Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review
Abstract
:1. Introduction
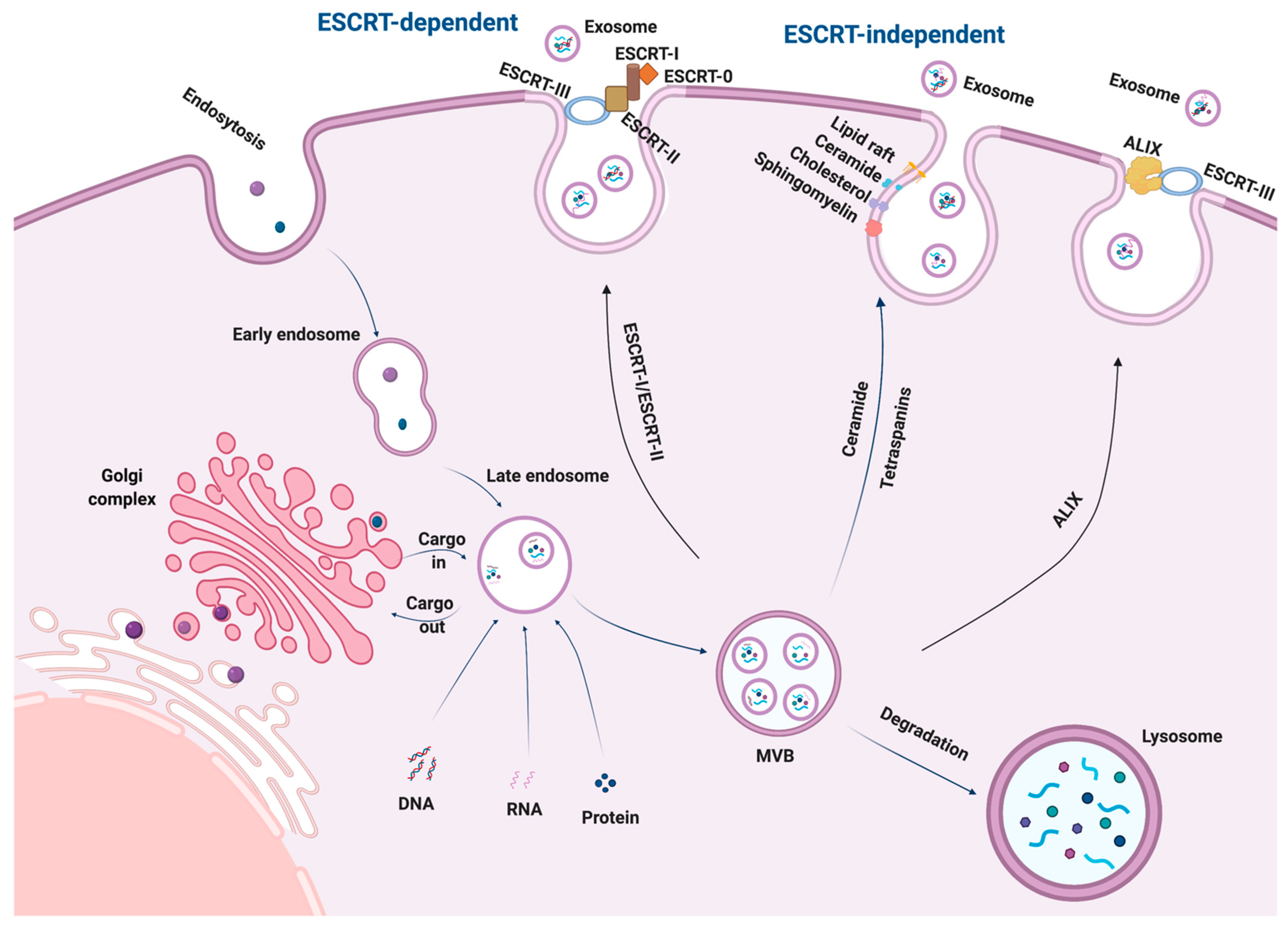
2. Biogenesis of Exosomes
3. Secretion and Uptake of Exosomes
4. Exosomal RNAs
4.1. Exosomal mRNAs
4.2. Exosomal miRNAs
4.3. Exosomal lncRNAs
4.4. Exosomal circRNAs
5. Exosome Isolation Techniques
5.1. Ultra-Speed Centrifugation
5.1.1. Differential Ultracentrifugation
5.1.2. Density-Gradient Ultracentrifugation
5.2. Immunoaffinity Capture
5.3. Ultrafiltration
5.4. Size-Exclusion Chromatography
5.5. Polymer Precipitation
5.6. Microfluidics-Based Techniques
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlotogorski-Hurvitz, A.; Dayan, D.; Chaushu, G.; Korvala, J.; Salo, T.; Sormunen, R.; Vered, M. Human saliva-derived exosomes: Comparing methods of isolation. J. Histochem. Cytochem. 2015, 63, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Adriano, B.; Cotto, N.M.; Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact. Mater. 2021, 6, 2479–2490. [Google Scholar] [CrossRef]
- Dini, L.; Tacconi, S.; Carata, E.; Tata, A.M.; Vergallo, C.; Panzarini, E. Microvesicles and exosomes in metabolic diseases and inflammation. Cytokine Growth Factor Rev. 2020, 51, 27–39. [Google Scholar] [CrossRef]
- Gehrmann, U.; Näslund, T.I.; Hiltbrunner, S.; Larssen, P.; Gabrielsson, S. Harnessing the exosome-induced immune response for cancer immunotherapy. Semin. Cancer Biol. 2014, 28, 58–67. [Google Scholar] [CrossRef]
- D’Anca, M.; Fenoglio, C.; Serpente, M.; Arosio, B.; Cesari, M.; Scarpini, E.A.; Galimberti, D. Exosome Determinants of Physiological Aging and Age-Related Neurodegenerative Diseases. Front. Aging Neurosci. 2019, 11, 232. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- Hosseini, K.; Ranjbar, M.; Pirpour Tazehkand, A.; Asgharian, P.; Montazersaheb, S.; Tarhriz, V.; Ghasemnejad, T. Evaluation of exosomal non-coding RNAs in cancer using high-throughput sequencing. J. Transl. Med. 2022, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhou, Q.; Qiao, B.; Shao, B.; Hu, S.; Wang, G.; Yuan, W.; Sun, Z. Exosome-derived noncoding RNAs: Function, mechanism, and application in tumor angiogenesis. Mol. Ther. Nucleic Acids 2022, 27, 983–997. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Moita, C.; Van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III–dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, e201904113. [Google Scholar] [CrossRef] [Green Version]
- Kenific, C.M.; Zhang, H.; Lyden, D. An exosome pathway without an ESCRT. Cell Res. 2021, 31, 105–106. [Google Scholar] [CrossRef]
- Lata, S.; Schoehn, G.; Jain, A.; Pires, R.; Piehler, J.; Gőttlinger Heinrich, G.; Weissenhorn, W. Helical Structures of ESCRT-III Are Disassembled by VPS4. Science 2008, 321, 1354–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hariharan, H.; Kesavan, Y.; Raja, N.S. Impact of native and external factors on exosome release: Understanding reactive exosome secretion and its biogenesis. Mol. Biol. Rep. 2021, 48, 7559–7573. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Grassart, A.; Cheng, A.T.; Hong, S.H.; Zhang, F.; Zenzer, N.; Feng, Y.; Briner, D.M.; Davis, G.D.; Malkov, D.; Drubin, D.G. Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J. Cell Biol. 2014, 205, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.-W.; Geuze, H.J.; Stoorvogel, W. Proteomic and Biochemical Analyses of Human B Cell-derived Exosomes: Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [Green Version]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [Green Version]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome Uptake Depends on ERK1/2-Heat Shock Protein 27 Signaling and Lipid Raft-mediated Endocytosis Negatively Regulated by Caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef] [Green Version]
- Phuyal, S.; Hessvik, N.P.; Skotland, T.; Sandvig, K.; Llorente, A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014, 281, 2214–2227. [Google Scholar] [CrossRef]
- Elsherbini, A.; Bieberich, E. Chapter Five—Ceramide and Exosomes: A Novel Target in Cancer Biology and Therapy. In Advances in Cancer Research; Chalfant, C.E., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 140, pp. 121–154. [Google Scholar]
- Kajimoto, T.; Okada, T.; Miya, S.; Zhang, L.; Nakamura, S.-I. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013, 4, 2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kummer, D.; Steinbacher, T.; Schwietzer, M.F.; Thölmann, S.; Ebnet, K. Tetraspanins: Integrating cell surface receptors to functional microdomains in homeostasis and disease. Med. Microbiol. Immunol. 2020, 209, 397–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular Endosome Biogenesis in the Absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Yang, H.; Wang, J.; Ru, W.; Wu, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020, 53, e12857. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Vicente Manzanares, M.; Sánchez-Madrid, F. Organizing polarized delivery of exosomes at synapses. Traffic 2015, 16, 327–337. [Google Scholar] [CrossRef] [Green Version]
