Abstract
MYB transcription factors (TFs) play an active role in plant responses to abiotic stresses, but they have not been systematically studied in kiwifruit (Actinidia chinensis). In this study, 181 AcMYB TFs were identified from the kiwifruit genome, unevenly distributed on 29 chromosomes. The high proportion (97.53%) of segmental duplication events (Ka/Ks values less than 1) indicated that AcMYB TFs underwent strong purification selection during evolution. According to the conservative structure, 91 AcR2R3-MYB TFs could be divided into 34 subgroups. A combination of transcriptomic data under drought and high temperature from four AcMYB TFs (AcMYB2, AcMYB60, AcMYB61 and AcMYB102) was screened out in response to stress and involvement in the phenylpropanoid pathway. They were highly correlated with the expression of genes related to lignin biosynthesis. qRT-PCR analysis showed that they were highly correlated with the expression of genes related to lignin biosynthesis in different tissues or under stress, which was consistent with the results of lignin fluorescence detection. The above results laid a foundation for further clarifying the role of MYB in stress.
1. Introduction
MYB is one of the largest transcription factor (TF) families in plants, and it is characterized by containing the MYB domain [1]. According to the number of repetitions of this domain and its similarity to prototype repetitions, it was divided into four categories, namely, 4R-MYB, R1R2R3-MYB/3R-MYB, R2R3-MYB and 1R-MYB/MYB-Related [2]. Among them, R2R3-MYB TF is one of the most abundant MYB proteins in plants, with two MYB domains at the N-terminal and most transcriptional activation domains at the C-terminal. It has a wide range of effects in plants, such as regulation of cell morphogenesis, meristem formation, hormone response, secondary metabolism and environmental stress response [3]. Many R2R3-MYB TFs have been reported to be involved in stress response, such as AtMYB12 [4], AtMYB20 [5], AtMYB75 [6] and so on. Notably, the expression of these R2R3-MYB TFs is often accompanied by the accumulation of phenylpropanoid, such as flavonoid, anthocyanin and lignin when induced by stress [4,5,6]. Moreover, the allocation of metabolic flux among the three is one of the means for plants to respond to changes in the external environment [6]. In particular, the lignin biosynthesis pathway, as the center of biosynthesis [7], is more significantly affected by the other two.
Kiwifruit belongs to Actinidiaceae (Actinidia Lindl.). Because of its large and thin leaves, thick stem pith and thick shallow roots, compared with other fruit trees, it is more sensitive to stress [8]. Fortunately, its fruit is rich in phenylpropanoid, such as anthocyanin and flavonoid, which can help plants resist stress [4,6]. However, more attention has been paid to the enrichment of fruit color by the two in the existing studies [9,10] without noticing its response to environmental stress, as well as the relationship between benzene and propane metabolism and stress response, especially lignin, which is the center of biosynthesis [7]. Although MYB TFs have been reported to be involved in the regulation of lignin biosynthesis [11,12], most of the existing studies related to lignin in kiwifruit have focused on its impact on post-harvest quality [13] or its prevention and treatment of cancer [14], neglecting the fact that its accumulation could alleviate the poor drought and moisture tolerance of kiwifruit [15]. This study aims to provide a reference for solving such problems, focusing on the identified 181 AcMYB TFs. Their chromosome distribution, gene duplication events, protein evolution, gene structure and conservative domain were analyzed. Moreover, AcMYB members’ responses to abiotic stresses were screened out based on the transcriptomic data. Quantitative real-time PCR (qRT-PCR) was used to explore the spatial and temporal expression differences and co-expression effects in different tissues under different stresses. The results will provide a reference for further study on the role of MYB TFs in stress.
2. Results
2.1. Identification and Chromosomal Distribution of MYBs TFs in Kiwifruit
According to the predictions made by Plant TFDB database, 138 AcMYB-related TFs and 97 AcMYB TFs were preliminarily identified from the kiwifruit genome. Then, TBtools was combined with NCBI bidirectional blast and hmmer 3.0 for retrieval, and 181 AcMYB TF genes were finally obtained, including 87 AcMYB-related sequences, 91 AcR2R3-MYB sequences and 3 3R-MYB sequences (Achn093361/Achn191781 and Achn365361/Achn029591 as redundant genes with completely consistent sequences). The basic physicochemical properties are shown in Table 1 (Tables S2–S4).

Table 1.
Classification and physicochemical properties of identified AcMYB TFs in kiwifruit.
