Google Scholar] [CrossRef]
- Zhao, C.; Saul, M.C.; Driessen, T.; Gammie, S.C. Gene expression changes in the septum: Possible implications for microRNAs in sculpting the maternal brain. PLoS ONE 2012, 7, e38602. [Google Scholar] [CrossRef]
- Zhao, C.; Eisinger, B.E.; Driessen, T.M.; Gammie, S.C. Addiction and reward-related genes show altered expression in the postpartum nucleus accumbens. Front. Behav. Neurosci. 2014, 8, 388. [Google Scholar] [CrossRef]
- Leko, A.H.; Cservenak, M.; Szabo, E.R.; Hanics, J.; Alpar, A.; Dobolyi, A. Insulin-like growth factor I and its binding protein-3 are regulators of lactation and maternal responsiveness. Sci. Rep. 2017, 7, 3396. [Google Scholar] [CrossRef]
- Dobolyi, A. Central amylin expression and its induction in rat dams. J. Neurochem. 2009, 111, 1490–1500. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Li, R.; Michal, J.J.; Zhang, S.; Dodson, M.V.; Zhang, Z.; Harland, R.M. Whole transcriptome analysis with sequencing: Methods, challenges and potential solutions. Cell. Mol. Life Sci. 2015, 72, 3425–3439. [Google Scholar] [CrossRef]
- Hitzemann, R.; Darakjian, P.; Walter, N.; Iancu, O.D.; Searles, R.; Mcweeney, S. Introduction to sequencing the brain transcriptome. Int. Rev. Neurobiol. 2014, 116, 1–19. [Google Scholar]
- Bendesky, A.; Kwon, Y.M.; Lassance, J.M.; Lewarch, C.L.; Yao, S.; Peterson, B.K.; He, M.X.; Dulac, C.; Hoekstra, H.E. The genetic basis of parental care evolution in monogamous mice. Nature 2017, 544, 434–439. [Google Scholar] [CrossRef]
- Leko, A.H.; Kumari, R.; Dora, F.; Keller, D.; Udvari, E.B.; Csikos, V.; Renner, E.; Dobolyi, A. Transcriptome Sequencing in the Preoptic Region of Rat Dams Reveals a Role of Androgen Receptor in the Control of Maternal Behavior. Int. J. Mol. Sci. 2021, 22, 1517. [Google Scholar] [CrossRef]
- Lopes, P.C.; De Bruijn, R. Neurotranscriptomic changes associated with chick-directed parental care in adult non-reproductive Japanese quail. Sci. Rep. 2021, 11, 15481. [Google Scholar] [CrossRef]
- Bentz, A.B.; Rusch, D.B.; Buechlein, A.; Rosvall, K.A. The neurogenomic transition from territory establishment to parenting in a territorial female songbird. BMC Genom. 2019, 20, 819. [Google Scholar] [CrossRef]
- Ye, P.; Li, M.; Liao, W.; Ge, K.; Jin, S.; Zhang, C.; Chen, X.; Geng, Z. Hypothalamic transcriptome analysis reveals the neuroendocrine mechanisms in controlling broodiness of Muscovy duck (Cairina moschata). PLoS ONE 2019, 14, e0207050. [Google Scholar]
- Lynch, K.S.; O’connell, L.A.; Louder, M.I.M.; Balakrishnan, C.N.; Fischer, E.K. Understanding the Loss of Maternal Care in Avian Brood Parasites Using Preoptic Area Transcriptome Comparisons in Brood Parasitic and Non-parasitic Blackbirds. G3 2019, 9, 1075–1084. [Google Scholar] [CrossRef]
- Volgyi, K.; Udvari, E.B.; Szabo, E.R.; Gyorffy, B.A.; Hunyadi-Gulyas, E.; Medzihradszky, K.; Juhasz, G.; Kekesi, K.A.; Dobolyi, A. Maternal alterations in the proteome of the medial prefrontal cortex in rat. J. Proteom. 2017, 153, 65–77. [Google Scholar] [CrossRef][Green Version]
- Udvari, E.B.; Volgyi, K.; Gulyassy, P.; Dimen, D.; Kis, V.; Barna, J.; Szabo, E.R.; Lubec, G.; Juhasz, G.; Kekesi, K.A.; et al. Synaptic proteome changes in the hypothalamus of mother rats. J. Proteom. 2017, 159, 54–66. [Google Scholar] [CrossRef]
