Abstract
We previously identified the aur1 biosynthetic gene cluster (BGC) in Streptomyces lavendulae subsp. lavendulae CCM 3239 (formerly Streptomyces aureofaciens CCM 3239), which is responsible for the production of the unusual angucycline-like antibiotic auricin. Auricin is produced in a narrow interval of the growth phase after entering the stationary phase, after which it is degraded due to its instability at the high pH values reached after the production phase. The complex regulation of auricin BGC is responsible for this specific production by several regulators, including the key activator Aur1P, which belongs to the family of atypical response regulators. The aur1P gene forms an operon with the downstream aur1O gene, which encodes an unknown protein without any conserved domain. Homologous aur1O genes have been found in several BGCs, which are mainly responsible for the production of angucycline antibiotics. Deletion of the aur1O gene led to a dramatic reduction in auricin production. Transcription from the previously characterized Aur1P-dependent biosynthetic aur1Ap promoter was similarly reduced in the S. lavendulae aur1O mutant strain. The aur1O-specific coactivation of the aur1Ap promoter was demonstrated in a heterologous system using a luciferase reporter gene. In addition, the interaction between Aur1O and Aur1P has been demonstrated by a bacterial two-hybrid system. These results suggest that Aur1O is a specific coactivator of this key auricin-specific positive regulator Aur1P. Bioinformatics analysis of Aur1O and its homologues in other BGCs revealed that they represent a new family of transcriptional coactivators involved in the regulation of secondary metabolite biosynthesis. However, they are divided into two distinct sequence-specific subclasses, each of which is likely to interact with a different family of positive regulators.
1. Introduction
Gram-positive soil bacteria of the genus Streptomyces (class Actinobacteria, order Actinomycetales, family Streptomycetaceae) are characterized by their ability to produce a wide range of bioactive secondary metabolites, including many known antibiotics. They undergo an exceptional process of morphological differentiation, initiated by germination of spores to form a network of branched multinucleoid hyphae (a so-called substrate mycelium). In response to various signals, it differentiates into white air-grown hyphae (a so-called aerial mycelium), which eventually undergo septation into unigenomic spore chains. Secondary metabolite production is coordinated with morphological differentiation and often occurs at the time of aerial mycelium formation or at the end of the exponential growth phase during growth in a liquid medium [1,2].
The genes responsible for the biosynthesis of antibiotics and other secondary metabolites are typically grouped into so-called biosynthetic gene clusters (BGCs) together with genes encoding pathway-specific or cluster-situated regulatory proteins. These regulatory genes located in BGCs are controlled by global regulators that integrate physiological and environmental signals to control the production of multiple secondary metabolites and have pleiotropic roles in stress response and morphological differentiation. In general, the production of secondary metabolites in streptomycetes is tightly and elaborately regulated by pyramidal regulatory cascades, including global regulators with the primary role of sensing various intracellular and extracellular signals and pathway-specific or cluster-situated regulatory proteins that are specifically regulated by these global regulators. In addition, feedback control of global regulators by pathway-specific regulators has been characterized in some regulatory cascades. These networks determine the production of antibiotic under specific culture condition. A number of global regulators have been characterized in Streptomyces (e.g., AbsA2, AdpA, AfsQ1, AtrA, DasR, GlnR, PhoP/PhoR, WblA) [3,4,5,6]. Among these global regulatory systems, those that use small diffuse hormone-like signaling molecules, including γ-butyrolactone (GBL) autoregulator, furans, γ-butenolides, PI factor, and N-methylphenylalanyl-dehydrobutyrine diketopiperazine have been most characterized. The best characterized prototype GBL system consists of a GBL synthase, responsible for GBL biosynthesis, and a related GBL receptor, which belong to the TetR family. In the absence of GBL, it binds a specific DNA sequence in front of its target genes (including pathway-specific regulators among others), thereby suppressing their transcription, usually during the exponential phase. Binding of a specific GBL autoregulator to the GBL receptor prevents this binding, thereby allowing induction target gene expression. Through this mechanism, GBLs regulate and coordinate production of secondary metabolites and, in some cases, also affect morphological differentiation [4,7].
Pathway-specific transcriptional regulators have been classified into different families based on sequence or structure similarities. The most common and best studied is the Streptomyces antibiotic regulatory protein (SARP) family. These members are activators characterized by an N-terminal winged helix-turn-helix (HTH) DNA-binding domain of the OmpR-type. The first prototype of this family studied was ActII-ORF4 for the aromatic polyketide actinorhodin in Streptomyces coelicolor. Another widespread family is LAL (large ATP-binding regulators of the LuxR family), whose genes encode large transcriptional activators and are located predominantly in the BGC for the type I PKSs, such as pikD for pikromycin in Streptomyces venezuelae. The transcriptional activator StrR, which controls the biosynthesis of aminoglycoside antibiotic streptomycin in S. griseus, is a prototype of another family designated by this regulator. It represents the first complete regulatory pathway leading to BGC activation. Interestingly, unusually, it belongs to the ParB-Spo0J family of DNA segregation proteins. Genes of this family are found in most glycopeptide BGCs. Another common family includes atypical response regulators (ARRs), which show homology to the OmpR family of response regulators of bacterial two-component signal transducing systems. However, they lack the residues necessary for phosphorylation as well as the accompanying sensor histidine kinase genes. They are characterized by an N-terminal receiver domain and a winged HTH C-terminal domain. The most studied member of this ARR family is the activator JadR1 for the angucycline antibiotic jadomycin in S. venezuelae. Interestingly, unlike classical response regulators, JadR1 is feedback-regulated with jadomycin. Genes for most other members of this ARR family have been found in angycycline BGCs and encoded transcriptional activators (e.g., LanI, LndI, Aur1P, GcnR, ChaI, SchA25, SimReg1), which are essential for the biosynthesis of the corresponding antibiotics in other Streptomyces spp. In addition, a similar feedback regulatory mechanism through end-product binding was found for two other homologues, Aur1P and SimgReg1. In addition to transcriptional activators, many BGCs are often negatively regulated by TetR family transcriptional regulators. Members of this family are widespread in various bacteria and act mainly as repressors to regulate genes for biosynthetic enzymes for antibiotics, drug-efflux pumps and other proteins. The structure of the TetR-family proteins consists of two domains: the N-terminal HTH DNA-binding domain and the C-terminal regulatory domain which binds the ligand, leading to the loss of its DNA-binding activity and the subsequent activation of transcription of the target gene [1,3,4,5,6,8].
Initial sequence analysis of aur1 BGC in Streptomyces aureofaciens CCM 3239 revealed 15 open reading frames that showed high similarity to several angucycline type II polyketide synthase (PKS) BGCs, which was responsible for the antibiotic auricin [9]. We later found that the cluster is not located in the chromosome, but on the large linear plasmid pSA3239 [10]. However, a recent genomic sequence of this strain (GenBank Acc. No. CP024985) revealed that it was incorrectly administrated by the Czech Collection of Microorganisms (CCM) and is, in fact, Streptomyces lavendulae subsp. lavendulae CCM 3239 [11]. The complete sequence of auricin BGC, including its contiguous regions, revealed its unusual organization. It consists of a central region (aur1A-aur1N), which contains polyketide aglycone biosynthetic genes that were very similar to the angucycline biosynthetic genes. However, a number of auricin-specific tailoring biosynthetic genes were scattered up to 30 kb from this central region. In addition, auricin BGC contains an unusually large number of genes encoding regulatory proteins of different families (Figure 1) [8,12,13].
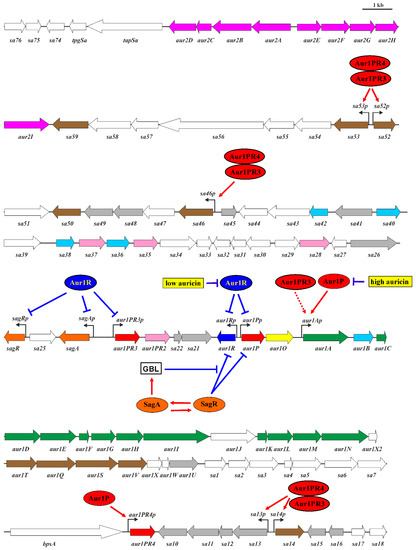
Figure 1.
