Abstract
Fanconi anemia (FA) is a rare genetic disorder caused by pathogenic variants (PV) in at least 22 genes, which cooperate in the Fanconi anemia/Breast Cancer (FA/BRCA) pathway to maintain genome stability. PV in FANCA, FANCC, and FANCG account for most cases (~90%). This study evaluated the chromosomal, molecular, and physical phenotypic findings of a novel founder FANCG PV, identified in three patients with FA from the Mixe community of Oaxaca, Mexico. All patients presented chromosomal instability and a homozygous PV, FANCG: c.511-3_511-2delCA, identified by next-generation sequencing analysis. Bioinformatic predictions suggest that this deletion disrupts a splice acceptor site promoting the exon 5 skipping. Analysis of Cytoscan 750 K arrays for haplotyping and global ancestry supported the Mexican origin and founder effect of the variant, reaffirming the high frequency of founder PV in FANCG. The degree of bone marrow failure and physical findings (described through the acronyms VACTERL-H and PHENOS) were used to depict the phenotype of the patients. Despite having a similar frequency of chromosomal aberrations and genetic constitution, the phenotype showed a wide spectrum of severity. The identification of a founder PV could help for a systematic and accurate genetic screening of patients with FA suspicion in this population.
1. Introduction
Fanconi anemia (FA) is a rare genetic disorder characterized by chromosomal instability, a high predisposition to physical developmental abnormalities, progressive bone marrow failure, solid tumors, and hematological malignancies [,]. Genetic heterogeneity is a striking feature of FA. Germline pathogenic variants (PV) in 22 FANC genes (FANCA to FANCW) have been so far associated with the FA phenotype. The protein products of the FANC genes participate in the Fanconi anemia/Breast Cancer (FA/BRCA) pathway, a biochemical network that regulates DNA damage repair in response to DNA interstrand crosslinks, maintains genomic stability during DNA replication, and assists other cellular processes [,,].
A large number of genes and hundreds of unique PV have been associated with the FA phenotype, making allelic and locus heterogeneity the rule. Worldwide, the most frequent disease-causing PV among patients with FA occur in FANCA (~64%), FANCC (~12%), and FANCG (~8%) [,]. Recurrent PV have been identified in specific ethnic backgrounds due to a founder effect []. Notable examples, explained by high rates of carrier individuals, include the FANCA c.295C>T in Spanish Gypsies [], FANCC c.456+4A>T in Ashkenazi Jews [], and FANCC c.67delG in the Mennonite Community [,].
Regarding FANCG (MIM#602956), founder PV have been reported in Portuguese-Brazilian (c.1077-2A>G), Korean-Japanese (c.307+1G>C), French-Acadian (c.1480+1G>C), and Black South African (c.637_643del) populations [,]. FANCG gene covers around 6 kilobases (kb) that are mapped to chromosome 9p13; it comprises 14 exons and encodes a 2.6 kb messenger RNA (mRNA) transcript that is translated to a 622 amino acid protein with a molecular weight of 68 kilodaltons [], this protein has seven tetratricopeptide repeat (TPR) motifs, which are required to mediate its protein-protein interactions [,]. FANCG protein is an integral component of the FA-core complex, which is a dynamic E3 ubiquitin ligase protein assembly that catalyzes the mono-ubiquitination of the FANCD2-FANCI heterodimer. This mono-ubiquitination is a critical event in the recruitment of the repair machinery to the DNA lesion as part of the FA/BRCA pathway activation [,,].
Here, we report the novel PV in FANCG:c.511-3_511-2delCA, detected in three patients from the Mixe indigenous community in Mexico. We used a comprehensive molecular and clinical approach to elucidate the consequences of this PV and we provide evidence for a founder effect.
2. Results
2.1. Chromosomal Breakage Analysis Confirmed a FA Diagnosis
Chromosomal breakage analysis with diepoxybutane (DEB) was performed in the three patients to support the suspicion of a clinical FA diagnosis. The number of spontaneous and DEB-induced aberrations, as well as the presence of radial chromosomal figures, were considered criteria to establish the diagnosis of FA. In all three cases, the frequency of radial exchange figures and chromatid breaks showed an increase in DEB-induced samples, compared to those recorded in the spontaneous aberration cultures. Hypersensitivity to DEB treatment in patients was similar to that observed in the FA-positive control (VU817 cell line), while induced aberration frequency observed in the patients was higher than the historical average reported in our laboratory for healthy individuals (negative controls). These results confirmed the FA diagnosis in the patients, who were identified as FANC32, FANC143, and FANC155 (Figure 1).
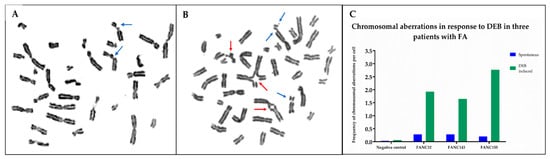
Figure 1.
DEB-induced chromosomal aberrations in lymphocytes from healthy and FA individuals. DEB: diepoxybutane. (A) Representative metaphase from a healthy individual (negative control); chromosome aberrations are mainly chromatid breaks. (B) Representative metaphase from one of the FA patients; increased chromosome aberrations are observed showing chromatid breaks and radial figures, characteristics of patients with FA. Blue arrows show chromatid and isochromatid breaks and red arrows show radial figures. (C) Frequency of chromosomal aberrations per cell for each FA patient analyzed in this study.
2.2. Pathogenic Variant Found in FANCG: c.511-3_511-2delCA Is Predicted to Induce Exon 5 Skipping
Targeted massively parallel DNA sequencing from the three patients with FA revealed a homozygous two-base deletion on FANCG (Supplementary Figure S1A and Table 1). This alteration is located 2 base pair (bp) downstream from the nearest acceptor splice site between intron 4 and exon 5. The genotype was subsequently validated via Sanger sequencing (Supplementary Figure S1B). According to gnomAD (https://gnomad.broadinstitute.org/, accessed on 6 December 2021) this variant (rs1491369358) has only been observed in 1/251,480 alleles, corresponding to a worldwide frequency of 0.00000398. Further analysis of the effect of this novel variant, following the recommended American College of Medical Genetics (ACMG) criteria, supports its pathogenicity.

Table 1.
Features of the pathogenic variant found in FANCG.
Inspection of the genomic region where the variant was found suggests that this change could disrupt mRNA processing. In silico analysis using The Human Splice Finder tool revealed that FANCG:c.511-3_511-2delCA impairs the wild-type acceptor site, most probably affecting splicing. MaxEntScan rated this PV with a score of 1.1, with respect to a reference score of 9.63, predicting a deleterious modification of this splice site. Finally, CRYP—SKYP tool indicated that the deletion may promote exon 5 skipping (probability of cryptic site activation = 0.40). These results consensually predict that this PV in the acceptor splice site leads to loss of FANCG exon 5. The consequence of exon 5 skipping would lead to reading frame shift, and generation of a premature stop codon at position p.S171Vfs5*, resulting in a truncated FANCG protein (Supplementary Figure S1C).