- Udvari, E.B.; Volgyi, K.; Kekesi, K.A.; Simon, D.; Hunyadi-Gulyas, E.; Dobolyi, A. Proteomic Analysis of the Maternal Preoptic Area in Rats. Neurochem. Res. 2019, 44, 2314–2324. [Google Scholar] [CrossRef]
- Bridges, R.S. Neuroendocrine regulation of maternal behavior. Front. Neuroendocrinol. 2015, 36, 178–196. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, X.; Ma, Y.; Li, X.K. The biological function of pigeon crop milk and the regulation of its production. Yi Chuan 2017, 39, 1158–1167. [Google Scholar]
- Zann, R.A. The Zebra Finch: A Synthesis of Field and Laboratory Studies; Oxford University Press: Oxford, UK, 1996. [Google Scholar]
- Morvai, B.; Nanuru, S.; Mul, D.; Kusche, N.; Milne, G.; Szekely, T.; Komdeur, J.; Miklosi, A.; Pogany, A. Diurnal and Reproductive Stage-Dependent Variation of Parental Behaviour in Captive Zebra Finches. PLoS ONE 2016, 11, e0167368. [Google Scholar] [CrossRef]
- Banerjee, A.; Chokkalla, A.K.; Shi, J.J.; Lee, J.; Venna, V.R.; Vemuganti, R.; Mccullough, L.D. Microarray Profiling Reveals Distinct Circulating miRNAs in Aged Male and Female Mice Subjected to Post-stroke Social Isolation. Neuromol. Med. 2020, 23, 305–314. [Google Scholar] [CrossRef]
- Ibi, D.; Takuma, K.; Koike, H.; Mizoguchi, H.; Tsuritani, K.; Kuwahara, Y.; Kamei, H.; Nagai, T.; Yoneda, Y.; Nabeshima, T.; et al. Social isolation rearing-induced impairment of the hippocampal neurogenesis is associated with deficits in spatial memory and emotion-related behaviors in juvenile mice. J. Neurochem. 2008, 105, 921–932. [Google Scholar] [CrossRef]
- D’amelio, P.B.; Klumb, M.; Adreani, M.N.; Gahr, M.L.; Ter Maat, A. Individual recognition of opposite sex vocalizations in the zebra finch. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Kohl, J.; Dulac, C. Neural control of parental behaviors. Curr. Opin. Neurobiol. 2018, 49, 116–122. [Google Scholar] [CrossRef]
- Dobolyi, A.; Grattan, D.R.; Stolzenberg, D.S. Preoptic inputs and mechanisms that regulate maternal responsiveness. J. Neuroendocrinol. 2014, 26, 627–640. [Google Scholar] [CrossRef]
- Zhang, G.W.; Shen, L.; Tao, C.; Jung, A.H.; Peng, B.; Li, Z.; Zhang, L.I.; Tao, H.W. Medial preoptic area antagonistically mediates stress-induced anxiety and parental behavior. Nat. Neurosci. 2021, 24, 516–528. [Google Scholar] [CrossRef]
- Wu, Z.; Autry, A.E.; Bergan, J.F.; Watabe-Uchida, M.; Dulac, C.G. Galanin neurons in the medial preoptic area govern parental behaviour. Nature 2014, 509, 325–330. [Google Scholar] [CrossRef]
- Curley, J.P.; Jensen, C.L.; Franks, B.; Champagne, F.A. Variation in maternal and anxiety-like behavior associated with discrete patterns of oxytocin and vasopressin 1a receptor density in the lateral septum. Horm. Behav. 2012, 61, 454–461. [Google Scholar] [CrossRef]
- Fazekas, E.A.; Morvai, B.; Zachar, G.; Dora, F.; Szekely, T.; Pogany, A.; Dobolyi, A. Neuronal activation in zebra finch parents associated with reintroduction of nestlings. J. Comp. Neurol. 2020, 528, 363–379. [Google Scholar] [CrossRef]
- Olah, S.; Cservenak, M.; Keller, D.; Fazekas, E.A.; Renner, E.; Low, P.; Dobolyi, A. Prolactin-induced and neuronal activation in the brain of mother mice. Brain Struct. Funct. 2018, 223, 3229–3250. [Google Scholar] [CrossRef]