Auricin BGC and the model of the regulation of auricin biosynthesis [8,12,20]. Details of individual genes and their products are provided in GenBank Acc. No. KJ396772. The green arrows correspond to the auricin aglycone biosynthetic genes; the grey arrows to putative polyketide biosynthetic genes; the brown arrows to D-forosamine biosynthetic and attachment genes; the yellow arrow to the aur1O gene; the red arrows to the positive regulatory genes, with red-dark color having a role in regulation of auricin biosynthesis and red-light color having no role in regulation; the blue arrows to the negative regulatory genes, with blue-dark color having a role in regulation of auricin biosynthesis and blue-light color having no role in regulation; the orange arrows to the GBL autoregulator-receptor system genes; the purple arrows to the other type II PKS BGC. Bent arrows indicate the position and direction of promoters. Red lines ending with an arrow indicate transcriptional activation, whereas blue lines ending with a perpendicular line indicate repression. Indirect activation is indicated by a broken arrow.
Interestingly, in the liquid medium, auricin is transiently produced during a narrow interval after entering the stationary phase, after which it degrades to inactive metabolites due to its instability at high pH values, which are reached later in the stationary phase [14]. Structural analysis revealed that auricin has interesting structural features that set it apart from all other known angucyclines. It is modified with d-forosamine and contains a unique aglycone similar to griseusin-like spiroketal pyranonaphthoquinones [15].
This unusual production of auricin is due to its strict and complex regulation of its biosynthesis, which involves both forward and feedback control by auricin intermediates upon interaction with several pathway-specific regulators [8,14]. Some of them have already been characterized. Aur1P, which belongs to the ARR family, is a key positive regulator. It activates the expression of the core auricin biosynthetic genes (aur1A-aur1U) (Figure 1) from the aur1Ap promoter [16]. This activation is abolished by some auricin intermediates to create feedback control of auricin production [14]. The Aur1R repressor belonging to the TetR family suppresses the expression of the aur1P gene by direct binding of its aur1Pp promoter. Some auricin intermediates break this bond [17]. Two other characterized positive regulators, Aur1PR3 and Aur1PR4, belong to the SARP family. They play a role in the regulation of auricin-tailoring biosynthetic genes. In addition, the expression of their genes is under different control with Aur1R and Aur1P [12,18,19]. At the global level, the SagA/SagR GBL autoregulator-receptor system controls the expression of pathway-specific aur1P and aur1R regulatory genes. In addition, this GBL system is regulated by a feedback mechanism with Aur1R [20]. Auricin regulation is summarized in Figure 1.
The aim of the present study was to characterize the role of another putative aur1O regulatory gene, located just behind the key auricin-specific regulatory gene aur1P, in the regulation of auricin. For this purpose, the aur1O gene was inactivated and auricin production was examined in the resulting mutant. Gene expression analysis using S1-nuclease mapping was used to characterize the expression of the aur1Ap biosynthetic promoter and its dependence upon aur1O. The putative role of the Aur1O as a co-activator of Aur1P was investigate in a heterologous system using the luciferase reporter gene. In addition, the interaction between Aur1O and Aur1P were investigated by a bacterial two-hybrid system. The results showed that Aur1O binds Aur1P and is a specific coactivator of this key auricin-specific positive regulator. Bioinformatics analysis of Aur1O in other BGCs revealed that Aur1O and its homologues represent a new family of transcriptional coactivators involved in the regulation of secondary metabolite biosynthesis, which have two different interacting positive regulators as partners. Their genes are present predominantly in Streptomyces spp. and are found mainly in angucycline BGCs.
2. Results and Discussion
2.1. Characterization of the aur1O Gene in Auricin Biosynthesis
We have previously identified the auricin-specific activator Aur1P required for auricin production [16]. Its gene is followed by the aur1O gene, and both genes form an operon driven by the aur1Pp promoter (Figure 1), because no other promoter has been identified before the aur1O gene. To investigate the role of the aur1O gene in auricin biosynthesis, the gene was inactivated in S. lavendulae subsp. lavendulae CCM 3239 using a PCR targeting system designed to disrupt Streptomyces genes [21]. The apramycin-resistance (AprR) marker gene was used to replace the entire aur1O gene. The strategy resulted in four independent mutant S. lavendulae ∆aur1O::AprR clones, which were verified by Southern blot hybridization analysis (Figure 2). All clones had similar phenotypes and the deletion of aur1O did not affect growth and differentiation (data not shown). One representative mutant strain was selected for further study.
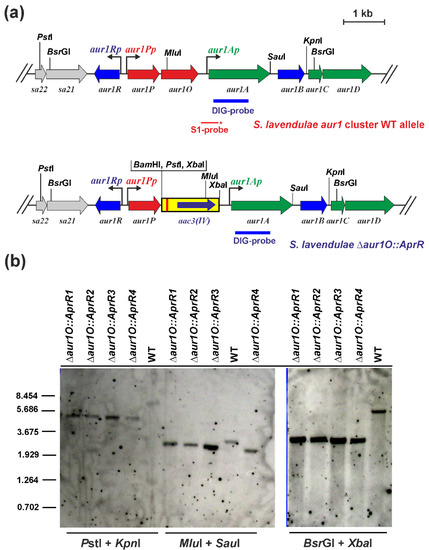
Figure 2.
(a) Physical map of the aur1 BGC around the aur1O gene in wild-type S. lavendulae subsp. lavendulae CCM 3239 [8] and a disrupted aur1O gene allele. Genes are indicated by arrows. The green arrows correspond to the auricin biosynthetic genes, the red arrows to the positive regulatory genes, and the blue-arrows to the negative regulatory genes. The yellow box indicates the AprR aac3(IV) gene with oriT origin of transfer from pIJ773 [21]. The bent arrows indicate the positions and directions of transcription from the indicated promoters. The thin red line below the map represents the DNA fragment used as a probe in S1 nuclease mapping (the 5′-labelled end is marked with an asterisk). The blue bar below the maps represents the probe used for Southern hybridization analysis. Relevant restriction sites are indicated. (b) Southern blot hybridization analysis of chromosomal DNA from four independently obtained S. lavendulae ∆aur1O::AprR clones and wild-type S. lavendulae subsp. lavendulae CCM 3239 (WT) as reference. 1 μg of DNA from the corresponding strain was digested with the indicated restriction endonucleases, separated by electrophoresis in a 0.8% (w/v) agarose gel, and transferred to a Hybond N membrane (Roche) as described in [22]. Hybridization was performed according to the standard DIG protocol (Roche, Germany) using a DIG-labeled probe covering the aur1A gene. Lambda DNA-BstEII digest was used as a size standard.
Auricin production in the prepared S. lavendulae ∆aur1O::AprR mutant strain was analyzed as described previously [12]. The S. lavendulae ∆aur1O::AprR mutant strain, as well as wild-type as a control, were cultured in liquid Bennet medium, and ethyl acetate extracts were prepared from several time points. Extracts were analyzed by TLC followed by biochromatography with Bacillus subtilis as the test strain. In parallel, the extracts were analyzed by HPLC. The deletion of aur1O had a dramatic effect on auricin production. The yellow spots and inhibition zones corresponding to auricin (Rf = 0.13) in the mutant were smaller than in the wild-type strain at all time points examined (Figure 3a,b). HPLC analysis similarly showed a significant reduction in the peak corresponding to auricin in the mutant extract, compared to the wild type (Figure 3c). Determination of the auricin level revealed a 4.6-fold decrease in the aur1O mutant, compared to the wild-type strain. To confirm that this reduced peak in the aur1O mutant corresponds to auricin, this peak was isolated and high resolution ESI MS analysis revealed a molecular ion [M + H]+ at m/z 542.2014. This value corresponded exactly to auricin [14]. To verify that this phenotype was due to deletion of aur1O, the mutant strain S. lavendulae ∆aur1O::AprR was complemented in trans by transformation with plasmid pAPHII15-aur1O, which contained the aur1O gene under the control of the ermEp* promoter [23]. The level of auricin in the complemented strain was similar to that of wild-type S. lavendulae subsp. lavendulae CCM 3239 (data not shown). It confirmed that the decrease of auricin in the mutant strain S. lavendulae ∆aur1O::AprR is indeed due to the deletion of aur1O. These results suggest that the aur1O gene has no role in auricin biosynthesis but plays an important role in the positive regulation of auricin biosynthesis.