- Saito, N.; Okada, Y.; Noda, Y.; Kinoshita, Y.; Kondo, S.; Hirokawa, N. KIFC2 Is a Novel Neuron-Specific C-Terminal Type Kinesin Superfamily Motor for Dendritic Transport of Multivesicular Body-Like Organelles. Neuron 1997, 18, 425–438. [Google Scholar] [CrossRef] [Green Version]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.m.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Extracellular vesicles: Exosomes, microvesicles, and friends. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pironti, G.; Strachan, R.T.; Abraham, D.; Mon-Wei Yu, S.; Chen, M.; Chen, W.; Hanada, K.; Mao, L.; Watson, L.J.; Rockman, H.A. Circulating Exosomes Induced by Cardiac Pressure Overload Contain Functional Angiotensin II Type 1 Receptors. Circulation 2015, 131, 2120–2130. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef] [Green Version]
- Miranda, K.C.; Bond, D.T.; McKee, M.; Skog, J.; Păunescu, T.G.; Da Silva, N.; Brown, D.; Russo, L.M. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010, 78, 191–199. [Google Scholar] [CrossRef] [Green Version]
- El Fekih, R.; Hurley, J.; Tadigotla, V.; Alghamdi, A.; Srivastava, A.; Coticchia, C.; Choi, J.; Allos, H.; Yatim, K.; Alhaddad, J.; et al. Discovery and Validation of a Urinary Exosome mRNA Signature for the Diagnosis of Human Kidney Transplant Rejection. J. Am. Soc. Nephrol. 2021, 32, 994. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Phatak, P.; Burger, A.M. Telomerase and its potential for therapeutic intervention. Br. J. Pharmacol. 2007, 152, 1003–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldvaser, H.; Gutkin, A.; Beery, E.; Edel, Y.; Nordenberg, J.; Wolach, O.; Rabizadeh, E.; Uziel, O.; Lahav, M. Characterisation of blood-derived exosomal hTERT mRNA secretion in cancer patients: A potential pan-cancer marker. Br. J. Cancer 2017, 117, 353–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Dong, X.; Chen, Y.; Wang, X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. CCLM 2018, 56, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. The Amyloid Beta Peptide: A Chemist’s Perspective. Role in Alzheimer’s and Fibrillization. Chem. Rev. 2012, 112, 5147–5192. [Google Scholar] [CrossRef]
- Mitsuhashi, M.; Taub, D.D.; Kapogiannis, D.; Eitan, E.; Zukley, L.; Mattson, M.P.; Ferrucci, L.; Schwartz, J.B.; Goetzl, E.J. Aging enhances release of exosomal cytokine mRNAs by Aβ1-42-stimulated macrophages. FASEB J. 2013, 27, 5141–5150. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.-L.; Cao, Y.-H.; Pan, M.-M.; Liu, H.; Tang, R.-N.; Ma, K.-L.; Chen, P.-S.; Liu, B.-C. CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clin. Chim. Acta 2014, 428, 26–31. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef] [Green Version]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [Green Version]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10 ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical utility of the exosome based ExoDx Prostate(IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2–10 ng/mL. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.L.; Yan, W.; Liu, Y.W.; Wang, Z.; Hu, Q.; Nie, E.; Zhou, X.; Li, R.; Wang, X.F.; Jiang, T.; et al. Tumour exosomes from cells harbouring PTPRZ1–MET fusion contribute to a malignant phenotype and temozolomide chemoresistance in glioblastoma. Oncogene 2017, 36, 5369–5381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, T.; Mizutani, K.; Kawakami, K.; Fujita, Y.; Ehara, H.; Ito, M. CD44v8-10 mRNA contained in serum exosomes as a diagnostic marker for docetaxel resistance in prostate cancer patients. Heliyon 2020, 6, e04138. [Google Scholar] [CrossRef] [PubMed]
- Pope, S.M.; Lässer, C. Toxoplasma gondii infection of fibroblasts causes the production of exosome-like vesicles containing a unique array of mRNA and miRNA transcripts compared to serum starvation. J. Extracell. Vesicles 2013, 2, 22484. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.X.; Li, Y.M.; Ye, M.; Guo, Y.Y.; Li, Q.W.; Peng, X.M.; Wang, Q.; Zhang, S.F.; Zhao, H.X.; Zhang, H.; et al. KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncol. Lett. 2017, 13, 3608–3616. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Sun, X.; Xu, J.; Han, X.; Xing, Z.; Wang, D.; Ge, J.; Meng, L.; Xu, X. Serum Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) mRNA Protected by Exosomes as a Potential Biomarker for Gastric Cancer. Med. Sci. Monit. 2019, 25, 7770–7783. [Google Scholar] [CrossRef]
- Del Re, M.; Biasco, E.; Crucitta, S.; Derosa, L.; Rofi, E.; Orlandini, C.; Miccoli, M.; Galli, L.; Falcone, A.; Jenster, G.W.; et al. The Detection of Androgen Receptor Splice Variant 7 in Plasma-derived Exosomal RNA Strongly Predicts Resistance to Hormonal Therapy in Metastatic Prostate Cancer Patients. Eur. Urol. 2017, 71, 680–687. [Google Scholar] [CrossRef]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol. Oncol. 2019, 13, 212–227. [Google Scholar] [CrossRef] [Green Version]