- Lonstein, J.S.; Greco, B.; De Vries, G.J.; Stern, J.M.; Blaustein, J.D. Maternal behavior stimulates c-fos activity within estrogen receptor alpha-containing neurons in lactating rats. Neuroendocrinology 2000, 72, 91–101. [Google Scholar] [CrossRef]
- Balakrishnan, C.N.; Mukai, M.; Gonser, R.A.; Wingfield, J.C.; London, S.E.; Tuttle, E.M.; Clayton, D.F. Brain transcriptome sequencing and assembly of three songbird model systems for the study of social behavior. PeerJ 2014, 2, e396. [Google Scholar] [CrossRef]
- Boss, J.; Liedvogel, M.; Lundberg, M.; Olsson, P.; Reischke, N.; Naurin, S.; Akesson, S.; Hasselquist, D.; Wright, A.; Grahn, M.; et al. Gene expression in the brain of a migratory songbird during breeding and migration. Mov. Ecol. 2016, 4, 4. [Google Scholar] [CrossRef]
- Gedman, G.; Haase, B.; Durieux, G.; Biegler, M.T.; Fedrigo, O.; Jarvis, E.D. As above, so below: Whole transcriptome profiling demonstrates strong molecular similarities between avian dorsal and ventral pallial subdivisions. J. Comp. Neurol. 2021, 529, 3222–3246. [Google Scholar] [CrossRef]
- Lovell, P.V.; Huizinga, N.A.; Friedrich, S.R.; Wirthlin, M.; Mello, C.V. The constitutive differential transcriptome of a brain circuit for vocal learning. BMC Genom. 2018, 19, 231. [Google Scholar] [CrossRef]
- Clayton, D.F. The genomics of memory and learning in songbirds. Annu. Rev. Genom. Hum. Genet. 2013, 14, 45–65. [Google Scholar] [CrossRef]
- Palazzo, A.; Marsano, R.M. Transposable elements: A jump toward the future of expression vectors. Crit. Rev. Biotechnol. 2021, 41, 792–808. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Treiber, C.D.; Waddell, S. Transposon expression in the Drosophila brain is driven by neighboring genes and diversifies the neural transcriptome. Genome Res. 2020, 30, 1559–1569. [Google Scholar] [CrossRef]
- Ferrari, R.; Grandi, N.; Tramontano, E.; Dieci, G. Retrotransposons as Drivers of Mammalian Brain Evolution. Life 2021, 11, 376. [Google Scholar] [CrossRef]
- Bodea, G.O.; Mckelvey, E.G.Z.; Faulkner, G.J. Retrotransposon-induced mosaicism in the neural genome. Open Biol. 2018, 8, 180074. [Google Scholar] [CrossRef]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef]
- Huang, Y.C.; Hessler, N.A. Social modulation during songbird courtship potentiates midbrain dopaminergic neurons. PLoS ONE 2008, 3, e3281. [Google Scholar] [CrossRef]
- Mattson, B.J.; Williams, S.; Rosenblatt, J.S.; Morrell, J.I. Comparison of two positive reinforcing stimuli: Pups and cocaine throughout the postpartum period. Behav. Neurosci. 2001, 115, 683–694. [Google Scholar] [CrossRef]
- Attwell, D.; Bouvier, M. Cloners quick on the uptake. Curr. Biol. 1992, 2, 541–543. [Google Scholar] [CrossRef]
- Rudnick, G.; Kramer, R.; Blakely, R.D.; Murphy, D.L.; Verrey, F. The SLC6 transporters: Perspectives on structure, functions, regulation, and models for transporter dysfunction. Pflugers Arch. 2014, 466, 25–42. [Google Scholar] [CrossRef]
- Larhammar, M.; Patra, K.; Blunder, M.; Emilsson, L.; Peuckert, C.; Arvidsson, E.; Ronnlund, D.; Preobraschenski, J.; Birgner, C.; Limbach, C.; et al. SLC10A4 is a vesicular amine-associated transporter modulating dopamine homeostasis. Biol. Psychiatry 2015, 77, 526–536. [Google Scholar] [CrossRef]