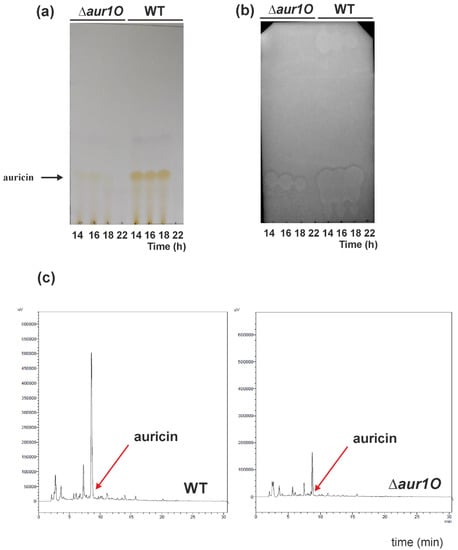
Figure 3.
Analysis of auricin production in wild-type (WT) and aur1O mutant by TLC (a) followed by biochromatography with B. subtilis test strain. (b) 10 μL of ethyl acetate extracts from each strain grown in Bennet medium to indicated time points were separated by TLC. For TLC biochromatography, 1 μL aliquots of the ethyl acetate extracts were separated by TLC and overlaid with B. subtilis as described in Material and Methods. Arrows indicate the yellow spot and the inhibition zone corresponding to auricin [14]. (c) HPLC analysis of auricin production by wild-type S. lavendulae subsp. Lavendulae CCM 3239 (WT) and aur1O mutant (∆aur1O) strains. 10 μL of the ethyl acetate extracts from each strain grown 14 h in Bennet medium were analyzed (details described in Material and Methods). The arrow indicates the position of the auricin peak.
2.2. Transcriptional Analysis of the aur1Ap Promoter in the aur1O Mutant
We have previously characterized the aur1Ap promoter (Figure 1), which directs the expression of the aur1A-aur1U operon containing the auricin biosynthetic genes of its BGC. During growth in liquid Bennet medium, the aur1Ap promoter in not active in the exponential phase and is induced upon entry into the stationary phase. In addition, the promoter is induced at the beginning of aerial mycelium formation during growth and differentiation on solid Bennet medium. The activity of the promoter was dependent upon the auricin-specific transcriptional activator Aur1P, which binds directly to the promoter to activate its transcription [8,9,16]. To investigate whether the deletion of aur1O affects the transcription of the aur1Ap promoter, S1-nuclease mapping was performed using RNA isolated from wild-type S. lavendulae subsp. lavendulae CCM 3239 and mutant S. lavendulae ∆aur1O::AprR strains at different growth stages in liquid Bennet medium. A single RNA-protected fragment was identified with the level of aur1Ap mRNA induced after entering the stationary phase of growth in the wild-type strain, similar to previous reports [9,16,18]. However, the level of aur1Ap mRNA from all time points was dramatically reduced in the aur1O mutant. No RNA-protected fragment was identified by tRNA control (Figure 4a). As an internal control of RNA quality, S1-nuclease mapping was performed with the same RNA samples using a probe for the hrdBp2 promoter, which is continuously expressed during growth [24]. RNA-protected fragments of similar intensities corresponding to the hrdBp2 promoter were identified in all RNA samples (Figure 4b). These results suggest that the aur1Ap promoter is dependent on the aur1O gene. Decreased transcription of this first aur1Ap biosynthetic promoter is probably responsible for reduced auricin production in the aur1O mutant.
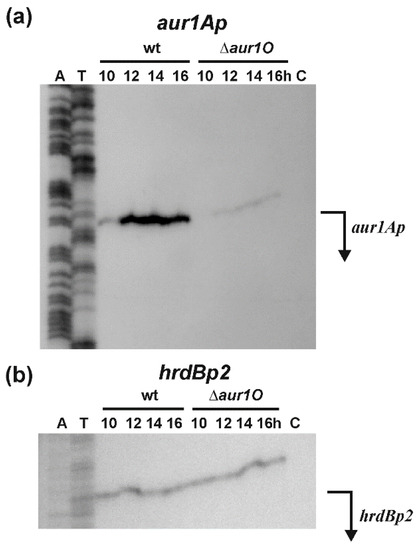
Figure 4.
(a) High-resolution S1-nuclease mapping of the transcription start site (TSS) for the aur1Ap promoter in wild-type S. lavendulae subsp. lavendulae CCM 3239 (WT) and aur1O mutant strains (∆aur1O). The 5′-labelled DNA fragment (Figure 2a) was hybridized to RNA isolated from cultures grown in liquid Bennet medium at the indicated time points (corresponding to different growth phases). E. coli tRNA was used as a control (lane C). (b) Control S1-nuclease mapping with the same RNA samples and a DNA probe for the hrdBp2 promoter [24]. The RNA-protected DNA fragments were analyzed on DNA sequencing gels together with G+A (lane A) and T+C (lane T) sequencing ladders derived from the end-labelled fragments [25]. Thick bent horizontal arrows indicate the positions of RNA-protected fragments corresponding to TSS of the promoters.
2.3. Activation of the aur1Ap Promoter by aur1P and aur1O in the Heterologous System
Because both aur1P and aur1O genes form an operon, it is possible that both are involved in activating the aur1Ap promoter. Aur1P contains the HTH DNA binding domain at the N-terminus and directly binds to and activates the aur1Ap promoter [16]. However, Aur1O does not contain any DNA-binding motif or other conserved domain. Therefore, it may affect the binding of Aur1P to the aur1Ap promoter. We used a heterologous system to test the effect of Aur1P and Aur1O on the transcription of the aur1Ap promoter. A DNA fragment containing the aur1Ap promoter, including its Aur1P-binding site [16], was cloned into the luciferase reporter vector pMU1s*. This PhiBT1 phage-based integration plasmid contains the synthetic luxCDABE operon and is able to integrate in a single copy into the Streptomyces chromosome [26]. The resulting recombinant plasmid pMU1s-aur1Ap (Figure 5a) was conjugated to the heterologous host strain Streptomyces coelicolor M1146 [27] and the luminescence of eight independent AprR clones was determined during differentiation on solid Bennet medium. The luminescence of all clones was at the background level of approximately 200 relative luminescence units (RLU), indicating that the aur1Ap promoter is not active in the heterologous S. coelicolor M1146 strain (Figure 5b). The aur1P gene, including its aur1Pp promoter, was inserted into pMU1s-aur1Ap, resulting in pMU1s-aur1PAp (Figure 5a), which was similarly conjugated to S. coelicolor M1146, and the luminescence of eight independent AprR clones was determined. The activity of the aur1Ap promoter in this construct increased during growth and its maximum (on average 118,030 RLU) coincided with the onset of aerial mycelium formation (Figure 5b). In general, the production of secondary metabolites coincides with the initiation of aerial mycelium formation, and the aur1Ap promoter was similarly induced in this stage in original S. lavendulae subsp. lavendulae CCM 3239 strain [9]. This result confirmed the activation of the aur1Ap promoter by the auricin-specific transcriptional activator Aur1P also in the heterologous S. coelicolor M1146 strain. In contrast to aur1Ap, the aur1Pp promoter is highly active in the S. coelicolor M1146 heterologous system with a similar time course as the activated aur1Ap promoter, with increasing activity from onset to maximum at 64 h (on average 65,050 RLU) (data not shown). In S. lavendulae subsp. lavendulae CCM 3239, both negative regulators, SagR (GBL receptor) and Aur1R (pathway-specific pseudo GBL receptor), bind the aur1Pp promoter to downregulate its activity prior to auricin production (Figure 1). The heterologous S. coelicolor M1146 strain contains the homologue of SagR (ScbR, SCO6265), but does not contain the homologue of Aur1R (ScbR2, SCO6286), because this strain contains a deletion of the cpk BGC from SCO6270 to SCO6286 [27]. The high increasing activity of the aur1Pp promoter in this S. coelicolor M1146 heterologous system suggests that the homologous GBL receptor ScbR did not significantly affect the activity of the aur1Pp promoter and consequently the level of Aur1P under these conditions. The aur1P-aur1O operon, including the aur1Pp promoter, was similarly inserted into pMU1s-aur1Ap, resulting in pMU1s-aur1POAp (Figure 5a). This construct was similarly conjugated to S. coelicolor M1146 and the luminescence of eight independent clones was determined. The activity of the aur1Ap promoter in this construct was 3.22-fold higher (on average 380,814 RLU) and its maximum similarly coincided with the onset of aerial mycelium formation (Figure 5b). This result showed a positive effect of Aur1O on the activation of the aur1Ap promoter by Aur1P.