2.3. Reduced Genetic Variation in Locus g.29754068-38771831 Supporting a Founder Effect for FANCG:c.511-3_511-2delCA in the Mixe Population
The three patients with FA described in this study come from an isolated mountainous region in the state of Oaxaca (southern Mexico). They self-identified as belonging to the Mixe indigenous group and shared a surname. Although a kin relationship was unknown, we investigated the possibility of a common haplotype using high-density microarrays (CytoScan 750K array). Several long-contiguous stretches of homozygosity (LCSH) > 3 Mb were shared in autosomal regions by these three patients with FA (Table 2). Some of these LCSH have previously been reported in the HapMap and the 1000 Genomes Project (1KGP) in populations of Asian (CHB), Northern European (CEU) and American (AMR) origin (Supplementary Table S1) [,,,]. We found two shared zones among the LCSH regions not previously reported. The first one in chromosome 5q spanned 2935 kb (g. 101035497-98100800) and the other one in chromosome 9p spanned 9018 kb (g.29754068-38771831); the latter, importantly, included the FANCG locus. Inbreeding coefficient (F), calculated with the total LCSH extension and used to estimate the degree of kinship, suggested a possible parental relationship between the three patients (Table 2).

Table 2.
Native component, homozygosity, and coefficient of inbreeding in the three Mixe patients with Fanconi anemia.
When we analyzed the g.29754068-38771831 region, we discovered that 439 out of 544 single nucleotide variants (SNV) (80.7%) included in the CytoScan 750K array were common among the three patients. We compared the allelic frequency of these SNVs with the frequency reported in the 1KGP for the global and the Mexican population (MXL). We found that 99.32% of the alleles of our patients are coincident frequencies, compared to the global frequencies and the MXL group, whereas the remaining 0.68% has a different frequency. These results confirm that patients share a common haplotype of 30 SNVs around FANCG, supporting the discovery of a founder effect (Supplementary Table S2).
Finally, to ascertain that our patients share the Mexican population (MXL) genetic background, as reported in the 1KGP, a global ancestry analysis was performed. Considering that Mexicans present a remarkable genetic diversity due to population admixture, we decided to contrast all SNVs data of these 3 Mixe patients against other populations beside the MXL. These populations included the following: Native-Americans (NAT), Utah residents with Northern and Western European ancestry (CEU), and Yoruba in Ibadan, Nigeria (YRI). Through similar approach it has been demonstrated that the MXL population presents an ancestry pattern with Native and European components predominance (NAT: 46.59% and CEU: 48.44%, respectively) [,]. Our results indicated that the patients have a proportion of the NAT as high as 79 to 88% (Table 2), with lower CEU and YRI contributions, when compared to the MXL population. Although diversity is observed, the predominance of CEU and NAT ancestry pattern is conserved in both MXL and Mixe patients (Figure 2).
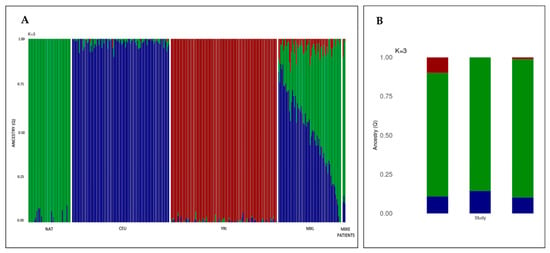
Figure 2.
Mixe patients with FA present an ancestry pattern that have the hallmarks of the genetic diversity of the Mexican population. (A) Barplot showing the inference of global ancestry for the Mixe and Mexican individuals using ADMIXTURE, population reference panels include NAT, CEU, and YRI (K = 3 model). Each individual is depicted as a vertical bar. Colors represent the percentage of ancestry assigned to each cluster for each individual. Green, blue, and red colors indicate Native-Americans (NAT), Northern Europeans (CEU), and Yorubas (YRI) populations, respectively. The two panels located at the extreme right of the figure show the three-way ancestry components for the admixed Mexican (MXL) population and the Mixe patients (last three bars). (B) Detailed bar plot of the three Mixe patients with FA, left to right FANC32, FANC143 and FANC155.
2.4. The Fraction of Pathogenic Variants Linked to a Founder Effect in FANCG Is the Highest among the Most Common FANC Genes
Most founder variants described so far in FA are clustered together in the three most frequently mutated FANC genes (i.e., FANCA, FANCC, and FANCG). The PV described here adds a new founder PV (FPV) in FANCG (Table 3, Figure 3). The literature analysis indicates that the number of FPV, validated through haplotype analysis, was eleven, three, and eight for FANCA, FANCC, FANCG, respectively; this study adds the ninth FPV in FANCG gene. FANCG has the lowest number of unique variants among the most frequently affected genes in FA. There were differences between the proportion of founder and non-founder variants among these genes (Figure 4). The Fisher’s exact test to compare the proportion of FPV between FANCG and FANCA was p < 0.0001, whereas between FANCG and FANCC was p < 0.0001 (Figure 4).

Table 3.
Founder pathogenic variants reported in FANCG.
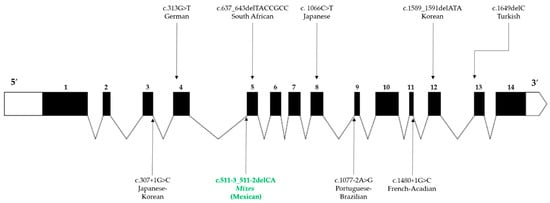
Figure 3.
Worldwide founder pathogenic variants reported in FANCG. The structure of FANCG is described in 5′-3′ orientation. The untranslated regions are illustrated at the ends of the gene (white areas), the exons are represented by numbered black boxes, and the angled lines correspond to the introns. The location of the variants is indicated by vertical arrows, those located at the bottom of the figure represent splicing variants, while deletions and missense variants are located at the top. The variant described in this study is represented by green letters.
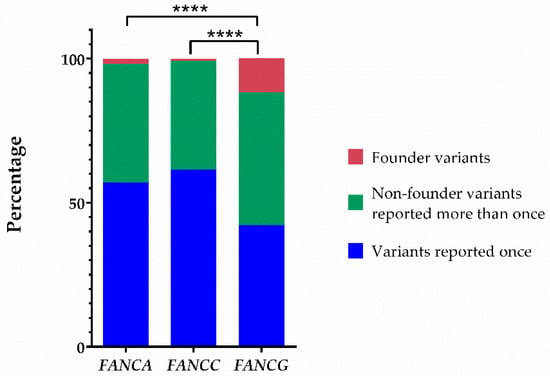
Figure 4.
The proportion of variants with founder effect in FANCG is the highest among the most frequently reported genotypes. Proportions of founder pathogenic variants, non-founder pathogenic variants reported more than once, and pathogenic variants reported just once in FANCA, FANCC, and FANCG. The significance level of Fisher’s exact test is represented by four asterisks (p < 0.0001).
2.5. Evaluation of the Phenotype of Mixe Patients with FA in the Framework of the Reported FANCG Pathogenic Variant
Medical records of each patient were analyzed to identify physical findings, mainly those described in VACTERL-H (Vertebral, Anal, Cardiac, Tracheo-esophageal fistula, Esophageal atresia, Renal, upper Limb, and Hydrocephalus) association (MIM#192350) and PHENOS (skin Pigmentation, small Head, small Eyes, Nervous system, Otology, Short stature) acronyms []. Likewise, hematologic data analysis was used to classify the severity of the bone marrow failure (BMF).