- Stolzenberg, D.S.; Numan, M. Hypothalamic interaction with the mesolimbic DA system in the control of the maternal and sexual behaviors in rats. Neurosci. Biobehav. Rev. 2011, 35, 826–847. [Google Scholar] [CrossRef]
- Freeman, M.E.; Kanyicska, B.; Lerant, A.; Nagy, G.M. Prolactin: Structure, function, and regulation of secretion. Physiol. Rev. 2000, 80, 1523–1631. [Google Scholar] [CrossRef]
- Grattan, D.R.; Kokay, I.C. Prolactin: A pleiotropic neuroendocrine hormone. J. Neuroendocrinol. 2008, 20, 752–763. [Google Scholar] [CrossRef]
- Dobolyi, A.; Olah, S.; Keller, D.; Kumari, R.; Fazekas, E.A.; Csikos, V.; Renner, E.; Cservenak, M. Secretion and Function of Pituitary Prolactin in Evolutionary Perspective. Front. Neurosci. 2020, 14, 621. [Google Scholar] [CrossRef]
- Crowley, W.R. Neuroendocrine regulation of lactation and milk production. Compr. Physiol. 2015, 5, 255–291. [Google Scholar]
- Scott, N.; Prigge, M.; Yizhar, O.; Kimchi, T. A sexually dimorphic hypothalamic circuit controls maternal care and oxytocin secretion. Nature 2015, 525, 519–522. [Google Scholar] [CrossRef]
- Walther, D.J.; Bader, M. A unique central tryptophan hydroxylase isoform. Biochem. Pharmacol. 2003, 66, 1673–1680. [Google Scholar] [CrossRef]
- Holschbach, M.A.; Lonstein, J.S. Motherhood and infant contact regulate neuroplasticity in the serotonergic midbrain dorsal raphe. Psychoneuroendocrinology 2017, 76, 97–106. [Google Scholar] [CrossRef]
- De-Miguel, F.F.; Trueta, C. Synaptic and extrasynaptic secretion of serotonin. Cell. Mol. Neurobiol. 2005, 25, 297–312. [Google Scholar] [CrossRef]
- Black, S.A.; Rylett, R.J. Choline transporter CHT regulation and function in cholinergic neurons. Cent. Nerv. Syst. Agents Med. Chem. 2012, 12, 114–121. [Google Scholar] [CrossRef]
- Fujimoto, H.; Liu, H.X.; Lopatina, O.; Brown, D.A.; Higashida, H. Scopolamine modulates paternal parental retrieval behavior in mice induced by the maternal mate. Neurosci. Lett. 2013, 546, 63–66. [Google Scholar] [CrossRef]
- Shea, S.D.; Margoliash, D. Basal forebrain cholinergic modulation of auditory activity in the zebra finch song system. Neuron 2003, 40, 1213–1226. [Google Scholar] [CrossRef]
- Cachope, R.; Mateo, Y.; Mathur, B.N.; Irving, J.; Wang, H.L.; Morales, M.; Lovinger, D.M.; Cheer, J.F. Selective activation of cholinergic interneurons enhances accumbal phasic dopamine release: Setting the tone for reward processing. Cell Rep. 2012, 2, 33–41. [Google Scholar] [CrossRef]
- Klandorf, H.; Lea, R.W.; Sharp, P.J. Thyroid function in laying, incubating, and broody bantam hens. Gen. Comp. Endocrinol. 1982, 47, 492–496. [Google Scholar] [CrossRef]
- Seko, D.; Ogawa, S.; Li, T.S.; Taimura, A.; Ono, Y. mu-Crystallin controls muscle function through thyroid hormone action. FASEB J. 2016, 30, 1733–1740. [Google Scholar] [CrossRef]
- Roberts, L.M.; Woodford, K.; Zhou, M.; Black, D.S.; Haggerty, J.E.; Tate, E.H.; Grindstaff, K.K.; Mengesha, W.; Raman, C.; Zerangue, N. Expression of the thyroid hormone transporters monocarboxylate transporter-8 (SLC16A2) and organic ion transporter-14 (SLCO1C1) at the blood-brain barrier. Endocrinology 2008, 149, 6251–6261. [Google Scholar] [CrossRef]