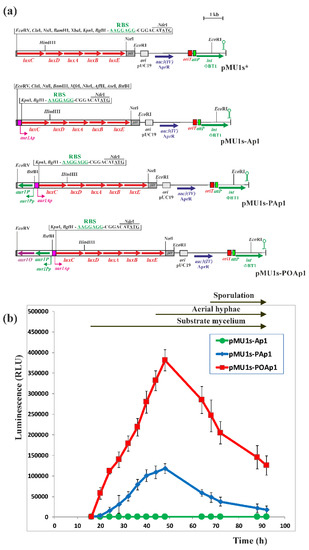
Figure 5.
Activation of the aur1Ap biosynthetic promoter by Aur1P and Aur1O in the heterologous S. coelicolor M1146 system. (a) Maps of the luxCDABE reporter plasmid pMU1s* [26] and recombinant plasmids containing the aur1Ap promoter with and without the aur1P and aur1O genes. Arrows indicate corresponding genes and bent arrows promoters. The plasmids contain the synthetic operon luxCDABE, the aac3(IV) AprR gene, the oriT transfer origin, the E. coli ColE1 replication origin from pUC19, phage fd, major transcription terminator, and attP attachment site and ΦBT1 phage integrase gene. Relevant restriction sites are indicated. (b) The activity of the luxCDABE operon after fusion with the corresponding promoter DNA fragments. The corresponding plasmids were conjugated to S. coelicolor M1146 and luminescence was measured in a Synergy HT microplate reader in RLU after growth and differentiation on solid Bennet medium in 96-well plates at the indicated time points. Each point represents the mean of eight assays, and error bar indicates the standard deviation from the mean. The arrows above the graph indicate the positions of developmental stages.
2.4. aur1O Interacts with aur1P
The results of the above experiments suggest that Aur1O together with Aur1P activate the aur1Ap promoter. Because Aur1O does not contain a DNA binding domain, it can exert its activation function on the aur1Ap promoter through interaction with Aur1P. A bacterial two-hybrid (BACTH) system was used to study the interaction between these proteins. This system is based on functional complementation between Bordetella pertussis adenylate cyclase fragments T18 and T25, expressed separately from the two compatible plasmids pKT25 and pUT18C [28]. The full-length aur1P and aur1O genes were fused as C-terminal fusions in plasmids pKT25 and pUT18C, resulting in pKT25-aur1P, pKT25-aur1O, pUT18C-aur1P, and pUT18C-aur1O. These constructs (including the pKT25+pUT18C negative control) were co-transformed into E. coli BTH101 and screened on LB medium with isopropyl-β-d-thiogalactopyranoside (IPTG) and X-Gal. The negative control correctly created white colonies. Both combinations showed a LacZ+ phenotype and formed blue colonies (Figure 6a). Measurement of β-galactosidase activity in sets of three independent experiments for each combination of plasmids was used to quantify protein interactions. The level of β-galactosidase activity was similar in both combinations and significantly higher than in the negative control (Figure 6b). The results showed that Aur1O interacts with Aur1P. Thus, the role of Aur1O may be to coactivate the auricin-specific activator Aur1P.
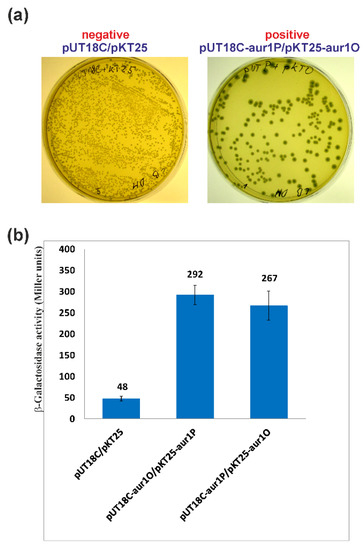
Figure 6.
Analysis of the Aur1O-Aur1P interaction using the BACTH system [28]. Combinations of the plasmids were transformed into E. coli BTH101 and screened on solid LB medium supplemented with ampicillin (Amp), kanamycin (Kan), IPTG, and X-Gal. (a) Example of plates with negative and positive combinations of plasmids. (b) β-galactosidase activity for each plasmid combination determined in triplicates as described in Materials and Methods. The error bars indicate standard deviations from the mean (given by the values above each bar).
2.5. Presence of aur1O Homologues in Other BGCs
The aur1O gene product showed high similarity to various proteins of unknown function (Figure 7a), whose genes were found in several antibiotic BGCs, especially in Streptomyces spp. (Figure 8). This suggests that these homologues could play a similar role in their corresponding BGCs. All, including Aur1O, do not contain any conserved domain. Most BGCs containing the gene encoding the aur1O homologue belong to the type II PKS for angucycline aromatic polyketides, including the aur1 for auricin; cha is responsible for the production of chattamycin in S. chattanoogensis [29], saq for saquayamycin in Micromonospora sp. Tu6368 [30], pga for gaudimycin in Streptomyces sp. PGA64 [31], sch for angycyclines Sch 47554 and Sch 47555 in Streptomyces sp. SCC-2136 [32], gcn for grincamycin in S. lusitanus SCSIO LR32 [33], ovm for oviedomycin in S. antibioticus ATCC 11891 [34], and lac for unknown angycycline in S. lavendulae FRI-5 [35]. However, some of the BGCs are responsible for the biosynthesis of secondary metabolites belonging to other structural types; med belongs to type II PKS for the aromatic polyketide medermycin in Streptomyces sp. AM-7161, which belongs to the class of pyranonaphthoquinones [36], tyl belongs to type I PKS responsible for the macrolide polyketide tylosin in S. fradiae [37], two BGCs, epa and phz, are responsible for the biosynthesis of phenazines in Kitasatospora sp. HKI 714 and S. tendae Tue1028 [38,39], and nap is responsible for napyradomycin in S. tendae Tue1028, which belongs to a specific family of chlorinated meroterpenoid dyhydroquinones [40].
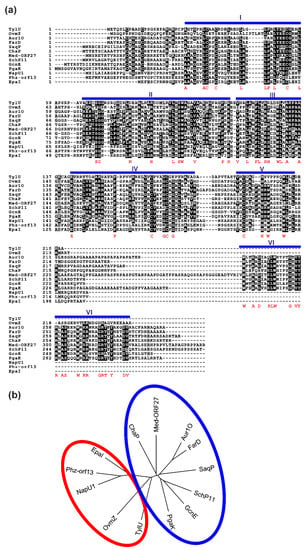
Figure 7.
(a) A comparison of Aur1O with similar proteins from BGCs. Protein sequences (and accession numbers) are as follows: Aur1O of S. lavendulae subsp. lavendulae CCM 3239 (AAK59995), TylU of S. fadiae T59235 tylosin BGC (AAD40806), OvmZ of S. antibioticus ATCC 11891 oviedomycin BGC (CAG14961), FarD of S. lavendulae FRI-5 lac BGC for unknown aromatic polyketide (BAG74709), SaqP of Micromonospora sp. Tu6368 saquayamycin BGC (ACP19350), ChaP of S. chattanoogensis L10 chattamycin BGC (AIU99193), Med-ORF27 of Streptomyces sp. AM-7161 medermycin BGC (BAC79023), SchP11 of Streptomyces sp. SCC-2136 sch BGC for angucyclines Sch 47554 and Sch 47555 (CAH10120), GcnE of S. lusitanus SCSIO LR32 grincamycin BGC (AGO50608), PgaK of Streptomyces sp. PGA64 gaudimycin BGC (AHW57770), NapU1 of Streptomyces sp. CNQ-525 napyradiomycin BGC (ABS50476), Phz-orf13 of S. tendae Tue1028 phenazine BGC (AFS18594), EpaI of Kitasatospora sp. HKI 714 endophenazine BGC (AHW81468). The conserved domains are indicated by blue bars above the sequences. Identical residues are highlighted in black. Similar residues are shaded. The highly conserved residues are listed below the sequences in red. The numbers refer to the deposited amino acid sequences in databases. (b) Phylogenetic tree of these Aur1O homologues; the separate branches are indicated by circles.
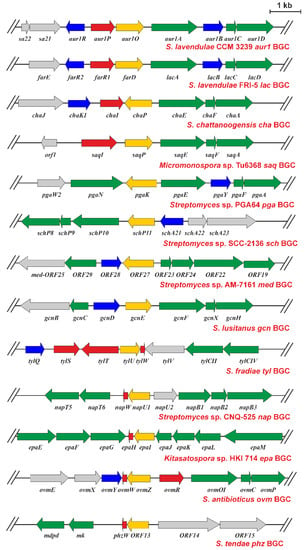
Figure 8.