Genomic localization analysis showed that all 29 chromosomes had AcMYB TFs distribution. Chromosome 13 had the highest number of AcMYB TFs (11). A minimum of two AcMYB TFs was detected on chromosomes 4, 9, 11, 12, 18, 20, 21 and 29 (Figure 1).
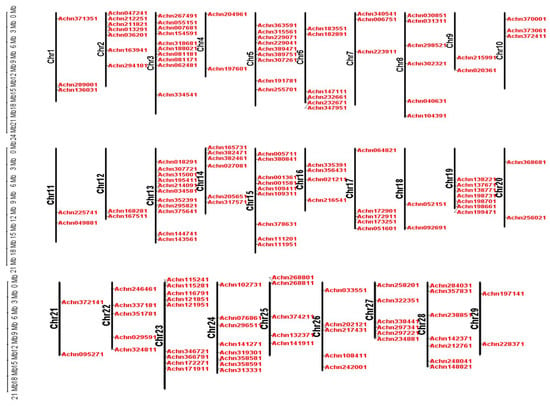
Figure 1.
Chromosome distribution of AcMYB TFs in kiwifruit.
2.2. Phylogeny Analysis of AcMYB TFs
To elucidate the phylogenetic relationship of the AcR2R3-MYB TFs in kiwifruit, 91 AcMYBs and 131 AtMYBs members were subjected to phylogenetic tree construction (Arabidopsis thaliana classification S1–S25 with reference to Dubos [2]). They were found to be divided into 34 subgroups, with all members of the A26 subgroup being 3R-MYBs (Figure 2). Previous studies have shown that the S1 (A19) [16], S2 (A13) [17], S11 (A15) [18], S20 (A24) [19] and S22 (A29) [20] subgroups in Arabidopsis thaliana are involved in the regulation of resistance to biotic and abiotic stresses. The S3 (A12) [21], S4 (A1), S5 (A3), S6 (A5) [22,23], S7 (A2) [24] and S12 (A14) subgroups participated in the regulation of primary and secondary metabolism in Arabidopsis thaliana. The S9 (A18), S14 (A21) [25,26], S15 (A7) [27,28], S18 (A25), S19 (A23) [29,30] and S21 (A30) subgroups affect the growth and development process of Arabidopsis thaliana. The gene functions of the subgroups S13 (A9), S16 (A20), S23 (A28), S24 (A17) and S25 (A27) have not been completely analyzed, and they are temporarily classified into a category of function to be determined. Notably, although the gene function of the S13 subgroups has not been fully resolved, it has also been reported that it could help plants resist drought stress [31].
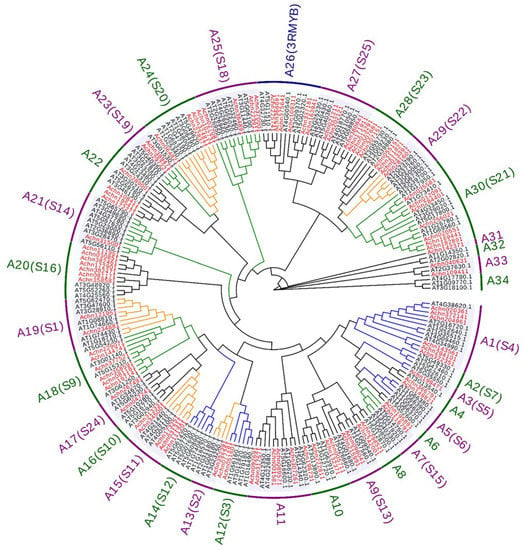
Figure 2.
Phylogenetic analysis of R2R3-MYB, Arabidopsis thaliana classification S1–S25 with reference to Dubos [2], with red as AcMYBs and black as AtMYBs, orange line as stress response-related MYBs, blue line as metabolism-related MYBs and green line as growth-related MYBs.
2.3. Conservative Motif and Gene Structure Analysis of AcR2-MYB and AcR3-MYB TFs
Using MEME to identify the conservative motifs in the protein sequences of AcR2-MYBs and AcR3-MYBs (R2R3-MYBs and 3R-MYBs protein sequences) in kiwifruit, it was found that most AcMYBs proteins had motif4/motif 6, motif 3, motif1 and motif 2, which formed the R2R3-repeat (Figure 3d). R2-MYB and R3-MYB are homologous to the subfamily R2R3-repeat, each of which contains a highly conservative tryptophan residue (W) in its repeating domain, and these motifs occur in almost all genes. Notably, Achn375641 and Achn295821, Achn163941 and Achn380251, Achn361141 and Achn368681, all contain motif 9 (including the moiety R1-reapeat), which can be classified as 3R-MYB. Based on the information of gene structure and phylogenetic analysis, we also found that AcMYBs in different subgroups contained different numbers of exons. The genes in the same subgroup showed highly similar gene structure and conservative motifs (Figure 3a–c).