- Naicker, M.; Naidoo, S. Expression of thyroid-stimulating hormone receptors and thyroglobulin in limbic regions in the adult human brain. Metab. Brain Dis. 2018, 33, 481–489. [Google Scholar] [CrossRef]
- Lea, R.W.; Klandorf, H.; Harvey, S.; Hall, T.R. Thyroid and adrenal function in the ring dove (Streptopelia risoria) during food deprivation and a breeding cycle. Gen. Comp. Endocrinol. 1992, 86, 138–146. [Google Scholar] [CrossRef]
- Bertagna, X. Proopiomelanocortin-derived peptides. Endocrinol. Metab. Clin. N. Am. 1994, 23, 467–485. [Google Scholar] [CrossRef]
- Gerets, H.H.; Peeters, K.; Arckens, L.; Vandesande, F.; Berghman, L.R. Sequence and distribution of pro-opiomelanocortin in the pituitary and the brain of the chicken (Gallus gallus). J. Comp. Neurol. 2000, 417, 250–262. [Google Scholar] [CrossRef]
- Zhan, C.; Zhou, J.; Feng, Q.; Zhang, J.E.; Lin, S.; Bao, J.; Wu, P.; Luo, M. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J. Neurosci. 2013, 33, 3624–3632. [Google Scholar] [CrossRef]
- Boswell, T.; Dunn, I.C. Regulation of Agouti-Related Protein and Pro-Opiomelanocortin Gene Expression in the Avian Arcuate Nucleus. Front. Endocrinol. 2017, 8, 75. [Google Scholar] [CrossRef]
- Strader, A.D.; Schioth, H.B.; Buntin, J.D. The role of the melanocortin system and the melanocortin-4 receptor in ring dove (Streptopelia risoria) feeding behavior. Brain Res. 2003, 960, 112–121. [Google Scholar] [CrossRef]
- Medina, F.; Siddiqui, A.; Scimonelli, T.; Fenske, C.; Wilson, C.A.; Celis, M.E. The inter-relationship between gonadal steroids and POMC peptides, beta-endorphin and alpha-MSH, in the control of sexual behavior in the female rat. Peptides 1998, 19, 1309–1316. [Google Scholar] [CrossRef]
- Soares, N.L.; Vieira, H.L.A. Microglia at the Centre of Brain Research: Accomplishments and Challenges for the Future. Neurochem. Res. 2022, 47, 218–233. [Google Scholar] [CrossRef]
- Murata, T.; Obiri, N.I.; Puri, R.K. Structure of and signal transduction through interleukin-4 and interleukin-13 receptors (review). Int. J. Mol. Med. 1998, 1, 551–557. [Google Scholar] [CrossRef]
- Amo-Aparicio, J.; Garcia-Garcia, J.; Francos-Quijorna, I.; Urpi, A.; Esteve-Codina, A.; Gut, M.; Quintana, A.; Lopez-Vales, R. Interleukin-4 and interleukin-13 induce different metabolic profiles in microglia and macrophages that relate with divergent outcomes after spinal cord injury. Theranostics 2021, 11, 9805–9820. [Google Scholar] [CrossRef]
- Bauer, M.E.; Teixeira, A.L. Neuroinflammation in Mood Disorders: Role of Regulatory Immune Cells. Neuroimmunomodulation 2021, 28, 99–107. [Google Scholar] [CrossRef]
- Forstmeier, W.; Segelbacher, G.; Mueller, J.C.; Kempenaers, B. Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol. Ecol. 2007, 16, 4039–4050. [Google Scholar] [CrossRef]
- Péter, A. Solomon Coder: A Simple Solution for Behavior Coding. v 16.06.26. 2015. Available online: http://solomoncoder.com/ (accessed on 15 June 2016).
- Nixdorf-Bergweiler Be, B.H. A Stereotaxic Atlas of the Brain of the Zebra Finch, Taeniopygia Guttata: With Special Emphasis on Telencephalic Visual And Song System Nuclei in Transverse and Sagittal Sections [Internet]; National Center for Biotechnology Information: Bethesda, MD, USA, 2007.