Physical maps of S. lavendulae subsp. lavendulae CCM 3239 aur1 BGC around the aur1O gene [8] and other BGCs containing the aur1O homologue. Genes are indicated by arrows. The green arrows correspond to the biosynthetic genes, the yellow arrows to the aur1O homologue, the red arrows to the positive regulatory genes, and the blue arrows to the negative regulatory genes. BGC accession numbers are: S. lavendulae FRI-5 lac BGC for unknown aromatic polyketide (AB434932, LC209815), S. chattanoogensis L10 chattamycin (cha) BGC (KM264312), Micromonospora sp. Tu6368 saquayamycin (saq) BGC (FJ670504), Streptomyces sp. PGA64 gaudimycin (pga) BGC (AY034378), Streptomyces sp. SCC-2136 angucyclines Sch 47554 and Sch 47555 (sch) BGC (AJ628018), Streptomyces sp. AM-7161 medermycin (med) BGC (AB103463), S. lusitanus SCSIO LR32 grincamycin (gcn) BGC (KC962511), S. fradiae T59235 tylosin (tyl) BGC (AF145049), Streptomyces sp. CNQ-525 napyradiomycin (nap) BGC (EF397639), Kitasatospora sp. HKI 714 endophenazine (epa) BGC (KJ207079), S. antibioticus ATCC 11891 oviedomycin (ovm) BGC (AJ632203), S. tendae Tue1028 phenazine (phz) BGC (JQ659263).
There are only two reports in which the function of the Aur1O homologue was partially characterized. In the first, the tylU gene was identified in type I PKS BGC for tylosin in S. fradiae. Its product has been shown to be important for tylosin production because deletion of tylU resulted in reduced tylosin yield of approximately 80%. Tylosin production is regulated by several regulators, and TylU is thought to play a regulatory role in the production of tylosin as a helper protein for the tylosin positive SARP-family regulator TylS. TylU is thought to promote the binding of TylS to a promoter that directs the expression of another tylosin-specific activator gene tylR, whose product is directly involved in the activation of tylosin biosynthesis genes [37]. These results of the role of TylU in tylosin production are similar to our case, with the difference that the auricin-specific activator Aur1P does not belong to the SARP family, but to the ARR family [14,16,41]. In addition, both Aur1P and Aur1O proteins directly activate the aur1Ap biosynthetic promoter. In two other reports, the ovmZ gene was identified in the type II PKS BGC for the angucycline antibiotic oviedomycin. The ovmZ gene identified in the S. antibioticus ATCC 118911 has not been characterized [34]. However, in homologous oviedomycin BGC in S. ansochromogenes, its product was shown to be important for oviedomycin production because deletion of ovmZ led to loss of oviedomycin production. Interestingly, ovmZ was co-transcribed with the small downstream ovmW gene (Figure 8), which was similarly necessary for oviedomycin production. Like OwmZ, OvmW does not contain any conserved domain. However, unlike OvmZ, OwmW contains the HTH DNA binding domain of the HTH_17 family (pfam12728). Both genes have been defined as positive regulators necessary for oviedomycin BGC activation. They are interdependent and cooperate as partners. As in our case, expression of both ovmZ/ovmW genes activated the ovmOp biosynthetic promoter fused to the gusA reporter gene in the S. coelicolor M1146 heterologous system [42].
Interestingly, unlike ovmZ, no ovmW orthologue was identified downstream of the aur1O gene, even in the entire auricin BGC, and in several other BGCs, except tyl, nap, epa, and phz (Figure 8). Alignment of the amino acid sequences of Aur1O homologues revealed several conserved domains (Figure 7a). Interestingly, all Aur1O homologues whose genes are translationally coupled with the ovmW homologue (TylU, OvmZ, NapU1, Phz-orf13, EpaI) (Figure 8) are shorter and do not have a conserved C-terminal domain VI containing many aromatic and basic amino acids (Figure 7a). In addition, a phylogenetic tree containing Aur1O homologues (Figure 7b) shows that all homologues that likely cooperatively function with the OvmW homologue belong to a separate branch.
Interestingly, almost all other BGCs lacking the homologous owmW gene contain a gene encoding ARR similar to the auricin-specific Aur1P positive regulator (Supplementary Figure S1). In two cases (chaI, farR1) [29,43], this gene is located near the aur1O homologue (Figure 7). In other cases, they are placed in a different position within the BGC; the homologous pgaR1 gene is located approximately 4.5 kb downstream of the pgaK gene in pga [31], the schA25 gene is located approximately 4.5 kb upstream of the schP11 gene in sch [32], the med-ORF30 gene is located approximately 4 kb downstream of the med-ORF27 gene in med [36], and the gcnR gene is located approximately 27 kb downstream of the gcnE gene in gcn [33]. There is an exception in saq BGC, where the putative transcriptional activator gene saqI, located near saqP (Figure 8), belongs to the StrR family [30]. Both genes were located near the beginning of BGC, so it cannot be rule out that a gene homologous to aur1P is located in a more upstream region, outside the sequenced region. Assuming that both Aur1O and Aur1P homologues can interact as in our case, it is possible that the conserved C-terminal domain specific for these Aur1O homologues may be involved in interacting with these transcriptional regulators. In addition to the C-terminal conserved DNA-binding domain, all these ARRs are conserved in the N-terminal effector domain, which may be involved in this interaction (Supplementary Figure S1).
All these results suggest that the Aur1O homologues represent a new family of transcriptional coregulators that play a role in activating their BGC in association with two different DNA binding regulators. Shorter Aur1O homologues (TylU, NapU1, EpaI, OvmZ, Phz-ORF13) may exert their positive function upon interaction with the product of small ovmW homologous gene, which contains a DNA binding domain, as previously described for oviedomycin BGC activation [42]. Longer Aur1O homologues (Aur1O, FarD, ChaP, SaqP, PgaK, SchP11, Med-ORF27, GcnN), which contain specific conserved C-terminal domain VI (Figure 7a), may similarly interact with their cognate DNA-binding activators (as in the case of Aur1O and Aur1P interaction) to activate the expression of biosynthetic genes in their respective BGCs. These transcriptional activators belong mainly to the ARR family (Supplementary Figure S1).
In conclusion, a new family of transcriptional coactivators was identified. This new family is divided into two distinct subclasses. The shorter coactivators contain only the first five domains (I, II, III, IV, V) (Figure 7a) and can interact with the short OvmW homologue [42]. The longer coactivators contain six conserved domains (I, II, III, IV, V, VI) (Figure 7a) and can interact with transcriptional activators mainly from the ARR family. This type of regulation of antibiotic BGCs is novel. Although many families of transcriptional regulators regulating antibiotic production have been characterized [1,5,6], such coactivation of a DNA binding transcriptional activator with some other associated protein has not been described, with the exception of Aur1O and its two homologues, TylU and OvmZ. Moreover, this type of regulation is likely to be widespread because Blast search in databases revealed homologous aur1O genes in many Streptomyces species sequenced, most of which are found in unknown secondary metabolite BGCs. Examination of the 99 most similar Aur1O homologues (Supplementary Figure S2) revealed that almost all of their genes were located in BGCs (one corresponding gene was not close to any BGC and the three corresponding nucleotide sequence contigs were very short), in most cases in close proximity to the gene encoding the putative ARR activator, as in the cases of aur1O/aur1P. Almost all BGCs belong to angucycline type II PKSs (80 cases) or other type II PKSs (14 cases). However, one BGC belongs to the non-ribosomal peptide synthases (NRPS). Therefore, although rarely, this system may be involved in the regulation of some other BGC classes.
In future, it would be interesting to study this interesting mechanism of this activation in both branches. Structural analysis of both protein pairs would help to elucidate this mechanism and domain interactions. This is a challenge not only for our group, but also other partners who are investigating similar BGCs.