Figure 3.
Subgroups of AcR2-MYB and AcR3-MYB genes in kiwifruit (a) protein evolution analysis, (b) conservative motif analysis, (c) gene structure analysis and (d) conservative domain sequence.
2.4. Gene Duplication and Synteny Analysis of AcMYB TFs in Kiwifruit
An estimation of gene duplication events in kiwifruit genome by collinearity analysis revealed that two tandem duplication events, located in Chr6 and Chr14, respectively, occurred in its evolution. A total of 78 segmental duplication events, including 99 genes, were distributed across the remaining 25 chromosomes, except for Chr12/17/20/29 (Figure 4a), and an ultrahigh proportion (97.53%) of segmental duplication was identified. At the same time, the Ka/Ks values of all collinearity pairs were less than 1 (Table S5), indicating that AcMYB TFs in kiwifruit underwent strong purification selection during evolution.
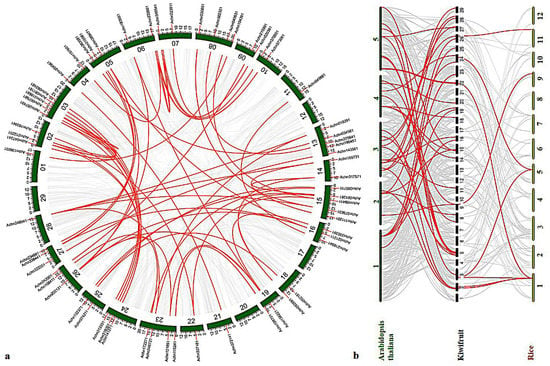
Figure 4.
Collinearity analysis of AcMYBs TF in kiwifruit. (a) Collinearity analysis and gene duplication in kiwifruit genome. The gray line represents all autosomal blocks in kiwifruit genome, the red line represents the AcMYB gene duplication events, and the AcMYB gene is represented by a vertical red line. (b) Collinearity analysis of kiwifruit and two model plants; the green chromosome is Arabidopsis thaliana, and the yellow chromosome is rice.
In order to further deduce the evolutionary mechanism of AcMYBs in kiwifruit, a comparative syntenic map of kiwifruit and two model plants, namely, Arabidopsis thaliana and rice, was constructed. The results showed that there were 39 orthologous genes between kiwifruit and Arabidopsis thaliana (Table S6) and only 9 orthologous genes between kiwifruit and rice (Table S7). Among them, four AcMYBs could be found in a syntenic relationship with both Arabidopsis thaliana and rice, suggesting that it might exist before the differentiation of monocotyledonous and dicotyledonous plants.
2.5. Expression Analysis Based on Transcriptomic Data
Transcriptional abundance of AcR2R3-MYB TFs members was extracted from the kiwifruit transcriptomic data under drought and high-temperature stresses previously sequenced by our group [32,33] (Tables S8 and S9). Among them, 31 members did not detect transcriptional abundance in the control or drought stress treatment group, and 26 members did not detect transcriptional abundance in the control or high-temperature stress treatment group. After the low-quality reading (padj > 0.05) was removed, the heatmap (Figure 5) was generated with corresponding FPKM values of the control or stress treatment group using the TBtools. Among them, there were 14 down-regulated AcMYBs in the high-temperature treatment group, where |log2fold change| > 2 was down-regulated by AcMYB3, AcMYB4, AcMYB61 and AcMYB66; 4 up-regulated AcMYBs, of which, AcMYB60 up-regulated |log2fold change| > 2. In the drought treatment group, there were 8 down-regulated AcMYBs, among which AcMYB14, AcMYB15, AcMYB16, AcMYB42 and AcMYB61 down-regulated |log2fold change| > 2; 10 up-regulated AcMYBs, of which |log2fold change| > 1.5 for AcMYB2, AcMYB78, AcMYB102 and AcMYB108 were up-regulated.