- Haque, S.; Vaiselbuh, S.R. Exosomal DNMT1 mRNA transcript is elevated in acute lymphoblastic leukemia which might reprograms leukemia progression. Cancer Genet. 2022, 260–261, 57–64. [Google Scholar] [CrossRef]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta BBA Mol. Cell Res. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Liz, J.; Esteller, M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2016, 1859, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Si, M.; Yang, J.; Sun, S.; Wu, H.; Cui, S.; Qu, X.; Yu, X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020, 474, 36–52. [Google Scholar] [CrossRef]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e785. [Google Scholar] [CrossRef]
- Pfeffer, S.; Zavolan, M.; Grässer Friedrich, A.; Chien, M.; Russo James, J.; Ju, J.; John, B.; Enright Anton, J.; Marks, D.; Sander, C.; et al. Identification of Virus-Encoded MicroRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; Van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; De Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-G.; Song, J.; Zhang, Y.-Q.; Wang, P.-C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014, 10, 2395–2400. [Google Scholar] [CrossRef] [Green Version]
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.D.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.T.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074.e2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, A.G.-E.; Cheng, K.; Marbán, E. Exosomes as Critical Agents of Cardiac Regeneration Triggered by Cell Therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsonov, R.; Burdakov, V.; Shtam, T.; Radzhabovа, Z.; Vasilyev, D.; Tsyrlina, E.; Titov, S.; Ivanov, M.; Berstein, L.; Filatov, M.; et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumor Biol. 2016, 37, 12011–12021. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 81. [Google Scholar] [CrossRef] [PubMed]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life Sci. 2018, 75, 3539–3551. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Yu, J.; Wang, J.; Li, H.; Che, J.; Cao, B. Isolation and Identification of miRNAs in exosomes derived from serum of colon cancer patients. J. Cancer 2017, 8, 1145–1152. [Google Scholar] [CrossRef] [Green Version]
- Santangelo, A.; Imbrucè, P.; Gardenghi, B.; Belli, L.; Agushi, R.; Tamanini, A.; Munari, S.; Bossi, A.M.; Scambi, I.; Benati, D.; et al. A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. J. Neuro-Oncol. 2018, 136, 51–62. [Google Scholar] [CrossRef]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Cai, G.; Liu, K.; Zhuang, Z.; Jia, K.; Pei, S.; Wang, X.; Wang, H.; Xu, S.; Cui, C.; et al. Microglia exosomal miRNA-137 attenuates ischemic brain injury through targeting Notch1. Aging 2021, 13, 4079–4095. [Google Scholar] [CrossRef] [PubMed]
- Zabegina, L.; Nazarova, I.; Nikiforova, N.; Slyusarenko, M.; Sidina, E.; Knyazeva, M.; Tsyrlina, E.; Novikov, S.; Reva, S.; Malek, A. A New Approach for Prostate Cancer Diagnosis by miRNA Profiling of Prostate-Derived Plasma Small Extracellular Vesicles. Cells 2021, 10, 2372. [Google Scholar] [CrossRef]
- Chen, L.; Cao, P.; Huang, C.; Wu, Q.; Chen, S.; Chen, F. Serum exosomal miR-7977 as a novel biomarker for lung adenocarcinoma. J. Cell. Biochem. 2020, 121, 3382–3391. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Z.; Chen, W.; Wang, X.; Cao, M.; Han, X.; Zhang, K.; Teng, B.; Cao, J.; Wu, W.; et al. M2 Macrophage-Derived Exosomes Promote Angiogenesis and Growth of Pancreatic Ductal Adenocarcinoma by Targeting E2F2. Mol. Ther 2021, 29, 1226–1238. [Google Scholar] [CrossRef]
- Liao, Z.; Chen, Y.; Duan, C.; Zhu, K.; Huang, R.; Zhao, H.; Hintze, M.; Pu, Q.; Yuan, Z.; Lv, L.; et al. Cardiac telocytes inhibit cardiac microvascular endothelial cell apoptosis through exosomal miRNA-21-5p-targeted cdip1 silencing to improve angiogenesis following myocardial infarction. Theranostics 2021, 11, 268–291. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, R.; Baghaei, K.; Amani, D.; Piccin, A.; Hashemi, S.M.; Asadzadeh Aghdaei, H.; Zali, M.R. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci. 2021, 269, 119035. [Google Scholar] [CrossRef]
- Cai, G.; Cai, G.; Zhou, H.; Zhuang, Z.; Liu, K.; Pei, S.; Wang, Y.; Wang, H.; Wang, X.; Xu, S.; et al. Mesenchymal stem cell-derived exosome miR-542-3p suppresses inflammation and prevents cerebral infarction. Stem Cell Res. Ther. 2021, 12, 2. [Google Scholar] [CrossRef]
- Zhou, C.; Wei, W.; Ma, J.; Yang, Y.; Liang, L.; Zhang, Y.; Wang, Z.; Chen, X.; Huang, L.; Wang, W.; et al. Cancer-secreted exosomal miR-1468-5p promotes tumor immune escape via the immunosuppressive reprogramming of lymphatic vessels. Mol. Ther. 2021, 29, 1512–1528. [Google Scholar] [CrossRef]