3. Material and Methods
3.1. Bacterial Strains, Plasmids, and Culture Conditions
S. lavendulae subsp. lavendulae CCM 3239 wild type strain [11] was obtained from the Czech Collection of Microorganisms (Brno, Czech Republic). For sporulation, the strain was grown on solid Bennet medium as described in [15]. For the detection of auricin, S. lavendulae subsp. lavendulae CCM 3239 wild type and ∆aur1O mutant strains were cultivated in liquid Bennet medium to various growth phases as previously described [15]. For RNA isolation, 5 × 108 colony-forming units (CFU) of S. lavendulae subsp. lavendulae CCM 3239 wild type and ∆aur1O mutant spores were inoculated into 50 mL Bennet medium in 250 mL Erlenmeyer flasks and the culture was grown on an orbital shaker at 270 r.p.m. and 28 °C to different growth stages. Escherichia coli DH5a (Invitrogen, Waltham, MA, USA) was used as a host for standard cloning experiments. E. coli BW25113/pIJ790 was used as a host for PCR-targeted gene disruption using the AprR plasmid pIJ773, and E. coli ET12567/pUZ8002 was used as a non-methylating host [21]. The Streptomyces integrative plasmid pAPHII15 [44] was used for the complementation studies. The luciferase reporter plasmid pMU1s*containing the synthetic luxCDABE operon was used for detection of promoters in Streptomyces [26]. The conditions for E. coli growth and transformation were described in [22]. Luria–Bertani (LB) medium was used for E. coli growth. If required, the media were supplemented with 100 μg/mL Amp (Sigma-Aldrich, Darmstadt, Germany), 50 μg/mL Apr (Sigma-Aldrich, Darmstadt, Germany), 50 μg/mL Kan (Sigma-Aldrich, Darmstadt, Germany), 40 μg/mL chloramphenicol (Sigma-Aldrich, Darmstadt, Germany), 50 μg/mL streptomycin (Sigma-Aldrich, Darmstadt, Germany).
3.2. Recombinant DNA Techniques
Standard DNA manipulation methods were performed as described in [22]. Chromosomal DNA from S. lavendulae subsp. lavendulae CCM 3239 wild type and ∆aur1O mutant strains was prepared according to [45]. Southern blot hybridization analysis was performed as described in [22]. 1 μg of DNA was digested with restriction enzymes, separated by electrophoresis in a 0.8% (w/v) agarose gel, and transferred to a Hybond N membrane (Roche, Mannheim, Germany). Hybridization was performed according to the standard DIG protocol (Roche, Mannheim, Germany) using a DIG-labelled probe (850-bp DNA fragment covering the aur1A gene, Figure 1a) prepared by PCR amplification using primers aur1AFw and aur1ARv (Supplementary Table S1). Signals were detected by DIG chemiluminescent detection kit using CSPD (Roche, Mannheim, Germany).
3.3. Disruption of the S. lavendulae subsp. lavendulae CCM 3239 aur1O Gene
The PCR targeted REDIRECT procedure [21] was used to delete the entire coding region of the aur1O gene in S. lavendulae subsp. lavendulae CCM 3239. The AprR cassette from template plasmid pIJ773 was PCR amplified using primers Aur1OdDir and Aur1OdRev (Supplementary Table S1). The resulting PCR product was used to electroporate E. coli BW25113/pIJ790 containing the cosmid pCosSA25 [17]. The correct replacement of the aur1O gene in the resulting cosmid pCosSA25-aur1O was verified by restriction mapping. The recombinant cosmid was transformed into the non-methylating E. coli ET12567/pUZ8002 strain and introduced into S. lavendulae subsp. lavendulae CCM 3239 by conjugation. Colonies were screened for AprR and Kan sensitivity, indicating a double crossover. Four such colonies were identified, resulting in mutant strains S. lavendulae ∆aur1O::AprR1, 2, 3, 4, and the correct double-cross event was confirmed by Southern-blot hybridization. All four clones had similar phenotypes. One representative strain was selected for further study. Plasmid pAPHII15-aur1O used to complement the aur1O mutation was prepared by inserting a PCR-amplified DNA fragment containing the entire aur1O gene, including its RBS site, into the integrative Streptomyces expression plasmid pAPHII15 [44]. A 900-bp ClaI-XbaI DNA fragment was prepared by PCR amplification using chromosomal DNA from S. lavendulae subsp. lavendulae CCM 3239 as a template and the primers aur1OCla and aur1OXba (Supplementary Table S1) to introduce a ClaI site upstream and an XbaI site downstream of the aur1O gene. Pfu DNA polymerase (Stratagene, La Jolla, CA, USA) was used to ensure a high fidelity of DNA synthesis during PCR. The DNA fragment was digested with ClaI and XbaI and cloned into pAPHII15 digested with the same enzymes, resulting in pAPHII15-aur1O, which was verified by nucleotide sequencing.
3.4. RNA Isolation and S1-Nuclease Mapping
Isolation of total RNA from S. lavendulae subsp. lavendulae CCM 3239 and high-resolution S1 nuclease mapping were performed according to [46]. RNA samples (40 μg) were hybridized to approximately 0.02 pmol DNA probe labelled at one 5′ end with [γ-32P] ATP (producing approximately 106 dpm/pmol of probe) and treated with 120 U of S1-nuclease (New England Biolabs, San Diego, CA, USA). The DNA probe for the aur1Ap promoter (274 bp DNA fragment) was prepared by PCR amplification using chromosomal DNA from S. lavendulae subsp. lavendulae CCM 3239 as a template and the 5′ end-labelled primer aur1AS1Rev from the aur1A coding region and the unlabeled primer aur1AS1Dir from the aur1A upstream region. The control hrdBp2 promoter DNA probe was described in [24]. Oligonucleotides were labelled at the 5′ end with [γ-32P] ATP (ICN, Costa Mesa, CA, USA, 4500 Ci/mmol) and T4 polynucleotide kinase (New England Biolabs, San Diego, CA, USA) as described in [22]. The protected DNA fragments were analyzed on DNA sequencing gels (6% polyacrylamide containing 8 M urea) along with G+A and T+C sequencing ladders derived from the end-labeled fragments [25].
3.5. Construction of luxCDABE-Based Luciferase Reporter Plasmids and Bioluminescence Measurement
The luciferase reporter plasmid pMU1s* containing the synthetic luxCDABE operon [26] was used to detect promoter activity. A 189-bp DNA fragment containing the aur1Ap promoter, including its Aur1P-binding site [16], was PCR amplified with high fidelity Pfu DNA polymerase (Stratagene, La Jolla, CA, USA), S. lavendulae subsp. lavendulae CCM 3239 chromosomal DNA as a template and the primers aur1ApDir and aur1ApRev (Supplementary Table S1). The fragment was digested with BamHI and KpnI and cloned into pMU1s* digested by the same restriction enzymes, resulting in pMU1s-Ap1. The aur1P gene, including its aur1Pp promoter [16], was similarly PCR amplified using the primers aur1PpDir and aur1PpRev (Supplementary Table S1). The resulting 900-bp DNA fragment was digested with EcoRV and BstBI and cloned into pMU1s-Ap1 digested with the same restriction enzymes, resulting in pMU1s-PAp1. The aur1P-aur1O operon, including the aur1Pp promoter, was similarly PCR amplified using the primers aur1PpDir and aur1OpRev (Supplementary Table S1). The resulting 1900-bp DNA fragment was digested with EcoRV and BstBI and cloned into pMU1s-Ap1 digested with the same restriction enzymes, resulting in pMU1s-POAp1. The nucleotide sequence of all the recombinant plasmids was checked by sequencing using primers luxCrev and pMU1fd (Supplementary Table S1). The resulting recombinant plasmids were conjugated to the heterologous S. coelicolor M1146 host strain [27], and eight independent AprR clones from each construct were selected to determine luminescence. Spores from each clone, approximately 105 CFU, were inoculated into each well of white 96-well plates (Sigma-Aldrich, Darmstadt, Germany) containing 0.25 mL of solid Bennet medium and grown to a confluent lawn. The plates were incubated at 28 °C and luminescence was measured every 4 h in a Synergy HT microplate reader (Bio-Tek Instruments, Winooski, VT, USA) in RLU. The background luminescence of an empty well without inoculated strain was approximately 200 RLU.