- Reiner, A.; Perkel, D.J.; Mello, C.V.; Jarvis, E.D. Songbirds and the revised avian brain nomenclature. Ann. N. Y. Acad. Sci. 2004, 1016, 77–108. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Szabo, E.R.; Cservenak, M.; Dobolyi, A. Amylin is a novel neuropeptide with potential maternal functions in the rat. FASEB J. 2012, 26, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- Palasca, O.; Santos, A.; Stolte, C.; Gorodkin, J.; Jensen, L.J. TISSUES 2.0: An integrative web resource on mammalian tissue expression. Database 2018, 2018, bay003. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.M.; Goodson, J.L. Hypothalamic oxytocin and vasopressin neurons exert sex-specific effects on pair bonding, gregariousness, and aggression in finches. Proc. Natl. Acad. Sci. USA 2014, 111, 6069–6074. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.; Vleck, C.M. Prolactin release and response to vasoactive intestinal peptide in an opportunistic breeder, the zebra finch (Taeniopygia guttata). Gen. Comp. Endocrinol. 2008, 157, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Zachar, G.; Montagnese, C.; Fazekas, E.A.; Kemecsei, R.G.; Papp, S.M.; Dora, F.; Renner, E.; Csillag, A.; Pogany, A.; Dobolyi, A. Brain Distribution and Sexually Dimorphic Expression of Amylin in Different Reproductive Stages of the Zebra Finch (Taeniopygia guttata) Suggest Roles of the Neuropeptide in Song Learning and Social Behaviour. Front. Neurosci. 2019, 13, 1401. [Google Scholar] [CrossRef] [PubMed]



| Sample_Name | Raw Reads (mlns) | Uniquely Mapped Reads Number (mlns) | Uniquely Mapped Reads % |
|---|---|---|---|
| Breeding 1 | 21.6 | 18.1 | 83.8% |
| Breeding 2 | 21.2 | 17.7 | 83.6% |
| Breeding 3 | 21.6 | 18.0 | 83.6% |
| Breeding 4 | 21.1 | 17.7 | 83.9% |
| Breeding 5 | 21.5 | 18.0 | 83.9% |
| Breeding 6 | 21.7 | 18.0 | 82.9% |
| Paired control 1 | 21.2 | 17.9 | 84.4% |
| Paired control 2 | 21.5 | 18.2 | 84.4% |
| Paired control 3 | 21.2 | 18.0 | 84.8% |
| Paired control 4 | 21.6 | 18.2 | 84.2% |
| Paired control 5 | 21.4 | 18.1 | 84.2% |
| Paired control 6 | 21.5 | 18.2 | 84.3% |
| Gene Name | Ensemble Gene ID | Protein Name | Log2FC | p-Value |
|---|---|---|---|---|
| SLC6A2 | ENSTGUG00000007218 | Solute carrier family 6 member 2 (sodium dependent noradrenalin transporter) | 3.366 | <0.001 |
| TH | ENSTGUG00000009295 | Tyrosine hydroxylase | 2.550 | <0.001 |
| TPH2 | ENSTGUG00000007300 | Tryptophan hydroxylase 2 | 2.400 | <0.001 |
| CHAT | ENSTGUG00000005959 | Choline-O-acetyltransferase | 1.979 | <0.001 |
| SLC10A4 | ENSTGUG00000008069 | Solute carrier family 10 member 4 (acidification of synaptic vesicles containing monoamines) | 1.896 | <0.001 |
| SLC5A7 | ENSTGUG00000009694 | Solute carrier family 5 member 7 (presynaptic sodium-dependent high-affinity choline transporter 1) | 1.656 | 0.044 |
| MIRLET7D, tgu-let-7a-3, tgu-let-7f | ENSTGUG00000017619ENSTGUG00000017688ENSTGUG00000017729 | NA | 1.525 | <0.001 |
| TG | ENSTGUG00000012562 | Thyroglobulin | −1.528 | 0.003 |
| ABCA10 | ENSTGUG00000003994 | ATP Binding Cassette Subfamily A Member 10 | −1.560 | 0.007 |