- Luo, T.; Liu, Q.; Tan, A.; Duan, L.; Jia, Y.; Nong, L.; Tang, J.; Zhou, W.; Xie, W.; Lu, Y.; et al. Mesenchymal Stem Cell-Secreted Exosome Promotes Chemoresistance in Breast Cancer via Enhancing miR-21-5p-Mediated S100A6 Expression. Mol. Ther. Oncolytics 2020, 19, 283–293. [Google Scholar] [CrossRef]
- Gao, M.; Yu, T.; Liu, D.; Shi, Y.; Yang, P.; Zhang, J.; Wang, J.; Liu, Y.; Zhang, X. Sepsis plasma-derived exosomal miR-1-3p induces endothelial cell dysfunction by targeting SERP1. Clin. Sci. 2021, 135, 347–365. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Ding, X.; Zhou, L.; Zhang, L.; Yang, X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene 2021, 40, 246–261. [Google Scholar] [CrossRef]
- Chen, F.; Xu, B.; Li, J.; Yang, X.; Gu, J.; Yao, X.; Sun, X. Hypoxic tumour cell-derived exosomal miR-340-5p promotes radioresistance of oesophageal squamous cell carcinoma via KLF10. J. Exp. Clin. Cancer Res. 2021, 40, 38. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2020, 121, 109553. [Google Scholar] [CrossRef] [PubMed]
- Farag, A.F.; Sabry, D.; Hassabou, N.F.; Alaa El-Din, Y. MicroRNA-134/MicroRNA-200a Derived Salivary Exosomes are Novel Diagnostic Biomarkers of Oral Squamous Cell Carcinoma. Egypt. Dent. J. 2021, 67, 367–377. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Cui, W.; Liu, Y.; Zhou, H.; Wang, Y.; Chen, X.; Chen, X.; Wang, Z. Serum Exosomal miRNA-1226 as Potential Biomarker of Pancreatic Ductal Adenocarcinoma. Onco Targets Ther. 2021, 14, 1441–1451. [Google Scholar] [CrossRef]
- Guo, X.; Gao, L.; Wang, Y.; Chiu, D.K.Y.; Wang, T.; Deng, Y. Advances in long noncoding RNAs: Identification, structure prediction and function annotation. Brief. Funct. Genom. 2016, 15, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Losko, M.; Kotlinowski, J.; Jura, J. Long Noncoding RNAs in Metabolic Syndrome Related Disorders. Mediat. Inflamm. 2016, 2016, 5365209. [Google Scholar] [CrossRef] [Green Version]
- Babak, T.; Blencowe, B.J.; Hughes, T.R. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genom. 2005, 6, 104. [Google Scholar] [CrossRef] [Green Version]
- Marques, A.C.; Ponting, C.P. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol. 2009, 10, R124. [Google Scholar] [CrossRef] [Green Version]
- Bermúdez, M.; Aguilar-Medina, M.; Lizárraga-Verdugo, E.; Avendaño-Félix, M.; Silva-Benítez, E.; López-Camarillo, C.; Ramos-Payán, R. LncRNAs as Regulators of Autophagy and Drug Resistance in Colorectal Cancer. Front. Oncol. 2019, 9, 1008. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-J.; Ying, X.-L.; Jiang, J.-H.; Xu, Y.-H. Prostate cancer antigen 3 as a biomarker in the urine for prostate cancer diagnosis: A meta-analysis. J. Cancer Res. Ther. 2014, 10, C218–C221. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Yang, H.; Yang, H. Diagnostic value of urine prostate cancer antigen 3 test using a cutoff value of 35 μg/L in patients with prostate cancer. Tumor Biol. 2014, 35, 8573–8580. [Google Scholar] [CrossRef]
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013, 333, 213–221. [Google Scholar] [CrossRef]
- Wang, J.; Yang, K.; Yuan, W.; Gao, Z. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Bladder Cancer Diagnosis and Prognosis. Med. Sci. Monit. 2018, 24, 9307–9316. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Zou, J.; Guo, Y.; Wei, L.; Yu, F.; Yu, B.; Xu, A. Long Noncoding RNA POU3F3 and α-Synuclein in Plasma L1CAM Exosomes Combined with β-Glucocerebrosidase Activity: Potential Predictors of Parkinson’s Disease. Neurotherapeutics 2020, 17, 1104–1119. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Han, C.-L.; Wang, K.-L.; Sui, Y.-P.; Li, Z.-B.; Chen, N.; Fan, S.-Y.; Shimabukuro, M.; Wang, F.; Meng, F.-G. Integrated analysis of exosomal lncRNA and mRNA expression profiles reveals the involvement of lnc-MKRN2-42:1 in the pathogenesis of Parkinson’s disease. CNS Neurosci. Ther. 2020, 26, 527–537. [Google Scholar] [CrossRef]
- Dong, A.; Preusch, C.B.; So, W.-K.; Lin, K.; Luan, S.; Yi, R.; Wong, J.W.; Wu, Z.; Cheung, T.H. A long noncoding RNA, LncMyoD, modulates chromatin accessibility to regulate muscle stem cell myogenic lineage progression. Proc. Natl. Acad. Sci. USA 2020, 117, 32464–32475. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, P.; Bian, X.; Xu, S.; Zhou, Q.; Zhang, Y.; Ding, M.; Han, M.; Huang, L.; Bi, J.; et al. Elevated plasma levels of exosomal BACE1-AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Mol. Med. Rep. 2020, 22, 227–238. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, H.; Chen, Y.; Li, K.; Li, T.; Chen, J.; Zhang, B.; Guo, C.; Qing, L.; Shen, J.; et al. Exosomal long noncoding RNA HOXD-AS1 promotes prostate cancer metastasis via miR-361-5p/FOXM1 axis. Cell Death Dis. 2021, 12, 1129. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Fang, Q.-Q.; Chen, W.-Z.; Jiang, J.-X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.-B.; Xia, W.