FANC32 patient was the third of four children, born to non-consanguineous parents. His older sister had FA diagnosis and died due to BMF. The patient’s pregnancy was uneventful; he was delivered vaginally at 40 weeks of gestation (WG). At birth his weight was 2500 g (<3rd percentile, HP:0001518) and he had hypotonia (HP:0001252). Developmental milestones were delayed (HP:0012758), beginning to walk at 36 months. He was sent to our clinic at seven years old (y.o.) due to BMF with a history of recurrent upper respiratory tract infections (HP:0002788). Bone marrow aspiration and biopsy performed at age nine y.o. reported 50% cellularity with erythroid series and megakaryocyte predominance and no signs of fibrosis. He was initially treated with steroids and transfusions when required. He later started evaluation for hematopoietic stem-cell transplantation (HSCT); unfortunately, the patient stopped attending follow-up visits. At 14 y.o. anthropometric evaluation showed low weight (−2.31 standard deviation score (SDS), HP:0004325), height (−2.73 SDS, HP:0004322), and head circumference (−2.07 SDS, HP:0000252) for his age and sex. Physical examination revealed thenar hypoplasia of both hands (HP:0001245), generalized hyperpigmentation (HP:0000953), café au lait spots (HP:0000957), and melanonychia (HP:0100644). Cardiac and renal structural abnormalities were not found through sonographic evaluation.
FANC143 patient was the first child of non-consanguineous parents. Her pregnancy was complicated by fetal distress (HP:0025116) and oligohydramnios (HP:0001562) which warranted pregnancy termination by C-section at 36 WG. The birth weight was 1590 g (<3rd percentile, HP:0001518), she remained hospitalized her first month of life. Bilateral radial ray alteration (HP:0410049) and left hip dysplasia (HP:0001385) were identified. She had developmental delay (HP:0012758), particularly affecting language (HP:0002463). The patient was referred to our clinic when she was nearly seven y.o. due to BMF. She was initially treated with transfusions when needed and steroids. Physical examination evidenced low weight (−2.15 SDS, HP:0004325), short stature (−2.90 SDS, HP:0004322), and microcephaly (−3.23 SDS, HP:0000252). She had up-slanted palpebral fissures (HP:0000582), bifid uvula (HP:0000193), bilateral microtia (HP:0008551), with folded pinnae of the right ear (HP:0000396), agenesis of the right thumb (HP:0009777), floating hypoplastic left thumb (HP:0009601), generalized skin hyperpigmentation (HP:0000953), and café au lait spots (HP:0000957). Tympanometry and brainstem auditory evoked potentials showed right unilateral hearing loss (HP:0000365). Spinal X-rays showed spina bifida in S1 (HP:0004614) and a lumbosacral hemivertebra (HP:0008439). Renal and cardiac malformations were ruled out with imaging methods. Brain magnetic resonance imaging (MRI) revealed cortico-subcortical atrophy (HP:0002120 and HP:0012157), supratentorial ventriculomegaly (HP:0002119), and encephalomalacia (HP:0040197). The patient underwent HSCT at 8.6 y.o. and died a month later due to graft versus host disease complications and septic shock.
FANC155 patient was the younger of two children, born to non-consanguineous parents. Her family health history showed that two great-aunts died of breast cancer in their early thirties. She had a maternal uncle with a history of infertility and vertebral defects. The mother reported an uneventful pregnancy. She was vaginally delivered after 40 weeks, with a birth weight of 2400 g (<3rd percentile, HP:0001518). She had developmental delay (HP:0012758) with language disability (HP:0002463). At six y.o. she was evaluated at a local hospital due to a history of recurrent epistaxis (HP:0004406) and pallor (HP:0000980). After evaluation, a diagnosis of aplastic anemia (HP:0001915) was reached. She received blood transfusions when required and steroid therapy. At 12 y.o. she was referred to our clinic for HSCT. Physical examination evidenced weight (−1.26 SDS) and head circumference (−0.86 SDS) within normal range but low height (−2.18 SDS, HP:0004322) for her age and sex. She had up-slanted palpebral fissures (HP:0000582), ptosis (HP:0000508), simple myopic astigmatism (HP:0500041), hypoplastic helix in both ears (HP:0011039), left hypoplastic thenar region (HP:0001245) with limited ipsilateral thumb flexion, generalized hyperpigmentation (HP:0000953), and café au lait spots (HP:0000957). Audiometric findings were unremarkable. Brain MRI, renal ultrasonography, and echocardiography did not show alterations. She underwent HSCT from a non-related donor but died a couple of days after the procedure due to septic shock.
Table 4 summarizes the three patients’ clinical phenotypes. The median age at FA diagnosis was 6.45 y.o. All patients had short stature, skin pigmentation changes, and radial ray anomalies. Respecting VACTERL-H abnormalities, one patient had vertebral and upper limb (radial ray) malformations, and the other two patients had only upper limb alterations; none of them met criteria for VACTERL-H association (≥3/8 features). All three patients had at least three PHENOS anomalies; however, only one had ≥4/6 PHENOS features. Regarding hematologic phenotype, the three patients had moderate to severe BMF.

Table 4.
Summary of the clinical phenotype of the patients described in this study.
We compared the physical manifestations observed in the patients reported here to those described in the literature for cases with PV in FANCG, we excluded reports in which the phenotypes were not mentioned or individually described. We identified 84 cases with PV in FANCG. Remarkably, none of these had any of the 8 founder-effect PV listed in Table 3. The most frequent physical characteristics were short stature (54%), skin pigmentation changes (38%), upper limb (radial ray) abnormalities (31%), microcephaly (29%), and renal malformations (20%). The rest of abnormalities were present in less than 20%. The type and frequency of phenotypic anomalies are depicted in Figure 5. Three out of 84 patients met criteria for VACTERL-H (>3/8 features), seven out of 84 had >4/6 PHENOS features, and only one patient had VACTERL-H plus PHENOS. The median age at diagnosis was 6 y.o. and the median age at report was 9 y.o. The male-to-female ratio was 1:0.64 (p = 0.5).
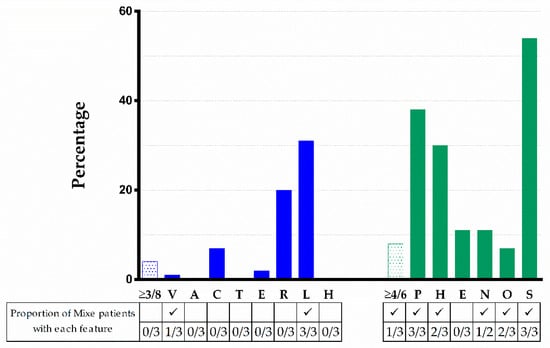
Figure 5.