3.6. BACTH System to Investigate Protein-Protein Interactions
A well-established BACTH system, which is based on functional complementation of the T18 and T25 adenylate cyclase fragments of Bordetella pertussis in a cya-deficient E. coli BTH101 strain [28], was used to assess the potential interaction between Aur1P and Aur1O proteins. The complete aur1P and aur1O genes were amplified by PCR with high fidelity Pfu DNA polymerase (Stratagene, La Jolla, CA, USA), S. lavendulae subsp. lavendulae CCM 3239 chromosomal DNA as a template and selected primes (Supplementary Table S1) to introduce XbaI and NdeI sites next to the translation initiation codon, and XhoI and KpnI sites downstream of the stop codon. The amplified DNA fragments were digested with XbaI and KpnI and then ligated into the plasmid pKT25 or pUT18C digested by the same restriction enzymes, resulting in the final recombinant plasmids. The aur1P gene was amplified with the primers aur1PDHdir and aur1PDHrev (Supplementary Table S1), and a 771-bp DNA fragment was cloned into pUT18C or pKT25, resulting in pUT18C-aur1P or pKT25-aur1P. The aur1O gene was amplified with the primers aur1ODHdir and aur1ODHrev (Supplementary Table S1), and an 882-bp DNA fragment was cloned into pUT18C or pKT25, resulting in pUT18C-aur1O or pKT25-aur1O. The nucleotide sequence of all the genes was checked by sequencing using primers KT25dir and KT25rev or UT18Cdir and UT18Crev (Supplementary Table S1). Combinations of the resulting pUT18C and pKT25 constructs were co-transformed into a cya-deficient E. coli BTH101 strain and screened on LB medium supplemented with 0.5 mM IPTG (Sigma-Aldrich, Darmstadt, Germany), 40 μg/mL X-Gal (Sigma-Aldrich, Darmstadt, Germany). Amp and Kan were added to select the plasmids and streptomycin to select the strain. Colonies were screened after two days at 30 °C. Quantitative measurements of β-galactosidase activities in sets of three independent transformants for each plasmid combination were performed according to [47]. The statistical significance of datasets was analyzed by two-tailed Student’s t test with p < 0.05 considered as significant.
3.7. Analysis of Auricin Production
Spores (4 × 109 CFU) of S. lavendulae subsp. lavendulae CCM 3239 wild type and ∆aur1O mutant strains were inoculated into 50 mL Bennet medium in 250 mL Erlenmeyer flasks and cultures were grown on an orbital shaker at 270 r.p.m. and 28 °C. 4 mL aliquots were taken from the cultures at various time points and centrifuged at 6000× g for 5 min. The supernatants were extracted twice with an equal volume of ethyl acetate (Merck, Darmstadt, Germany), the residual water was removed with sodium sulfate (Sigma-Aldrich, Darmstadt, Germany) and sodium chloride (Sigma-Aldrich, Darmstadt, Germany), and the extracts were evaporated in vacuo. Such samples were dissolved in 100 μL of 96% (v/v) ethanol (Merck, Darmstadt, Germany) and analyzed by TLC and HPLC as described previously [14]. Briefly, 10 μL aliquots were subjected to TLC analysis on silica gel 60 F254 plates (Merck, Darmstadt, Germany) with n-butanol saturated with water. Dried TLC plates were analyzed by biochromatography, covered with soft nutrient agar [48] containing fresh B. subtilis culture. The plates were incubated at 37 °C for 16 h and visually screened for growth inhibitory zones. HPLC analysis was performed using a Shimadzu LC-10A HPLC system with a SPD-20A UV-VIS detector (Shimadzu, Kyoto, Japan) and detection at 254 nm. Samples of 10 μL were applied to an OmmiSpher 5 C18 column (5 μm, 250 × 4.6 mm, Varian, Lake Forest, CA, USA) and eluted with a linear gradient of acetonitrile (solvent B) and 0.5 % (v/v) acetic acid in water (solvent A) at a flow rate of 1 mL/min. A 30-min linear gradient from 20% to 100% solvent B was used. Auricin showed a retention time of 8.7 min under these conditions and was confirmed by ESI-MS analysis as a molecular ion [M + H] of m/z = 542.2 with positive mode detection [14]. The ESI spectrum was measured using a Velos Pro Hybrid Ion Trap-Orbitrap Mass Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23052455/s1. Reference [49] is cited in the Supplementary Materials.
Author Contributions
J.K. conceived and designed research. E.M., B.S., B.R., D.C., D.H., R.N., R.J. and L.F. conducted experiments. E.M., R.N. and J.K. analyzed data. R.N. and J.K. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Slovak Research and Development Agency under contracts No. APVV-0203-11 and APVV-19-0009 and by VEGA 2/0026/20 grant from the Slovak Academy of Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available on request.
Acknowledgments
We are grateful to Bertold Gust (John Innes Centre, Norwich, UK) for kindly providing all the plasmids and strains used in the PCR-targeting system; the system itself was supplied by Plant Bioscience Ltd. (Norwich, UK). We thank Mervyn Bibb (John Innes Centre, Norwich, UK) for the strain Streptomyces coelicolor M1146, Justin Nodwell (McMaster University, Ontario, Canada) for the plasmid pMU1s*, and Daniel Labant (Institut Pasteur, Paris, France) for the bacterial two-hybrid system strains and plasmids.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Amp | ampicillin |
| Apr | apramycin |
| AprR | apramycin resistance |
| ARR | atypical response regulator |
| B | Bacillus |
| BACTH | bacterial two-hybrid |
| BGC | biosynthetic gene cluster |
| CCM | Czech Collection of Microorganisms |
| CFU | colony forming units |
| GBL | γ-butyrolactone |
| HPLC | high pressure liquid chromatography |
| HTH | helix-turn-helix |
| Kan | kanamycin |
| E | Escherichia |
| LB | Luria–Bertani (medium) |
| IPTG | isopropyl-β-d-thiogalactopyranoside |
| PCR | polymerase chain reaction |
| PKS | polyketide synthase |
| RLU | relative luminescence units |
| S | Streptomyces |
| SARP | Streptomyces antibiotic regulatory protein |
| TLC | thin layer chromatography |
| TSS | transcription start site |
| X-Gal | 5-bromo-4-chloro-3-indolyl-β-d-galactopyranosid |
References
- Bibb, M.J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef]
- Hopwood, D.A. Streptomyces in Nature and Medicine. The Antibiotic Makers; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Wei, J.; He, L.; Niu, G. Regulation of antibiotic biosynthesis in actinomycetes: Perspectives and challenges. Synth. Syst. Biotechnol. 2018, 3, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, X.; Zhan, X.; Mao, X.; Li, Y. The application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front. Microbiol. 2020, 11, 406. [Google Scholar] [CrossRef] [PubMed]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actino-615 bacteria: New persepctives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Wang, X.; Nie, J.; Niu, G. Regulation of Antibiotic Production by Signaling Molecules in Streptomyces. Front. Microbiol. 2019, 10, 2927. [Google Scholar] [CrossRef] [PubMed]
- Kormanec, J.; Novakova, R.; Mingyar, E.; Feckova, L. Intriguing properties of the angucycline antibiotic auricin and complex regulation of its biosynthesis. Appl. Microbiol. Biotechnol. 2014, 98, 45–60. [Google Scholar] [CrossRef]
- Novakova, R.; Bistakova, J.; Homerova, D.; Rezuchova, B.; Kormanec, J. Cloning and characterization of a polyketide synthase gene cluster involved in biosynthesis of a proposed angucycline-like polyketide auricin in Streptomyces aureofaciens CCM3239. Gene 2002, 297, 197–208. [Google Scholar] [CrossRef]
- Novakova, R.; Knirchova, R.; Farkasovsky, M.; Feckova, L.; Rehakova, A.; Mingyar, E.; Kormanec, J. The gene cluster aur1 for the angucycline antibiotic auricin is located on a large linear plasmid pSA3239 in Streptomyces aureofaciens CCM 3239. FEMS Microbiol. Lett. 2013, 342, 130–137. [Google Scholar] [CrossRef][Green Version]
- Busche, T.; Novakova, R.; Al’Dilaimi, A.; Homerova, D.; Feckova, L.; Rezuchova, B.; Mingyar, E.; Csolleiova, D.; Bekeova, C.; Winkler, A.; et al. Complete genome sequence of Streptomyces lavendulae subsp. lavendulae CCM 3239 (formerly “Streptomyces aureofaciens CCM 3239”), a producer of the angucycline-type antibiotic auricin. Genome Announc. 2018, 6, e00103-18. [Google Scholar]