-J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Gu, J.; Zhang, J.; Shi, H.; Hou, S.; Xu, X.; Chen, Y.; Zhang, Y.; Mao, F.; Qian, H.; et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020, 11, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Chen, C.; Luo, Y.; Yu, M.; He, W.; An, M.; Gao, B.; Kong, Y.; Ya, Y.; Lin, Y.; et al. Tumor-derived exosomal BCYRN1 activates WNT5A/VEGF-C/VEGFR3 feedforward loop to drive lymphatic metastasis of bladder cancer. Clin. Transl. Med. 2021, 11, e497. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Investig. 2020, 130, 404–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bo, X.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L.; Li, B. Exosome-transferred LINC01559 promotes the progression of gastric cancer via PI3K/AKT signaling pathway. Cell Death Dis. 2020, 11, 723. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Ru, Y.; Zhou, F.; Wang, N.; Xi, H.; Zhang, K.; Li, J.; Chang, R.; Xie, T.; et al. Circulating Exosomal Gastric Cancer-Associated Long Noncoding RNA1 as a Biomarker for Early Detection and Monitoring Progression of Gastric Cancer: A Multiphase Study. JAMA Surg. 2020, 155, 572–579. [Google Scholar] [CrossRef]
- Piao, H.Y.; Guo, S.; Wang, Y.; Zhang, J. Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin. Transl. Oncol. 2021, 23, 246–256. [Google Scholar] [CrossRef]
- Chen, C.; Shang, A.; Sun, Z.; Gao, Y.; Huang, J.; Ping, Y.; Chang, W.; Gu, C.; Sun, J.; Ji, P.; et al. Urinary Exosomal Long Noncoding RNA TERC as a Noninvasive Diagnostic and Prognostic Biomarker for Bladder Urothelial Carcinoma. J. Immunol. Res. 2022, 2022, 9038808. [Google Scholar] [CrossRef] [PubMed]
- Mi, X.; Xu, R.; Hong, S.; Xu, T.; Zhang, W.; Liu, M. M2 Macrophage-Derived Exosomal lncRNA AFAP1-AS1 and MicroRNA-26a Affect Cell Migration and Metastasis in Esophageal Cancer. Mol. Ther. Nucleic Acids 2020, 22, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Chen, H.; Xie, C.; Hou, M. Long-noncoding RNA MALAT1 sponges microRNA-92a-3p to inhibit doxorubicin-induced cardiac senescence by targeting ATG4a. Aging 2020, 12, 8241–8260. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pei, X.; Guo, G.; Qian, X.; Dou, D.; Zhang, Z.; Xu, X.; Duan, X. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell. Physiol. 2020, 235, 6896–6904. [Google Scholar] [CrossRef]
- Wilusz Jeremy, E.; Sharp Phillip, A. A Circuitous Route to Noncoding RNA. Science 2013, 340, 440–441. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, Y.; Nie, L.; Ding, X.; Zhang, X.; Zhao, W.; Xu, X.; Kyei, B.; Dai, D.; Zhan, S.; et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2019, 1862, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Cao, X.; Dong, D.; Shen, X.; Cheng, J.; Jiang, R.; Yang, Z.; Peng, S.; Huang, Y.; Lan, X.; et al. Circular RNA TTN Acts As a miR-432 Sponge to Facilitate Proliferation and Differentiation of Myoblasts via the IGF2/PI3K/AKT Signaling Pathway. Mol. Ther. Nucleic Acids 2019, 18, 966–980. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Zhang, X.; Odame, E.; Xu, X.; Chen, Y.; Ye, J.; Zhou, H.; Dai, D.; Kyei, B.; Zhan, S.; et al. CircRNA-Protein Interactions in Muscle Development and Diseases. Int. J. Mol. Sci. 2021, 22, 3262. [Google Scholar] [CrossRef]
- Pandey, P.R.; Yang, J.-H.; Tsitsipatis, D.; Panda, A.C.; Noh, J.H.; Kim, K.M.; Munk, R.; Nicholson, T.; Hanniford, D.; Argibay, D.; et al. circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res. 2020, 48, 3789–3805. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Xie, Q.; Xu, H.; Li, J.; Li, Y. Circular RNAs and cancer. Cancer Lett. 2017, 396, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in Cancer. Cancer Res. 2013, 73, 5609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Wang, K.; Wu, F.; Wang, W.; Zhang, K.; Hu, H.; Liu, Y.; Jiang, T. circRNA disease: A manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Harland, R.; Misher, L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development 1988, 102, 837–852. [Google Scholar] [CrossRef]
- Danan, M.; Schwartz, S.; Edelheit, S.; Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012, 40, 3131–3142. [Google Scholar] [CrossRef]
- He, J.; Ren, M.; Li, H.; Yang, L.; Wang, X.; Yang, Q. Exosomal Circular RNA as a Biomarker Platform for the Early Diagnosis of Immune-Mediated Demyelinating Disease. Front. Genet. 2019, 10, 860. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, H.; Yang, Y.; Wang, X.; Deng, T.; Liu, R.; Ning, T.; Bai, M.; Li, H.; Zhu, K.; et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics 2020, 10, 8211–8226. [Google Scholar] [CrossRef]
- Dou, Y.-Q.; Kong, P.; Li, C.-L.; Sun, H.-X.; Li, W.-W.; Yu, Y.; Nie, L.; Zhao, L.-L.; Miao, S.-B.; Li, X.-K.; et al. Smooth muscle SIRT1 reprograms endothelial cells to suppress angiogenesis after ischemia. Theranostics 2020, 10, 1197–1212. [Google Scholar] [CrossRef]