VACTERL-H and PHENOS features in 84 patients from the literature with a FANCG genotype and comparison with the three patients with FA here described. Upper limb includes abnormal thumb +/− abnormal radius. Nervous system includes structural brain malformations other than hydrocephalus. Otology comprises ear malformations and/or hearing loss. Solid blue or green bars: VACTERL-H or PHENOS; dotted bars: individual findings. Horizontal axis: abnormalities analyzed; vertical axis: percent of total cases with that abnormality. VACTERL-H association (≥3/8 features) was present in 4% and PHENOS (≥4/6 features) in 8% of patients. The proportion of Mixe patients with each feature is shown in the table below the figure. The character ✓ means that this finding was found among Mixe patients. Imaging brain evaluation was performed in only two patients.
3. Discussion
FA is a disorder characterized by high genetic, allelic, and locus heterogeneity. The simultaneous study of several genes using next generation sequencing (NGS)-based targeted panels has advanced the recognition of variants not previously reported within known FANC genes. Applying this NGS strategy, we diagnosed three patients from the same geographical region with a classic FA phenotype. We found a non-previously reported PV: FANCG:c.511-3_511-2delCA, consisting of a two base-pair deletion affecting the splicing acceptor site of FANCG exon 5. This deletion is predicted to lead to an aberrant splicing site and a stop codon, generating a truncated FANCG protein lacking TPR motifs and affecting the function of the FA core complex. Confirmation of a founder effect was obtained by haplotype and ancestry analysis. Overall, our results represent an integrative approach to evaluate the effect of this change in FANCG and provide useful information to guide a potential targeted FA screening on the Mixe population, a well-defined ethnic group in Mexico.
Besides having the same variant, these patients also share the same geographic and ethnic origin, as well as one of their surnames. These data, in addition to the very low reported frequency of this change (rs1491369358), which has been found in a heterozygous state only in one non-FA individual, led us to explore the possibility of a founder effect for PV FANCG:c. 511-3_511-2delCA. The analysis of the degree of inbreeding (F value), the percentage of homozygosity and the regions with LCSH allowed us to corroborate this inference, since all the patients have large stretches of homozygosity in their genomes due to inheritance of identical ancestral genomic segments from both parents. A consequence of this genetic structure is an increased incidence of recessive diseases, like FA. Additionally, the inferred haplotype in the boundaries of the FANCG locus supports the hypothesis of a founder effect in this novel PV. Moreover, the high contribution of the native component of around 80% in these three patients contrasts with the average 46% found in the MXL group and supports the idea that these subjects who belong to the Mixe group have reduced genetic diversity (Figure 2). While only two of these patients have a native contribution of over 85%, sufficient to name them Mexican natives [], the third one who does not reach this threshold also has the lower autosomic homozygosity and the more distant familial relationship, reflecting more mixture and illustrating the origin of mestizo-Mexicans [].
FANCG:c.511-3_511-2delCA is the first FPV reported in Mexican patients with FA and is consistent with the demographic history of the Mixes; an original population that has resisted several attempts to occupy their lands since the pre-Hispanic era and the Spanish conquest. Even though changes have taken place in the political-administrative organization of the region in which they reside since then, this isolated ethnic group has preserved its language, social structure, and territory []. These facts also highlight their sedentary nature, which has led to limited genetic admixture []. Considering the appraised size of the community where they come from [], the calculated prevalence of FA is 7.5/1,000,000, which falls in the upper rank of the estimated worldwide FA prevalence in the general population (1-9/1,000,000) []. Of note, FPV constitute more than 10% of the total PV discovered so far in FANCG, the highest proportion among the most prevalent FANC genes (Figure 4). Although most of the PV in FA are concentrated in FANCA, FANCC, and FANCG, a study suggests that they do not appear to be intrinsically more mutable than the other FANC genes []; therefore, another explanation should be sought to understand the higher prevalence of PV in these genes. On the other hand, it is unlikely that the rate of FPV in FANCG could confer any advantage to carriers, since half of them generate a truncated protein and others a dysfunctional one, so it is likely that they may occur due to chance. Certainly, this observation deserves further investigation.
The patients presented here have a classic FA phenotype, consisting of BMF and physical abnormalities []. The acronyms VACTERL-H and PHENOS include the most frequently reported physical abnormalities in patients with FA [,]. Among these 14 features, only five are present in over 25% of patients: short stature (43%), radial ray defects (40%), changes in skin pigmentation (37%), microcephaly (27%), and renal malformations (27%) []. All three patients described here share height, radial, and skin abnormalities; two also had microcephaly and ultrasonographic evaluations ruled out renal malformations.
Figure 5 shows that 4% of patients reported in the literature with PV in FANCG met criteria for VACTERL-H; if we consider all genotypes, this percentage rises to 12% []. The percentage of patients reported in the literature with PV in FANCG who had ≥4/6 PHENOS features was similar considering all genotypes (8.3 vs. 9%). None of our patients had VACTERL-H and only one had ≥4/6 PHENOS features. None of our patients had evident small eyes, but we did not measure palpebral fissure length, and ocular imaging was not performed. Yet, we did find other ophthalmologic manifestations such as ptosis, myopia, and astigmatism (FANC155). Small eyes are described in 11% of patients with FA regardless of genotype [], this feature stands for the “E” in the PHENOS acronym. However, the ocular phenotype in FA is wider as recently demonstrated [] and broadening the features that conform the “E” in PHENOS should be considered.
As expected, although these three patients share the same pathogenic FANCG genotype and a similar genetic background, their phenotypic presentation was not homogeneous, as evidenced in other cases []. Yet, they illustrate that clinical presentation in FA has a broad spectrum of severity that in this case goes from a florid presentation in FANC143, which includes five PHENOS features and uncommon findings like bifid uvula, to a more discreet presentation in patient FANC155 in whom her unilateral radial abnormality was only recorded after intentional evaluation by a dysmorphologist (Table 4). Concerning the hematologic phenotype, patients with FANCG PV have been found to have more severe cytopenia as well as a higher frequency and an earlier diagnosis of acute myeloid leukemia or myelodysplastic syndrome than patients with FANCA or FANCC genotypes []. All three patients have a hematologic phenotype that merited HSCT, yet platelets and neutrophils appeared to be more severely affected than red cells. Oncologic manifestations were not found in these patients, but this observation is certainly limited not only because of the small number of individuals but also due to their young age.
To our knowledge, the only available phenotypic description of a patient cohort with a FPV FANCG genotype refers to Black South African individuals, with the homozygous variant c.637_643del []. The analysis of 35 patients with this FPV could not definitely establish a particular phenotype for this genotype, yet it was recognized that they had a high frequency of skin pigmentation alterations (97%) and that abnormalities of the upper limbs were frequent (>70%) and subtle (without radial hypoplasia). Also, the frequency of renal abnormalities (37%) was higher than reported elsewhere []. A second study that included 24 patients and focused in endocrinologic data, showed that 33%, 46%, and 42% had microcephaly, short stature, and low weight for age, respectively []. When comparing the phenotype of the three Mixe patients to the Black South African patients, in whom the genotype also leads to a truncated protein (p.Tyr213Lysfs*6), the anthropometric alterations appear to be less severe in the South African patients []. We take these observations with caution, both because of the small size of the Mixe cohort and the influences of environmental factors and additional loci in these multifactorial traits cannot be overlooked. Therefore, a study in this geographical region should be warranted.