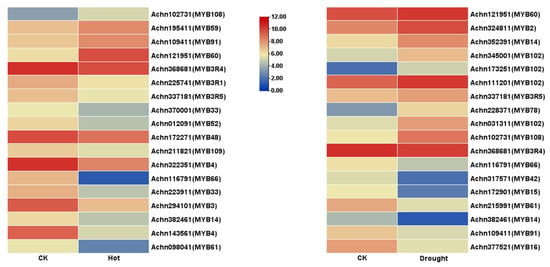
Figure 5.
Heatmap of AcR2R3-MYB TFs’ transcriptional profiles in kiwifruit under drought and high-temperature stress.
2.6. Screening of Stress-Responsive AcMYB TFs and Prediction Regulatory Network
Since homologous genes usually have similar functions, the function of the corresponding orthologous gene in kiwifruit can be deduced from Arabidopsis thaliana. In addition, the spatial–temporal expression pattern of specific stress-responsive genes is an important part of plant stress response [34]. Therefore, the AcMYBs, which is in the same subgroup as the AtMYBs reported in phylogenetic analysis and which changes significantly in stress-treated transcriptional profiles, is comprehensively analyzed to screen R2R3-MYB TFs in kiwifruit that respond to stress. The results show that Achn121951, which was clustered into one branch with AtMYB60 (AT1G08810.1) in the S1 subgroup, was significantly up-regulated under high-temperature stress; Achn173251, which was clustered into one branch with AtMYB102 (AT4G21440.1) in the S11 subgroup, significantly increased under drought stress; Achn215991, which was clustered into one branch with AtMYB61 (AT1G09540.1) in the S13 subgroup, was significantly down-regulated under drought stress; Achn324811, which was clustered into one branch with AtMYB2 (AT2G47190.1) in subgroup S20, was significantly down-regulated under drought stress. All of them might be involved in the plant stress response.
Relevant studies have shown that AtMYB102 [18] is involved in trauma and osmotic stress responses. AtMYB60 is only expressed in guard cells, which is necessary for photoinduced stomatal opening. At the same time, the pore size of its mutant is reduced, helping to limit water loss during drought [16]. As for AtMYB61, it can coordinate a small network of downstream target genes required for several aspects of plant growth and development, such as xylem formation and xylem cell differentiation, as well as lateral root formation, and act as a transcriptional regulator of stomatal closure [35]. AtMYB2 is involved in the regulation of ABA-induced genes under drought stress, and at the same time, it forms a complex with all known glucosinolate-related MYBs to regulate the biosynthesis of glucosinolate [19]. Notably, the responses of AtMYB61 and AtMYB2 to stress involved the regulation of phenylpropanoid pathway. Studies have shown that the response of R2R3-MYB gene to stress is often accompanied by changes in the content of phenylpropanoid pathway, such as flavonoid, anthocyanin and lignin [4,5,6]. Moreover, their distribution of metabolic flux is one of the means for plants to cope with changes in the external environment [6]. In particular, the lignin biosynthesis pathway [7], which is a biosynthesis center, is more significantly affected. Therefore, it is speculated that it may affect lignin biosynthesis and thus respond to stress.
The String website was used to predict the functional relationship between MYBs proteins of MYB2, MYB60, MYB61 and MYB102 in kiwifruit and related genes from the phenylpropanoid pathway (Arabidopsis thaliana as the species model). The connected genes in this network may have close functional ties. MYB60, MYB2 and MYB102 could be indirectly associated with caffeoyl-CoA O-methyltransferase (CCoAOMT), cinnamate 4-hydroxylase (C4H), ferulate-5-hydroxylase (F5H), 4-coumarate-CoA ligase (4CL), caffeic acid O-methyltransferase (COMT), phenylalanine ammonia-lyase (PAL), coumarate 3-hydroxylase (C3′H), cinnamyl alcohol dehydrogenase (CAD), shikimate O-hydroxycinnamoyltransferase (HCT), p-coumaryl alcohol by cinnamoyl-CoA reductase (CCR) and peroxidase (POD) through flavanone 3-hydroxylase (F3H), dihydro-flavonol 4-reductase (DFR), flavonol synthase (FLS), leucoanthocyanidin dioxygenase (LDOX), UDP flavonoid glucosyl transferase (UFGT) and other anthocyanin pathway-related genes (Figure 6a). There were direct functional relations or interaction relations between MYB61 and CCoAOMT, C4H, F5H, 4CL, COMT, PAL, C3′H, CAD, HCT, CCR and POD (Figure 6b). Therefore, it was speculated that the screened MYB2, MYB60, MYB61 and MYB102 might be related to a certain function of lignin biosynthesis. It was suggested that the lignin biosynthesis genes might be regulated by MYBs.