- Sun, R.; Liu, W.; Zhao, Y.; Chen, H.; Wang, Z.; Zhang, Y.; Sun, X.; Cui, X. Exosomal circRNA as a novel potential therapeutic target for multiple myeloma-related myocardial damage. Cancer Cell Int. 2021, 21, 311. [Google Scholar] [CrossRef]
- Xiong, L.; Chen, L.; Wu, L.; He, W.; Chen, D.; Peng, Z.; Li, J.; Zhu, X.; Su, L.; Li, Y.; et al. Lipotoxicity-induced circGlis3 impairs beta cell function and is transmitted by exosomes to promote islet endothelial cell dysfunction. Diabetologia 2022, 65, 188–205. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, Y.-H.; Yoon, C.; Huang, X.-Y.; Xu, Y.; Xie, J.-W.; Wang, J.-B.; Lin, J.-x.; Chen, Q.-Y.; Cao, L.-L.; et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877–3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020, 471, 38–48. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Huang, Z.-L.; Huang, J.; Xu, B.; Huang, X.-Y.; Xu, Y.-H.; Zhou, J.; Tang, Z.-Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Zhong, X.; Lin, Z.; Lin, J.; Qiu, M.; Li, X.; Hu, Z. Plasma exo-hsa_circRNA_0056616: A potential biomarker for lymph node metastasis in lung adenocarcinoma. J. Cancer 2020, 11, 4037–4046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Zhou, W.; Pan, X.; Sun, Z.; Sun, Y.; Xu, H.; Shi, P.; Li, J.; Gao, L.; Tian, X. Circular RNA Profiling Reveals Exosomal circ_0006156 as a Novel Biomarker in Papillary Thyroid Cancer. Mol. Ther Nucleic Acids 2020, 19, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, Z.; Liu, T.; Liu, Q.; Zhou, H.; Meng, S.; Feng, Z.; Tang, Y.; Liu, C.; Feng, J.; et al. Circular RNA Sequencing Reveals Serum Exosome Circular RNA Panel for High-Grade Astrocytoma Diagnosis. Clin. Chem. 2021, 68, hvab254. [Google Scholar] [CrossRef]
- Tian, C.; Liu, J.; Di, X.; Cong, S.; Zhao, M.; Wang, K. Exosomal hsa_circRNA_104484 and hsa_circRNA_104670 may serve as potential novel biomarkers and therapeutic targets for sepsis. Sci. Rep. 2021, 11, 14141. [Google Scholar] [CrossRef]
- Yu, M.; Yu, J.; Zhang, Y.; Sun, X.; Sun, R.; Xia, M.; Li, S.; Cui, X. A novel circRNA-miRNA-mRNA network revealed exosomal circ-ATP10A as a biomarker for multiple myeloma angiogenesis. Bioengineered 2022, 13, 667–683. [Google Scholar] [CrossRef]
- Zhong, A.-N.; Yin, Y.; Tang, B.-J.; Chen, L.; Shen, H.-W.; Tan, Z.-P.; Li, W.-Q.; He, Q.; Sun, B.; Zhu, Y.; et al. CircRNA Microarray Profiling Reveals hsa_circ_0058493 as a Novel Biomarker for Imatinib-Resistant CML. Front. Pharmacol. 2021, 12, 2479. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Zhao, R.; Li, B.; Qi, Y.; Qiu, W.; Guo, Q.; Zhang, S.; Zhao, S.; Xu, H.; Li, M.; et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 2022, 21, 16. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Zhang, D.; Zhu, Z.; Dela Cruz, C.S.; Jin, Y. Epithelial cell-derived microvesicles activate macrophages and promote inflammation via microvesicle-containing microRNAs. Sci. Rep. 2016, 6, 35250. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Hong, C.-S.; Stolz, D.B.; Watkins, S.C.; Whiteside, T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 2014, 411, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiemstra, T.F.; Charles, P.D.; Gracia, T.; Hester, S.S.; Gatto, L.; Al-Lamki, R.; Floto, R.A.; Su, Y.; Skepper, J.N.; Lilley, K.S.; et al. Human Urinary Exosomes as Innate Immune Effectors. J. Am. Soc. Nephrol. 2014, 25, 2017. [Google Scholar] [CrossRef] [Green Version]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Bano, R.; Ahmad, F.; Mohsin, M. A perspective on the isolation and characterization of extracellular vesicles from different biofluids. RSC Adv. 2021, 11, 19598–19615. [Google Scholar] [CrossRef]
- Cvjetkovic, A.; Lötvall, J.; Lässer, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef]
- Zhang, M.; Jin, K.; Gao, L.; Zhang, Z.; Li, F.; Zhou, F.; Zhang, L. Methods and Technologies for Exosome Isolation and Characterization. Small Methods 2018, 2, 1800021. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef] [PubMed]
- Carnino, J.M.; Lee, H.; Jin, Y. Isolation and characterization of extracellular vesicles from Broncho-alveolar lavage fluid: A review and comparison of different methods. Respir. Res. 2019, 20, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 2019, 9, 1015–1028. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Tong, C.; Chen, Q.; Zhao, L.; Ma, J.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of long intergenic noncoding RNAs in bovine mammary glands. BMC Genom. 2017, 18, 468. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- McNamara, R.P.; Caro-Vegas, C.P.; Costantini, L.M.; Landis, J.T.; Griffith, J.D.; Damania, B.A.; Dittmer, D.P. Large-scale, cross-flow based isolation of highly pure and endocytosis-competent extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1541396. [Google Scholar] [CrossRef] [Green Version]