In conclusion, in this study we present the ninth FPV in the FANCG gene, thereby contributing to reaffirm that this gene has the highest proportion of FPV among the most frequently affected FANC genes. Despite having similar genetic and environmental backgrounds, the phenotype of these three Mixe patients agrees with the highly variable dysmorphological phenotype of patients with PV in FANCG described in the literature. The identification of the founder FANCG:c.511-3_511-2delCA PV in these patients constitutes a fundamental step for the systematic and accurate genetic screening of patients with FA in the Mixe indigenous group. This could lead to carrier identification, appropriate genetic counseling for families with FA, and optimization of the search for potential bone marrow transplant donors. Finally, the identification of a PV in a population with a high prevalence of the disease represents an opportunity to study the phenotypic effect of the variant in a population with a homogeneous genetic background.
4. Materials and Methods
4.1. Editorial Policies and Ethical Considerations
This study was approved by the Ethics and Research Committees of the Instituto Nacional de Pediatría (INP) in Mexico City. All patients provided written informed consent to participate in this study (INP 041-2014) and blood samples of each participant were collected by peripheral venipuncture.
4.2. Patients
Three patients with cytopenias were independently referred for a medical evaluation to the INP. These patients came from the same geographic region in the southern part of Mexico (Oaxaca State). All patients underwent a comprehensive physical examination by trained medical geneticists. Clinical data including age, sex, parental consanguinity, family history, and anthropometric measurements (weight, height, and head circumference) were recorded. Hematological information, as full blood counts at presentation and subsequent monitoring, bone marrow biopsy results, age of development of bone marrow failure, and transfusions were documented through retrospective review of each patient’s medical file. Available imaging studies were evaluated. For the phenotype description, the Human Phenotype Ontology was used [].
4.3. Chromosomal Breakage Test
A diepoxybutane (DEB) test was performed in all patients. Two-paired lymphocyte cultures per blood sample were cultured in RPMI-1640 medium (Gibco, BRL, Grand Island, NY, USA) and stimulated with phytohemagglutinin (Gibco, BRL, Grand Island, NY, USA). To induce DNA damage, 0.1 μg/mL of DEB (Sigma, St Louis, MO, USA) was added from the start of the incubation to half of the cultures; the untreated cultures were used for the analysis of spontaneous chromosomal breakage. In parallel, cultures of blood samples from healthy individuals as negative controls and the FANCA cell line FA VU817 as a positive control were performed. Harvesting of all cultures was carried out after of 72 h at 37 °C in a 5% CO2 incubator. Metaphase spreads and staining were performed according to standard protocols []. We evaluated the frequency of chromosome aberrations including breaks, fragments, dicentrics, rings, and radial figures. The frequency of aberrations per cell, as well as the percentage of aberrant cells, were calculated.
4.4. Genotyping
4.4.1. Genomic DNA Extraction
The Gentra Puregene Kit (Qiagen, Venlo, Limburg, NL, USA) was used for the extraction of total genomic DNA from peripheral blood samples. The concentrations of the DNA were determined on a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and DNA integrity was verified by agarose gel electrophoresis.
4.4.2. Targeted Next-Generation Sequencing
A customized HaloPlex panel (Agilent Technologies, Santa Clara, CA, USA) was designed to target 16 FA genes (FANC—A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P and Q). The final design included all coding exons and 50 bp of their flanking 5′ and 3′ intronic regions, with a coverage of 99.5%. Agilent’s HaloPlex Enrichment System protocol was followed for library preparation and paired-end sequencing was performed in the MiSeq System platform (Illumina, San Diego, CA, USA). Raw reads were filtered and mapped to the GRCh37/hg19 reference genome with the Burrows-Wheeler Aligner (BWA). The overall mean sequencing depth of the samples was 1000X. Alignment and variant calling were performed using the Genome Analysis Toolkit (GATK), version 4.0.3. A pipeline was designed to maximize the accuracy of variant calls, according to GATK Best Practices recommendations [,]. Variants were described following the guidelines proposed by the HGVS nomenclature []. The classification of variants, according to the five-tier criteria of the ACMG guidelines [] as well as the estimated global frequency, were assessed using the online tool VarSome [].
4.4.3. Sanger Sequencing
FANCG variant-specific primers were designed to encompass the splicing acceptor site between intron 4 and exon 5, using the Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/, accessed on 22 August 2019). The selected primers were as follows: forward 5′- GACCTTGGCGGTAGGCAAA; and reverse 5′- ATTGGGGGAAACTACAGGCA. PCR amplification was carried out from 100 ng of DNA template, according to standard protocols []. PCR products were purified with QIAquick kit (QIAGEN, Venlo, Limburg, NL, USA) according to manufacturer instructions. Purified amplicons were bidirectionally sequenced using the Big Dye Terminator sequencing kit and resolved on an Applied Biosystems 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The electropherograms were analyzed with Chromas V2.6.6 software (www.technelysium.com.au, accessed on 24 January 2020) and target sequences were compared to the corresponding reference sequences FANCG (NG_007312.1) from GenBank (https://www.ncbi.nlm.nih.gov, accessed on 24 January 2020).
4.5. In Silico Splice Site Analysis
Human Splicing Finder (HSF) (http://www.umd.be/HSF/, accessed on 5 February 2020, MaxEntScan (http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html, accessed on 5 February 2020) and CRYP-SKIP (https://cryp-skip.img.cas.cz/, accessed on 7 February 2020) were the three bioinformatics tools used to predict the effect of the variant on splicing signals.
4.6. High-Resolution Microarray Analysis
We performed the analysis of the gDNA with Affymetrix CytoScan 750K Arrays (Affymetrix, Santa Clara, CA, USA) on the three patients. This array provides genome-wide coverage, including 550,000 markers for detecting copy number variation and 200,436 SNP probes. We performed the procedures for DNA digestion, ligation, PCR amplification, fragmentation, labelling, denaturing and hybridization into the array according to the protocols and QC guidelines provided by the supplier. Arrays were then stained and washed in the affymetrix GeneChip Fluidics Station 450 and scanned using an Affymetrix GeneChip Scanner 3000 7G (Affymetrix, Santa Clara, CA, USA); we analyzed the files obtained with the appropriate bioinformatics tools.
4.6.1. Analysis of Long-Contiguous Stretch of Homozygosity
We visualized the LCSH in the software Chromosome Analysis Suite (ChAS) software version 4.1, provided by Affymetrix (Affymetrix, Santa Clara, CA, USA). For the analysis, we used the NetAffx 33 hg19 annotation files (https://www.affymetrix.com/analysis/index.affx, accessed on 20 January 2021). For LCSH > 3Mb, the analysis configuration was set at LOH with marker count = 50 and size = 3000 kb, and for LCSH > 5Mb was set at marker count = 50 and size = 5000 kb.
4.6.2. Estimation of Coefficient of Inbreeding
Individual inbreeding coefficients (F) were estimated using LCSH > 3 Mb data; F was the total length of autosomal LCSH in kb divided by the total autosomal size covered by the Cytoscan 750 array (2,881,033,286 kb for hg19). The F value of 0.25 could reflect a first-degree parental relationship, 0.125 a second-degree, 0.0625 a third-degree and 0.03125 a fourth-degree [].