- Bekeova, C.; Rehakova, A.; Feckova, L.; Vlckova, S.; Novakova, R.; Mingyar, E.; Kormanec, J. Characterisation of the genes involved in the biosynthesis and attachment of the aminodeoxysugar D-forosamine in the auricin gene cluster of Streptomyces aureofaciens CCM3239. Appl. Microbiol. Biotechnol. 2016, 100, 3177–3195. [Google Scholar] [CrossRef] [PubMed]
- Mingyar, E.; Novakova, R.; Knirschova, R.; Feckova, L.; Bekeova, C.; Kormanec, J. Unusual features of the large linear plasmid pSA3239 from Streptomyces aureofaciens CCM 3239. Gene 2018, 642, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Kutas, P.; Feckova, L.; Rehakova, A.; Novakova, R.; Homerova, D.; Mingyar, E.; Rezuchova, B.; Sevcikova, B. Strict control of auricin production in Streptomyces aureofaciens CCM 3239 involves a feedback mechanism. Appl. Microbiol. Biotechnol. 2013, 97, 2413–2421. [Google Scholar] [CrossRef] [PubMed]
- Matulova, M.; Feckova, L.; Novakova, R.; Mingyar, E.; Csolleiova, D.; Zduriencikova, M.; Sedlak, J.; Patoprsty, V.; Sasinkova, V.; Uhliarikova, I.; et al. A structural analysis of the angucycline-like antibiotic auricin from Streptomyces lavendulae subsp. lavendulae CCM 3239 revealed its high similarity to griseusins. Antibiotics 2019, 8, 102. [Google Scholar] [CrossRef]
- Novakova, R.; Homerova, D.; Feckova, L.; Kormanec, J. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 2005, 151, 2693–2706. [Google Scholar] [CrossRef] [PubMed]
- Novakova, R.; Kutas, P.; Feckova, L.; Kormanec, J. The role of the TetR-family transcriptional regulator Aur1R in negative regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 2010, 156, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Novakova, R.; Rehakova, A.; Kutas, P.; Feckova, L.; Kormanec, J. The role of two SARP-family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 2011, 157, 1629–1639. [Google Scholar] [CrossRef]
- Rehakova, A.; Novakova, R.; Feckova, L.; Mingyar, E.; Kormanec, J. A gene determining a new member of the SARP family contributes to transcription of genes for the synthesis of the angucycline polyketide auricin in Streptomyces aureofaciens CCM3239. FEMS Microbiol. Lett. 2013, 346, 45–55. [Google Scholar] [CrossRef]
- Mingyar, E.; Feckova, L.; Novakova, R.; Bekeova, C.; Kormanec, J. A γ-butyrolactone autoregulator-receptor system involved in the regulation of auricin production in Streptomyces aureofaciens CCM 3239. Appl. Microbiol. Biotechnol. 2015, 99, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 18, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingstone, R.E.; Moore, D.O.; Seidman, J.S.; Smith, J.A.; Struh, K. Current Protocols in Molecular Biology; Wiley: New York, NY, USA, 1995. [Google Scholar]
- Bibb, M.J.; White, J.; Ward, J.M.; Janssen, G.R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol. Microbiol. 1994, 14, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Kormanec, J.; Farkasovsky, M. Differential expression of principal sigma factor homologues of Streptomyces aureofaciens correlates with the developmental stage. Nucleic Acids Res. 1993, 21, 3647–3652. [Google Scholar] [CrossRef]
- Maxam, A.M.; Gilbert, W. Sequencing end-labelled DNA with base specific chemical cleavages. Methods Enzymol. 1980, 65, 449–560. [Google Scholar]
- Craney, A.; Hohenauer, T.; Xu, Y.; Navani, N.K.; Li, Y.; Nodwell, J. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res. 2007, 35, e46. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D.A. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef]
- Zhou, Z.; Xu, Q.; Bu, Q.; Guo, Y.; Liu, S.; Liu, Y.; Du, Y.; Li, Y. Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. Chembiochem 2015, 16, 496–502. [Google Scholar] [CrossRef]
- Erb, A.; Luzhetskyy, A.; Hardter, U.; Bechthold, A. Cloning and sequencing of the biosynthetic gene cluster for saquayamycin Z and galtamycin B and the elucidation of the assembly of their saccharide chains. ChemBioChem 2009, 10, 1392–1401. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Xiang, S.; Li, L.; Wang, B.; Rajasarkka, J.; Grondahl-Yli-Hannuksela, K.; Ai, G.; Metsa-Ketela, M.; Yang, K. Targeted activation of silent natural product biosynthesis pathway by reporter-gided mutant selection. Metab. Engineer. 2015, 29, 134–142. [Google Scholar] [CrossRef]
- Basnet, D.B.; Oh, T.-J.; Vu, T.T.H.; Sthapit, B.; Liou, K.; Lee, H.C.; Yoo, J.-C.; Sohng, J.K. Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC-2136: Cloning, sequencing, and characterization. Mol. Cells 2006, 22, 154–162. [Google Scholar]
- Zhang, Y.; Huang, H.; Chen, Q.; Luo, M.; Sun, A.; Song, Y.; Ma, J.; Ju, J. Identification of the Grincamycin gene cluster unveils divergent roles for GcnQ in different hosts, tailoring the L-rhodinose moiety. Org. Lett. 2013, 15, 3254–3257. [Google Scholar] [CrossRef] [PubMed]
- Lombo, F.; Brana, A.F.; Salas, J.A.; Mendez, C. Genetic Organization of the Biosynthetic Gene Cluster for the Antitumor Angucycline Oviedomycin in Streptomyces antibioticus ATCC 11891l. ChemBioChem 2004, 5, 1181–1187. [Google Scholar] [CrossRef]
- Pait, I.G.; Kitani, S.; Kurniawan, Y.N.; Asa, M.; Iwai, T.; Ikeda, J.; Nihira, T. Identification and characterization of lbpA, an indigoidine biosynthetic gene in the γ-butyrolactone signaling system of Streptomyces lavendulae FRI-5. J. Biosci. Bioeng. 2017, 124, 369–375. [Google Scholar] [CrossRef]
- Ichinose, K.; Ozawa, M.; Itou, K.; Kunieda, K.; Ebizuka, Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: Towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 2003, 149, 1633–1645. [Google Scholar] [CrossRef]
- Bate, N.; Bignell, D.R.D.; Cundliffe, E. Regulation of tylosin biosynthesis involving ‘SARP-helper’ activity. Mol. Microbiol. 2006, 62, 148–156. [Google Scholar] [CrossRef]
- Heine, D.; Martin, K.; Hertweck, C. Genomics-guided discovery of endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 2014, 77, 1083–1097. [Google Scholar] [CrossRef]
- Saleh, O.; Bonitz, T.; Flinspach, K.; Kulik, A.; Barkard, N.; Muhlenweg, A.; Vente, A.; Polnick, S.; Lammerhofer, M.; Gust, B.; et al. Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Med. Chem. Commun. 2012, 3, 1009. [Google Scholar] [CrossRef]
- Winter, J.M.; Moffitt, M.C.; Zazopoulos, E.; McAlpine, J.B.; Dorrestein, P.C.; Moore, B.S. Molecular basis for chloronium-mediated meroterpene cyclization. J. Biol. Chem. 2007, 282, 16362–16368. [Google Scholar] [CrossRef]
- Wang, L.; Tian, X.; Wang, J.; Yang, H.; Fan, K.; Xu, G.; Yang, K.; Tan, H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Nat. Acad. Sci. USA 2009, 106, 8617–8622. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, J.; Zhuo, J.; Li, Y.; Tian, Y.; Tan, H. Activation and molecular mechanism of a cryptic oviedomycin biosynthetic gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017, 292, 19708–19720. [Google Scholar] [CrossRef]
- Kitani, S.; Iida, A.; Izumi, T.; Maeda, A.; Yamada, Y.; Nihira, T. Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene 2008, 425, 9–16. [Google Scholar] [CrossRef]
- Novakova, R.; Rehakova, A.; Feckova, L.; Kutas, P.; Knirschova, R.; Kormanec, J. Genetic manipulation of pathway regulation for overproduction of angucycline-like antibiotic auricin in Streptomyces aureofaciens CCM 3239. Folia Microbiol. 2011, 56, 278–282. [Google Scholar] [CrossRef]
- Kormanec, J.; Rezuchova, B.; Novakova, R. Screening systems for stable markerless genomic deletions/integrations in Streptomyces species. In Antimicrobial Therapies. Methods in Molecular Biology; Barreiro, C., Barredo, J.L., Eds.; Humana: New York, NY, USA, 2021; Volume 2296, pp. 91–141. [Google Scholar]
- Kormanec, J. Analyzing the developmental expression of sigma factors with S1-nuclease mapping. In Nuclease Methods and Protocols. Methods in Molecular Biology; Chein, C.H., Ed.; Humana Press: Totowa, NJ, USA, 2001; Volume 160, pp. 481–494. [Google Scholar]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Martinez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).