- Merchant, M.L.; Powell, D.W.; Wilkey, D.W.; Cummins, T.D.; Deegens, J.K.; Rood, I.M.; McAfee, K.J.; Fleischer, C.; Klein, E.; Klein, J.B. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteom. Clin. Appl. 2010, 4, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Lucchetti, D.; Fattorossi, A.; Sgambato, A. Extracellular Vesicles in Oncology: Progress and Pitfalls in the Methods of Isolation and Analysis. Biotechnol. J. 2019, 14, 1700716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.l.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef] [Green Version]
- Gheinani, A.H.; Vögeli, M.; Baumgartner, U.; Vassella, E.; Draeger, A.; Burkhard, F.C.; Monastyrskaya, K. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci. Rep. 2018, 8, 3945. [Google Scholar] [CrossRef]
- Guerreiro, E.M.; Vestad, B.; Steffensen, L.A.; Aass, H.C.D.; Saeed, M.; Øvstebø, R.; Costea, D.E.; Galtung, H.K.; Søland, T.M. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS ONE 2018, 13, e0204276. [Google Scholar] [CrossRef] [Green Version]
- Soares Martins, T.; Catita, J.; Martins Rosa, I.; Da Cruz Silva, O.A.B.E.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef] [Green Version]
- García-Romero, N.; Madurga, R.; Rackov, G.; Palacín-Aliana, I.; Núñez-Torres, R.; Asensi-Puig, A.; Carrión-Navarro, J.; Esteban-Rubio, S.; Peinado, H.; González-Neira, A.; et al. Polyethylene glycol improves current methods for circulating extracellular vesicle-derived DNA isolation. J. Transl. Med. 2019, 17, 75. [Google Scholar] [CrossRef]
- Samsonov, R.; Shtam, T.; Burdakov, V.; Glotov, A.; Tsyrlina, E.; Berstein, L.; Nosov, A.; Evtushenko, V.; Filatov, M.; Malek, A. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic. Prostate 2016, 76, 68–79. [Google Scholar] [CrossRef]
- Salieb-Beugelaar, G.B.; Simone, G.; Arora, A.; Philippi, A.; Manz, A. Latest Developments in Microfluidic Cell Biology and Analysis Systems. Anal. Chem. 2010, 82, 4848–4864. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.L.; Lu, H. Advances in microfluidic cell separation and manipulation. Curr. Opin. Chem. Eng. 2013, 2, 398–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gholizadeh, S.; Shehata Draz, M.; Zarghooni, M.; Sanati-Nezhad, A.; Ghavami, S.; Shafiee, H.; Akbari, M. Microfluidic approaches for isolation, detection, and characterization of extracellular vesicles: Current status and future directions. Biosens. Bioelectron. 2017, 91, 588–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Skog, J.; Hsu, C.-H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef] [Green Version]
| Exosome Sources | Diseases | Potential Biomarkers (mRNA) | References |
|---|---|---|---|
| Serum & glioblastoma CCM | Glioblastoma | EGFRvIII | [42] |
| Urine | Tubulointerstitial fibrosis & glomerular sclerosis | CD2AP | [48] |
| Serum & GBM CCM | Temozolomide resistance in GBM | MGMT & APNG | [49] |
| Urine | Prostate cancer | ERG, and SPDEF | [50] |
| U87 & A172 CCM | Temozolomide chemoresistance in glioblastoma | PTPRZ1-MET | [53] |
| Serum | Hepatocellular carcinoma | hnRNPH1 | [45] |
| Serum | Docetaxel resistance in prostate cancer | CD44v8-10 | [54] |
| HFF CCM | Toxoplasma-infected HFFs | RAB-13, EEF1A1, TMSB4X & LLPH | [55] |
| Serum | Colorectal cancer | KRAS mutation & BRAF mutation | [56] |
| Serum | Gastric cancer | MT1-MMP | [57] |
| Plasma | Resistance to hormonal therapy in prostate cancer | AR-V7 | [58] |
| Serum | Pancreatic ductal adenocarcinoma | WASF2, ARF6, SNORA74A & SNORA25 | [59] |
| Serum & CCM | Acute lymphoblastic leukemia | DNMT1 | [60] |
| Exosome Sources | Potential Biomarkers | Diseases | Target Genes/Pathways | Effects | References |
|---|---|---|---|---|---|
| Serum | miR-193b | AD | APP | Inhibits AD development | [72] |
| Glioblastoma stem CCM | miR-9 | Antiangiogenic therapy for glioblastoma | RGS5, SOX7 & ABCB1 | Promotes angiogenesis | [73] |
| Plasma | miR-146a | Heart failure | IRAK-1, TRAF6, NOX-4 SMAD4 & TGF-β | Promotes the proliferation and inhibit the apoptosis of cardiomyocytes | [74] |
| Plasma | miR-21 & miR-181a-5p | Thyroid cancer | N/A | Distinguishes between follicular and papillary thyroid cancer | [75] |
| HCT116 CCM & serum | miR-25, miR-130b, and miR-425 | Colorectal cancer | PTEN/PI3K/AKT pathway | Promotes the liver metastasis of colorectal cancer | [67] |
| CCM & serum | miR-1247-3p | Liver cancer | B4GALT3 | Promotes the lung metastasis of liver cancer | [76] |
| A2780 CCM | miR-223 | Epithelial ovarian cancer | PTEN/PI3K/AKT pathway | Promotes chemoresistance | [77] |
| Multiple sources | miR-21 | Various cancers | Multiple targets | Promotes cancer development | [78,79,80,81,82] |
| Microglia culture media | miRNA-137 | Ischemic brain injury | NOTCH1 | Promotes neuroprotection | [83] |
| Plasma | miR-125a-5p/miR-141-5p | Prostate cancer | N/A | N/A | [84] |
| Serum | miR-7977 | Lung adenocarcinoma | N/A | Promotes proliferation and invasion, and inhibits apoptosis of A549 cells | [85] |
| Pan02 CCM | miR-155-5p & miR-221-5p | PDAC | E2F2 | Promotes PDAC progression | [86] |