4.6.3. Haplotype Inference
We used the LSCH > 5 Mb data to perform a haplotype inference approach, specifically the LCSH region shared by the three patients that include FANCG. The number of SNVs identical in the three patients was calculated, and the allele frequency was reviewed to infer the haplotype. The SNVs was compared with those reported in global population and the unrelated Mexican individuals (86 samples, MXL population) found in the 1000 Genomes Project (phase 3) database (1KGP) (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data, accessed on 16 December 2021), using the LDhap tool (https://ldlink.nci.nih.gov/?tab=ldhap, accessed on 16 December 2021).
4.7. Ancestry Analysis
For the ancestry study, CEL files derived from the arrays were analyzed with the ChAS software to obtain the SNV genotyping calls, generated from the use of the BRLMM-P-plus algorithm. Subsequently, these results were contrasted with genotype information derived from Affymetrix high-density arrays present in the 1KGP. PLINK 1.9 software (https://zzz.bwh.harvard.edu/plink/download.shtml/, accessed on 25 January 2021) was used to filter and map the SNVs. SNVs with discordant strings or genotypes were corrected or removed. Finally, the global ancestry of the MXL population and the three patients was deducted through a supervised maximum likelihood ADMIXTURE approach, from K = 2 to K = 3 ancestral components. For this analysis, genotypic frequencies of Northern European (CEU), Yoruba (YRI), and Native American (NAT), reported in the 1KGP, were considered as reference parental populations.
4.8. Fanconi Anemia Variant Database Analysis
We searched the Fanconi anemia Mutation Database (displayed using Leiden Open Variation Database [LOVD, v.3.0], https://www2.rockefeller.edu/fanconi/genes/, accessed on 30 November 2021) for FANCA, FANCC, and FANCG. We first categorized the variant according to the number of times it has been reported. In those where five or more patients were acknowledged, we reviewed the cited publications in order to establish if a haplotype analysis demonstrated a founder effect. We complemented this with a PubMed/MEDLINE search using the terms: “FANCA OR FANCC OR FANCG” AND “mutation OR variant” AND “founder OR haplotype”. We classified the variants as founder PV (FPV), when demonstrated by haplotype analysis, repeatedly reported non-founder PV (recurrent, but not demonstrated as FPV by haplotype analysis), and those reported just once. We compared the proportion of these variant categories between FANCG and the other two genes FANCA and FANCC. Fisher’s exact test (p-values < 0.05) was used to statistically test these differences.
4.9. Phenotype Analysis
We searched PubMed/MEDLINE for publications limited to human subjects through 1 December 2020, using the terms “Fanconi” AND “anemia”. We collected and analyzed all cases with a FANCG genotype in whom individual patient’s phenotypes were detailed. We described frequencies of VACTERL-H (Vertebral, Anal, Cardiac, Tracheo-esophageal fistula, Esophageal atresia, Renal, upper Limb, and Hydrocephalus) and PHENOS (skin Pigmentation, small Head, small Eyes, Nervous system, Otology, Short stature) features. VACTERL-H association was considered if the patient had ≥3/8 features, and for PHENOS ≥ 4/6 features were needed. We extracted clinical information from publications reporting in FANCG FPV (as described in Section 4.8) to compare it to the phenotype of the patients reported here. The statistical analyses were performed using Microsoft Excel Office 365 (Microsoft, Redmond, WA, USA) and R (R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/, accessed on 16 December 2021. We used Fisher’s exact test and p-values < 0.05 were significant.
Supplementary Materials
Supplementary Materials can be found at: https://www.mdpi.com/article/10.3390/ijms23042334/s1.
Author Contributions
Conceptualization, B.G.-d.T., L.T. and S.F.; Data curation, B.G.-d.T., U.J., M.O.F.-R., L.T. and S.F.; Formal analysis, P.R., B.G.-d.T., U.J., F.P.-V., M.O.F.-R., A.R., B.M., M.T.V.-M., J.M.-Z., L.T. and S.F.; Funding acquisition, A.C., L.T. and S.F.; Investigation, S.F.; Methodology, P.R., M.O.F.-R., B.M., M.T.V.-M. and L.T.; Project administration, L.T. and S.F.; Resources, L.T. and S.F.; Software, U.J., F.P.-V. and L.T.; Supervision, S.F.; Validation, P.R.; Writing—original draft, P.R. and L.T.; Writing—review & editing, B.G.-d.T., U.J., F.P.-V., M.O.F.-R., A.R., B.M., M.T.V.-M., J.M.-Z., A.C., L.T. and S.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by Consejo Nacional de Ciencia y Tecnología (CONACyT), grant number SALUD-2014-1-233721, and by Instituto Nacional de Pediatría, Recursos Fiscales E022 project INP 2014/41.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Instituto Nacional de Pediatría (INP), Ciudad de Mexico, Mexico (protocol code: INP 2014/041; date of approval: 17 July 2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study to publish this paper.
Data Availability Statement
Available upon request, following the approved ethics committee guidelines.
Acknowledgments
We would like to thank the patients for their consent to participate in this study. We would like to acknowledge Yañez J., Paul Gaytan P., Lopez E., and Becerra S. from the Sequencing and Synthesis Unit of the Instituto de Biotecnologia, UNAM for primer synthesis and sequencing services. Pedro Reyes acknowledges the support of CONACYT, through the scholarship 899784, to develop his PhD in Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| 1KGP | 1000 Genomes Project |
| ACMG | American College of Medical Genetics |
| AMR | Ad Mixed American (1KGP super population) |
| ANC | Absolut Neutrophils Counts |
| bp | base pair |
| BMF | Bone marrow failure |
| CNS | Central nervous system |
| CEU | Utah residents (CEPH) with Northern and Western European ancestry (1KGP population) |
| ChAS | Chromosome Analysis Suite |
| CHB | Han Chinese in Beijing, China (1KGP population) |
| DEB | Diepoxibutane |
| FA | Fanconi anemia |
| FA/BRCA | Fanconi anemia/Breast Cancer pathway |
| FPV | Founder pathogenic variant |
| Hb | Hemoglobin |
| HGVS | Human Genome Variation Society |
| HSCT | Hematopoietic stem-cell transplantation |
| HSF | Human Splicing Finder |
| Hg19 | Homo sapiens (human) genome assembly GRCh37 |
| INP | Instituto Nacional de Pediatría |
| kb | kilobases |
| LCSH | Long-contiguous stretches of homozygosity |
| MCV | Mean Corpuscular Volume |
| MRI | Magnetic resonance imaging |
| mRNA | messenger RNA |
| MXL | Mexican Ancestry in Los Angeles CA, USA (1 KGP population) |
| MT | Mutant |
| NAT | Native Americans (1KGP population) |
| NGS | Next generation sequencing |
| NMD | Non-sense mediated decay |
| OMIM | Online Mendelian Inheritance in Man |
| PCR | Polymerase chain reaction |
| PHENOS | Skin Pigmentation, small Head, small Eyes, Nervous system, Otology, Short stature |
| PLT | Platelets |
| PVS1 | Pathogenic Very Strong 1 |
| PM2 | Pathogenic Moderate 2 |
| PP3 | Pathogenic Supporting 3 |
| PDB | Protein Data Bank |
| PV | Pathogenic variants |
| SNV | Single nucleotide variant |
| SDS | Standard deviation score |
| TPR | Tetratricopeptide repeat |
| URTI | Upper respiratory tract infection |
| VACTERL-H | Vertebral, Anal, Cardiac, Tracheo-esophageal fistula, Esophageal atresia, Renal, upper Limb, and Hydrocephalus |
| WG | Weeks of gestation |
| WT | Wild type |
| y.o. | Year old |
| YRI | Yoruba in Ibadan, Nigeria (1KGP population) |
References
- Auerbach, A.D. Fanconi anemia and its diagnosis. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2009, 668, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Alter, B.P.; Giri, N.; Savage, S.; Rosenberg, P. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica 2017, 103, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Savage, S.A.; Walsh, M.F. Myelodysplastic Syndrome, Acute Myeloid Leukemia, and Cancer Surveillance in Fanconi Anemia. Hematol. Clin. N. Am. 2018, 32, 657–668. [Google Scholar] [CrossRef]
- Kottemann, M.C.; Smogorzewska, A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 2013, 493, 356–363. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.; D’Andrea, A. Fanconi anemia pathway. Curr. Biol. 2017, 27, R986–R988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sroka, I.; Frohnmayer, L.; Van Ravenhorst, S.; Wirkkula, L. Fanconi Anemia Clinical Care Guidelines, 5th ed. Available online: https://www.fanconi.org/images/uploads/other/Fanconi_Anemia_Clinical_Care_Guidelines_5thEdition_web.pdf (accessed on 24 December 2021).