| ANLN | ENSTGUG00000005487 | Anillin Actin Binding Protein | −1.571 | <0.001 |
| PMP2 | ENSTGUG00000011663 | Peripheral Myelin Protein 2 | −1.585 | 0.002 |
| ALDH1A3 | ENSTGUG00000008854 | Aldehyde dehydrogenase 1 family, member A3 | −1.649 | 0.005 |
| CILP | ENSTGUG00000009096 | Cartilage Intermediate Layer Protein | −1.677 | 0.003 |
| JAK3 | ENSTGUG00000017135 | Janus kinase 3 | −1.697 | 0.032 |
| OLFML1 | ENSTGUG00000005163 | Olfactomedin Like 1 | −1.704 | 0.035 |
| CRYM | ENSTGUG00000009095 | Crystallin mu | −1.741 | 0.003 |
| C1S | ENSTGUG00000013265 | Complement C1s | −1.783 | 0.001 |
| VIM | ENSTGUG00000001298 | Vimentin | −1.821 | <0.001 |
| CLDN1 | ENSTGUG00000009337 | Claudin 1 | −1.824 | 0.037 |
| SEMA3B | ENSTGUG00000006529 | Semaphorin-3B | −1.949 | 0.005 |
| ZIC3 | ENSTGUG00000002155 | Zic family member 3 | −1.951 | 0.015 |
| MSX1 | ENSTGUG00000009947 | Msh homeobox 1 | −2.056 | 0.003 |
| SOCS3 | ENSTGUG00000003345 | Suppressor of cytokine signalling 3 | −2.121 | 0.018 |
| VILL | ENSTGUG00000000377 | Villin Like protein | −2.166 | 0.036 |
| RBPMS2 | ENSTGUG00000004722 | RNA Binding Protein, MRNA Processing Factor 2 | −2.261 | 0.006 |
| ADGRD2 | ENSTGUG00000007153ENSTGUG00000007166 | Adhesion G Protein-Coupled Receptor D2 | −2.275 | <0.001 |
| LOXL2 | ENSTGUG00000017286 | Lysyl oxidase homolog 2 | −2.503 | 0.005 |
| HDC | ENSTGUG00000007569 | Histidine decarboxylase | −2.601 | 0.002 |
| SLCO1C1 | ENSTGUG00000012325 | Solute carrier organic anion transporter family member 1C1 | −2.672 | <0.001 |
| TMEM255B | ENSTGUG00000009312 | Transmembrane protein 255B | −2.812 | <0.001 |
| POMC | ENSTGUG00000017017 | Pro-opiomelanocortin | −5.168 | <0.001 |
| Upregulated | ||||||
| Source | Term_Name | Term Id | Adjusted pValue | Term Size | Query Size | Intersection Size |
| KEGG | Tryptophan metabolism | KEGG:00380 | 0.026 | 32 | 9 | 1 |
| KEGG | Tyrosine metabolism | KEGG:00350 | 0.026 | 23 | 9 | 1 |
| GO:CC | Synapse | GO:0045202 | 0.003 | 584 | 9 | 4 |
| GO:CC | Integral component of membrane | GO:0016021 | 0.019 | 3080 | 9 | 5 |
| GO:MF | Neurotransmitter transmembrane transporter activity | GO:0005326 | 0.001 | 32 | 9 | 2 |
| GO:MF | Monooxygenase activity | GO:0004497 | 0.002 | 66 | 9 | 2 |
| GO:BP | Regulation of neurotransmitter levels | GO:0001505 | 0.002 | 166 | 9 | 3 |
| GO:BP | Transmembrane transport | GO:0055085 | 0.006 | 986 | 9 | 4 |
| Downregulated | ||||||
| Source | Term_Name | Term Id | Adjusted pValue | Term Size | Query Size | Intersection Size |
| KEGG | Neuroactive ligand-receptor interaction | KEGG:04080 | 0.048 | 233 | 36 | 3 |
| KEGG | Melanogenesis | KEGG:04916 | 0.048 | 70 | 36 | 2 |
| GO:BP | Mesenchymal cell differentiation | GO:0048762 | 0.030 | 129 | 36 | 4 |
| GO:BP | Thyroid hormone transport | GO:0070327 | 0.030 | 6 | 36 | 2 |
| Upregulated Genes | ||
| Ensemble ID | Gene Names | Protein Names |
| ENSTGUG00000007300 | TPH2 | Tryptophan hydroxylase 2 |
| ENSTGUG00000009295 | TH | Tyrosine hydroxylase |
| ENSTGUG00000005959 | CHAT | Choline-O-acetyl transferase |