| Cardiac telocyte CCM | miR-21-5p | Myocardial infarction | CDIP1 | Promotes angiogenesis | [87] |
| HT-29/SW480 CCM | miR-375-3p | Colon cancer | N/A | Regulates EMT of colon cancer cells | [88] |
| MSC CCM | miR-542-3p | Cerebral infarction | TLR4 | Inhibits inflammation and cerebral infarction | [89] |
| CCa CCM & serum | miR-1468-5p | Cervical cancer | HMBOX1 & JAK2/STAT3 pathway | Promotes tumor immune escape | [90] |
| MSC CCM | miR-21-5p | Breast cancer | S100A6 | Promotes chemoresistance | [91] |
| Plasma | miR-1-3p | Sepsis | SERP1 | Induces endothelial cell dysfunction | [92] |
| Plasma | miR-451a & miR-21-5p | AD | N/A | N/A | [93] |
| hUCMSC CCM & serum | miR-139-5p | Bladder cancer | PRC1 | Inhibits tumorigenesis | [94] |
| OSCC CCM & blood | miR-340-5p | OSCC | KLF10 | Promotes radioresistance | [95] |
| Saliva | miR-24-3p | OSCC | PER1 | Maintains the proliferation of OSCC cells | [96] |
| Saliva | miR-134 & miR-200a | OSCC | N/A | N/A | [97] |
| Serum | miR-1226 | PDAC | N/A | N/A | [98] |
| Exosome Sources | Potential Biomarkers | Diseases | Effects | Mechanistic Approaches | References |
|---|---|---|---|---|---|
| Plasma | Linc-POU3F3 | PD | N/A | N/A | [110] |
| Plasma | lnc-MKRN2-42:1 | PD | Affects the occurrence and development of PD | N/A | [111] |
| Various PC CCM & serum | lncRNA-UCA1 | PC | Promotes angiogenesis | miR-96-5p/AMOTL2 axis | [112] |
| Plasma | BACE1-AS | AD | N/A | N/A | [113] |
| Serum | HOXD-AS1 | Prostate cancer | Promotes metastasis | miR-361-5p/FOXM1 axis | [114] |
| Serum | SNHG16 | Breast cancer | Inhibits immunity | miR-16–5p/SMAD5 axis | [115] |
| Serum | lncUFC1 | NSCLC | Promotes proliferation, migration, and invasion | Inhibits PTEN expression via EZH2-mediated epigenetic silencing | [116] |
| Urine | lncBCYRN1 | Bladder cancer | Promotes lymphatic metastasis | Activates WNT5A/VEGF-C/VEGFR3 feedforward loop | [117] |
| Urine | lncLNMAT2 | Bladder cancer | Promotes lymphatic metastasis | N/A | [118] |
| Primary MSCs CCM | LINC01559 | GC | Promotes progression | Multiple approaches | [119] |
| GC CCM & serum | lncRNA-GC1 | GC | N/A | N/A | [120] |
| GC CCM | lncPCGEM1 | GC | Promotes invasion and metastasis | Maintains the stability of SNAI1 | [121] |
| Urine | TERC | BLCA | N/A | N/A | [122] |
| M1/M2 macrophage CCM | lncAFAP1-AS1 | Esophageal cancer | Promotes migration and metastasis | miR-26a/ATF2 axis | [123] |
| MSCs CCM | MALAT1 | DICS | Promotes mitochondrial metabolism and rejuvenation | miR-92a-3p/ATG4a axis | [124] |
| Serum | H19 | Breast cancer | Reduce DOX resistance | N/A | [125] |
| Exosome Sources | Potential Biomarkers | Diseases | Effects | Mechanistic Approaches | References |
|---|---|---|---|---|---|
| Serum | circ-G042080 | Myeloma-related myocardial damage | Promotes autophagy | miR-4268/TLR4 axis | [143] |
| Serum | circGlis3 | Type 2 diabetes | Regulates islet EC function | Regulates GMEB1 degradation & HSP27 phosphorylation | [144] |
| Plasma | circ-RanGAP1 | Gastric cancer | Promotes metastasis and development | miR-877–3p | [145] |
| HCC CCM | circRNA-100338 | HCC | Promotes angiogenesis and invasion | N/A | [146] |
| Plasma | circRNA_0056616 | Lymph node metastasis in lung adenocarcinoma | N/A | N/A | [147] |
| Serum | circ_0006156 | Thyroid cancer | Promotes tumorigenesis | miR-1178/TLR4 axis | [148] |
| Serum | circ_0075828, circ_0003828 & circ_0002976 | HGA | N/A | N/A | [149] |
| Serum | circRNA_104484 & circRNA_104670 | Sepsis | N/A | N/A | [150] |
| Serum | circ-ATP10A | Multiple myeloma | Promotes angiogenesis | Multiple axises | [151] |
| K562 & K562/G01 CCM | circ_0058493 | CML | Drug resistance | miR-548b-3p | [152] |
| GBM CCM | circNEIL3 | Glioma | Promotes progression | Stabilizing IGF2BP3 | [153] |
| Isolation Techniques | Advantages | Disadvantages |
|---|---|---|
|
|
|
| ||
|
| |
|
| |
|
|
|
|
| |
| ||
| ||
|
|
|
| ||
|
| |
| ||
|
| |
|
|
|
| ||
|
| |
|
|
|
| ||
|
| |
| ||
| ||
|
|
|
| ||
|
| |
|
| |
| ||
|
|
|
| ||
| ||
| ||
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yue, B.-L.; Huang, Y.-Z.; Lan, X.-Y.; Liu, W.-J.; Chen, H. Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review. Int. J. Mol. Sci. 2022, 23, 2461. https://doi.org/10.3390/ijms23052461
Wang J, Yue B-L, Huang Y-Z, Lan X-Y, Liu W-J, Chen H. Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review. International Journal of Molecular Sciences. 2022; 23(5):2461. https://doi.org/10.3390/ijms23052461
Chicago/Turabian StyleWang, Jian, Bing-Lin Yue, Yong-Zhen Huang, Xian-Yong Lan, Wu-Jun Liu, and Hong Chen. 2022. "Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review" International Journal of Molecular Sciences 23, no. 5: 2461. https://doi.org/10.3390/ijms23052461
APA StyleWang, J., Yue, B.-L., Huang, Y.-Z., Lan, X.-Y., Liu, W.-J., & Chen, H. (2022). Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review. International Journal of Molecular Sciences, 23(5), 2461. https://doi.org/10.3390/ijms23052461