- Niraj, J.; Färkkilä, A.; D’Andrea, A.D. The Fanconi Anemia Pathway in Cancer. Annu. Rev. Cancer Biol. 2019, 3, 457–478. [Google Scholar] [CrossRef]
- Gille, J.J.P.; Floor, K.; Kerkhoven, L.; Ameziane, N.; Joenje, H.; De Winter, J.P. Diagnosis of Fanconi Anemia: Mutation Analysis by Multiplex Ligation-Dependent Probe Amplification and PCR-Based Sanger Sequencing. Anemia 2012, 2012, 603253. [Google Scholar] [CrossRef] [PubMed]
- Callén, E.; Casado, J.A.; Tischkowitz, M.D.; Bueren, J.A.; Creus, A.; Marcos, R.; Dasí, A.; Estella, J.M.; Muñoz, A.; Ortega, J.J.; et al. A common founder mutation in FANCA underlies the world’s highest prevalence of Fanconi anemia in Gypsy families from Spain. Blood 2005, 105, 1946–1949. [Google Scholar] [CrossRef] [Green Version]
- Whitney, M.A.; Saito, H.; Jakobs, P.M.; Gibson, R.; Moses, R.E.; Grompe, M. A common mutation in the FACC gene causes Fanconi anaemia in Ashkenazi Jews. Nat. Genet. 1993, 4, 202–205. [Google Scholar] [CrossRef] [PubMed]
- De Vries, Y.; Lwiwski, N.; Levitus, M.; Kuyt, B.; Israels, S.; Arwert, F.; Zwaan, M.; Greenberg, C.R.; Alter, B.P.; Joenje, H.; et al. A Dutch Fanconi AnemiaFANCCFounder Mutation in Canadian Manitoba Mennonites. Anemia 2012, 2012, 865170. [Google Scholar] [CrossRef] [Green Version]
- Teresa, B.G.; Frias, S.; Molina, T.V.; Villarreal, M.T.; Rodriguez, A.; Carnevale, A.; López-Hernández, G.; Vollbrechtshausen, L.; Olaya-Vargas, A.; Torres, L. FANCC Dutch founder mutation in a Mennonite family from Tamaulipas, México. Mol. Genet. Genom. Med. 2019, 7, e710. [Google Scholar] [CrossRef] [Green Version]
- Auerbach, A.D.; Greenbaum, J.; Pujara, K.; Batish, S.D.; Bitencourt, M.A.; Kokemohr, I.; Schneider, H.; Lobitzc, S.; Pasquini, R.; Giampietro, P.F.; et al. Spectrum of sequence variation in theFANCG gene: An International Fanconi Anemia Registry (IFAR) study. Hum. Mutat. 2003, 21, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Morgan, N.V.; Essop, F.; Demuth, I.; de Ravel, T.; Jansen, S.; Tischkowitz, M.; Lewis, C.M.; Wainwright, L.; Poole, J.; Joenje, H.; et al. A common Fanconi anemia mutation in black populations of sub-Saharan Africa. Blood 2005, 105, 3542–3544. [Google Scholar] [CrossRef]
- Wilson, J.B.; Blom, E.; Cunningham, R.; Xiao, Y.; Kupfer, G.M.; Jones, N.J. Several tetratricopeptide repeat (TPR) motifs of FANCG are required for assembly of the BRCA2/D1-D2-G-X3 complex, FANCD2 monoubiquitylation and phleomycin resistance. Mutat. Res. Mol. Mech. Mutagen. 2010, 689, 12–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blom, E.; van de Vrugt, H.J.; de Vries, Y.; de Winter, J.P.; Arwert, F.; Joenje, H. Multiple TPR motifs characterize the Fanconi anemia FANCG protein. DNA Repair 2003, 3, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.; Rajendra, E.; Alcón, P.; O’Reilly, F.J.; Chorev, D.; Maslen, S.; Degliesposti, G.; Russo, C.J.; He, S.; Hill, C.H.; et al. Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature 2019, 575, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef]
- García-De-Teresa, B.; Rodríguez, A.; Frias, S. Chromosome Instability in Fanconi Anemia: From Breaks to Phenotypic Consequences. Genes 2020, 11, 1528. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [Green Version]
- Li, L.-H.; Ho, S.-F.; Chen, C.-H.; Wei, C.-Y.; Wong, W.-C.; Li, L.-Y.; Hung, S.-I.; Chung, W.-H.; Pan, W.-H.; Lee, M.T.M.; et al. Long contiguous stretches of homozygosity in the human genome. Hum. Mutat. 2006, 27, 1115–1121. [Google Scholar] [CrossRef]
- Pajusalu, S.; Žilina, O.; Yakoreva, M.; Tammur, P.; Kuuse, K.; Mölter-Väär, T.; Nõukas, M.; Reimand, T.; Õunap, K. The Diagnostic Utility of Single Long Contiguous Stretches of Homozygosity in Patients without Parental Consanguinity. Mol. Syndr. 2015, 6, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-C.; Ross, L.; Mahon, L.W.; Owen, R.; Hemmat, M.; Wang, B.T.; El Naggar, M.; A Kopita, K.; Randolph, L.M.; Chase, J.M.; et al. Regions of homozygosity identified by oligonucleotide SNP arrays: Evaluating the incidence and clinical utility. Eur. J. Hum. Genet. 2014, 23, 663–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, T.F.; Oliveira, L.F.; Ocampos, M.; Barbato, I.T.; De Luca, G.R.; Filho, J.H.B.; Pinto, L.L.D.C.; Bernardi, P.; Maris, A.F. Long contiguous stretches of homozygosity detected by chromosomal microarrays (CMA) in patients with neurodevelopmental disorders in the South of Brazil. BMC Med. Genom. 2019, 12, 50. [Google Scholar] [CrossRef]
- Silva-Zolezzi, I.; Hidalgo-Miranda, A.; Estrada-Gil, J.; Fernandez-Lopez, J.C.; Uribe-Figueroa, L.; Contreras, A.; Balam-Ortiz, E.; del Bosque-Plata, L.; Velazquez-Fernandez, D.; Lara, C.; et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. USA 2009, 106, 8611–8616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spear, M.L.; Diaz-Papkovich, A.; Ziv, E.; Yracheta, J.M.; Gravel, S.; Torgerson, D.G.; Hernandez, R.D. Recent shifts in the genomic ancestry of Mexican Americans may alter the genetic architecture of biomedical traits. eLife 2020, 9, e56029. [Google Scholar] [CrossRef]
- Yagasaki, H.; Hamanoue, S.; Oda, T.; Nakahata, T.; Asano, S.; Yamashita, T. Identification and characterization of novel mutations of the major Fanconi anemia gene FANCA in the Japanese population. Hum. Mutat. 2004, 24, 481–490. [Google Scholar] [CrossRef]