| ENSTGUG00000011058 | SLC18A2 | Solute carrier family 18 member A2 (synaptic vesicular monamine transporter) |
| ENSTGUG00000009694 | SLC5A7 | Choline:sodium symporter (presynaptic sodium-dependent high-affinity choline transporter 1) |
| ENSTGUG00000005989 | SLC18A3 | Solute carrier family 18 member A3 (synaptic vesicular acetylcholine transporter) |
| ENSTGUG00000007218 | SLC6A2 | Monoamine transmembrane transporter |
| ENSTGUG00000008069 | SLC10A4 | Solute carrier family 10 member A4 (acidification of synaptic vesicles containing monoamines) |
| Downregulated Genes | ||
| Ensemble ID | Gene Names | Protein Names |
| ENSTGUG00000017017 | POMC | Pro-opiomelanocortin |
| ENSTGUG00000002625 | EDNRA | Endothelin receptor |
| ENSTGUG00000013478 | P2RY2 | Purinergic receptor P2Y2 |
| ENSTGUG00000010805 | TCF7L2 | RNA polymerase II proximal promoter sequence-specific DNA binding protein (Transcription factor 7 like 2) |
| ENSTGUG00000009947 | MSX1 | RNA polymerase II-specific DNA-binding transcription activator |
| ENSTGUG00000009095 | CRYM | Crystallin mu |
| ENSTGUG00000012325 | SLCO1C1 | Thyroid hormone transporter |
| Name of Genes | Gene Symbol | NCBI Gene ID | Forward Primer | Range (bp) | Reverse Primer | Range (bp) |
|---|---|---|---|---|---|---|
| Actin beta | ACTB | 751978 | CGTGCTGTCTTCCCATCCAT | 82–101 | CTCTCTGTTGGCTTTGGGGT | 351–370 |
| Crystallin mu | CRYM | 100229846 | AGGACTCCTCTGTGCCTTCT | 206–225 | CTTCACTGCCCTCTCCTTGG | 480–499 |
| Glycerinaldehyde-3-phosphate dehydrogenase | GAPDH | 100190636 | GAGGGTAGTGAAGGCTGCTG | 795–814 | AGAGCTAAGCGGTGGTGAAC | 1113–1132 |
| Hypoxanthine phosphoribosyl-transferase 1 | HPRT1 | 100231349 | GTGTGATCAGTGAGACGGGG | 590–609 | CAAACAGCACAACCCAACCA | 907–926 |
| Pro-opiomelanocortin | POMC | - | TACGTCATGAGCCACTTCCG | 97–116 | CCTCATCCTCCTCCTCCTCC | 394–413 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, R.; Fazekas, E.A.; Morvai, B.; Udvari, E.B.; Dóra, F.; Zachar, G.; Székely, T.; Pogány, Á.; Dobolyi, Á. Transcriptomics of Parental Care in the Hypothalamic–Septal Region of Female Zebra Finch Brain. Int. J. Mol. Sci. 2022, 23, 2518. https://doi.org/10.3390/ijms23052518
Kumari R, Fazekas EA, Morvai B, Udvari EB, Dóra F, Zachar G, Székely T, Pogány Á, Dobolyi Á. Transcriptomics of Parental Care in the Hypothalamic–Septal Region of Female Zebra Finch Brain. International Journal of Molecular Sciences. 2022; 23(5):2518. https://doi.org/10.3390/ijms23052518
Chicago/Turabian StyleKumari, Rashmi, Emese A. Fazekas, Boglárka Morvai, Edina B. Udvari, Fanni Dóra, Gergely Zachar, Tamás Székely, Ákos Pogány, and Árpád Dobolyi. 2022. "Transcriptomics of Parental Care in the Hypothalamic–Septal Region of Female Zebra Finch Brain" International Journal of Molecular Sciences 23, no. 5: 2518. https://doi.org/10.3390/ijms23052518
APA StyleKumari, R., Fazekas, E. A., Morvai, B., Udvari, E. B., Dóra, F., Zachar, G., Székely, T., Pogány, Á., & Dobolyi, Á. (2022). Transcriptomics of Parental Care in the Hypothalamic–Septal Region of Female Zebra Finch Brain. International Journal of Molecular Sciences, 23(5), 2518. https://doi.org/10.3390/ijms23052518