- Demuth, I.; Wlodarski, M.; Tipping, A.J.; Morgan, N.; De Winter, J.P.; Thiel, M.; Gräsl, S.; Schindler, D.; D’Andrea, A.D.; Altay, C.; et al. Spectrum of mutations in the Fanconi anaemia group G gene, FANCG/XRCC9. Eur. J. Hum. Genet. 2000, 8, 861–868. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, M.; Jang, W.; Chae, H.; Kim, Y.; Chung, N.-G.; Lee, J.-W.; Cho, B.; Jeong, D.-C.; Park, I.Y.; et al. Founder Haplotype Analysis of Fanconi Anemia in the Korean Population Finds Common Ancestral Haplotypes for a FANCG Variant: Ancestral Haplotypes for AFANCGVariant. Ann. Hum. Genet. 2015, 79, 153–161. [Google Scholar] [CrossRef]
- Alter, B.P.; Giri, N. Thinking of VACTERL-H? Rule out Fanconi Anemia According to PHENOS: VACTERL-H, PHENOS, and Fanconi Anemia. Am. J. Med. Genet. Part A 2016, 170, 1520–1524. [Google Scholar] [CrossRef]
- Aguilar-Ordoñez, I.; Pérez-Villatoro, F.; García-Ortiz, H.; Barajas-Olmos, F.; Ballesteros-Villascán, J.; González-Buenfil, R.; Fresno, C.; Garcíarrubio, A.; Fernández-López, J.C.; Tovar, H.; et al. Whole genome variation in 27 Mexican indigenous populations, demographic and biomedical insights. PLoS ONE 2021, 16, e0249773. [Google Scholar] [CrossRef] [PubMed]
- Torres-Cisneros, G. Mixes. In Pueblos Indigenas del Mexico Contemporaneo; Comisión Nacional para el Desarrollo de los Pueblos Indígenas, PNUD: Mexico City, Mexico, 2004; pp. 1–43. [Google Scholar]
- Quinto-Cortés, C.D.; Arriola, L.A.; García-Hughes, G.; García-López, R.; Molina, D.P.; Flores, M.; Palacios, R.; Piñero, D. Genetic Characterization of Indigenous Peoples from Oaxaca, Mexico, and Its Relation to Linguistic and Geographic Isolation. Hum. Biol. 2010, 82, 409–432. [Google Scholar] [CrossRef]
- Cuentame INEGI. Available online: http://www.cuentame.inegi.org.mx/monografias/informacion/oax/territorio/div_municipal.aspx?tema=me&e=20 (accessed on 8 December 2021).
- Orphanet: Anemia de Fanconi. Available online: https://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=ES&Expert=84 (accessed on 19 December 2021).
- Rogers, K.J.; Fu, W.; Akey, J.M.; Monnat, J.R., Jr. Global and disease-associated genetic variation in the human Fanconi anemia gene family. Hum. Mol. Genet. 2014, 23, 6815–6825. [Google Scholar] [CrossRef] [Green Version]
- Shimamura, A.; Alter, B.P. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010, 24, 101–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiesco-Roa, M.O.; Giri, N.; McReynolds, L.J.; Best, A.F.; Alter, B.P. Genotype-phenotype associations in Fanconi anemia: A literature review. Blood Rev. 2019, 37, 100589. [Google Scholar] [CrossRef] [PubMed]
- Graf, C.M.; Nichele, S.; Siviero, R.B.; Loth, G.; Trennepohl, J.P.; Zinher, M.T.; Grandinetti, A.; Pilonetto, D.V.; Pasquini, R.; Moreira, A.T.R.; et al. Ocular Manifestations in Patients with Fanconi Anemia: A Single Center Experience Including 106 Patients. J. Pediatr. 2021. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Mehta, P.A.; Jiang, C.S.; Rosti, R.O.; Usleaman, G.; da Rosa, J.M.C.; Lach, F.P.; Goodridge, E.; Auerbach, A.D.; Davies, S.M.; et al. Comparison of the clinical phenotype and haematological course of siblings with Fanconi anaemia. Br. J. Haematol. 2020, 193, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Faivre, L.; Guardiola, P.; Lewis, C.; Dokal, I.; Ebell, W.; Zatterale, A.; Altay, C.; Poole, J.; Stones, D.; Kwee, M.L.; et al. Association of complementation group and mutation type with clinical outcome in fanconi anemia. European Fanconi Anemia Research Group. Blood 2000, 96, 4064–4070. [Google Scholar]
- Feben, C.; Kromberg, J.; Wainwright, R.; Stones, D.; Sutton, C.; Poole, J.; Haw, T.; Krause, A. Phenotypic consequences in black South African Fanconi anemia patients homozygous for a founder mutation. Genet. Med. 2014, 16, 400–406. [Google Scholar] [CrossRef] [Green Version]
- Dillon, B.; Feben, C.; Segal, D.; Du Plessis, J.; Reynders, D.; Wainwright, R.; Poole, J.; Krause, A. Endocrine profiling in patients with Fanconi anemia, homozygous for a FANCG founder mutation. Mol. Genet. Genom. Med. 2020, 8, e1351. [Google Scholar] [CrossRef]
- Köhler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021, 49, D1207–D1217. [Google Scholar] [CrossRef]
- Esmer, C.; Sánchez, S.; Ramos, S.; Molina, B.; Frias, S.; Carnevale, A. DEB test for Fanconi anemia detection in patients with atypical phenotypes. Am. J. Med. Genet. Part A 2005, 124A, 35–39. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Van Der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.-F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E.; et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef] [Green Version]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. The Basic Polymerase Chain Reaction (PCR). Cold Spring Harb. Protoc. 2018, 5, pdb-prot095117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearney, H.M.; Kearney, J.B.; Conlin, L.K. Diagnostic Implications of Excessive Homozygosity Detected by SNP-Based Microarrays: Consanguinity, Uniparental Disomy, and Recessive Single-Gene Mutations. Clin. Lab. Med. 2011, 31, 595–613. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).