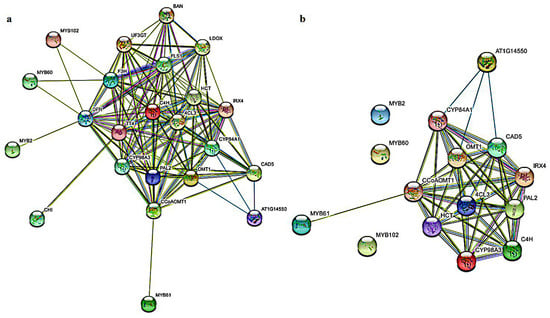
Figure 6.
Protein regulatory network of AcMYBs protein in kiwifruit. (a) Protein regulatory networks of MYBs and structural genes of phenylpropanoid pathway. (b) Protein regulatory networks of MYBs and structural genes of lignin metabolic pathway. The purple line represents that interaction of the experimental results. Yellow-green, black and blue lines represent text mining, co-expression and protein homology, respectively.
2.7. The Analysis of the MYB-Binding Site of the Candidate Genes from the Phenylpropanoid Pathway
Studies of MYB-binding sites have shown that cis elements that are generally rich in adenine and cytosine residues can be recognized by MYB TFs, such as MYB-binding site ((T/C) AAC (G/T) G (A/C/T) (A/C/T)), MBSI ((C/T) NGTT (A/G)), AC elements (ACC (A/T) A (A/C) (T/C) and ACC (A/T) (A/C/T) (A/C/T)) [36,37]. To identify whether the candidate genes from the phenylpropanoid pathway are regulated by the AcMYBs, the cis-acting elements of the 2 kb promoter upstream of the start codon of related genes were analyzed, and genes with the MYB-binding sites were identified (Figure 7). The results showed that most flavonoid and lignin-biosynthesis-related genes in kiwifruit have MYB-binding sites, such as MYB-binding site, AC- element and so on, except for CCR, CAD, FLS and LDOX.
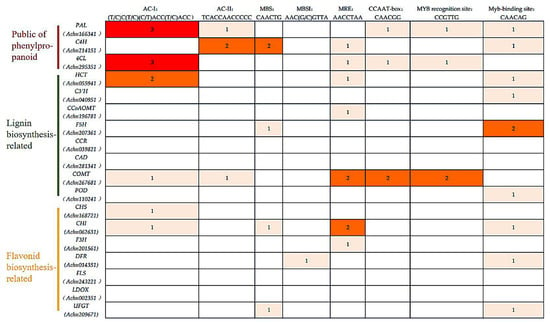
Figure 7.
MYB-binding site of the candidate genes from the phenylpropanoid pathway.
It was remarkable that ‘ACC’ was the core recognition motif in the AC elements [38], which has been found to be closely related to the benzene–propane metabolic pathway. Studies showed that SlMYB75 [39] could bind to the ‘ACCTACCC’ motif of the target gene promoter and effectively induce anthocyanin accumulation in tomato tissues. Meanwhile, both PtMYB4 and EgMYB2 can bind to the AC elements to regulate lignin biosynthesis [37]. Moreover, BplMYB46 protein [40] can also directly bind to the ‘ACCACCT’ motif of downstream target gene promoter to promote lignin deposition and secondary cell wall biosynthesis. In this study, the promoter regions of lignin-biosynthesis-related genes in kiwifruit were rich in AC elements, suggesting that these genes might be regulated by MYB TFs and involved in lignin biosynthesis.
2.8. Expression Profiles of AcMYBs and Lignin-Biosynthesis-Related Genes in Different Tissues of Kiwifruit
The expression profiles of AcMYBs and lignin-biosynthesis-related genes in different tissues of kiwifruit were analyzed (Figure 8). The results showed that the screened MYBs generally had high expression in stem or stem tip, petiole, young leaf or leaf, but low expression in fruit. As for the related genes from lignin biosynthesis, except for the high expression of AcC4H and AcCCR in flowers, the rest were generally expressed in stems or stem tips, petioles, leaves or young leaves, and it was noted that AcHCT, AcCAD and AcPOD also had high expression in roots.
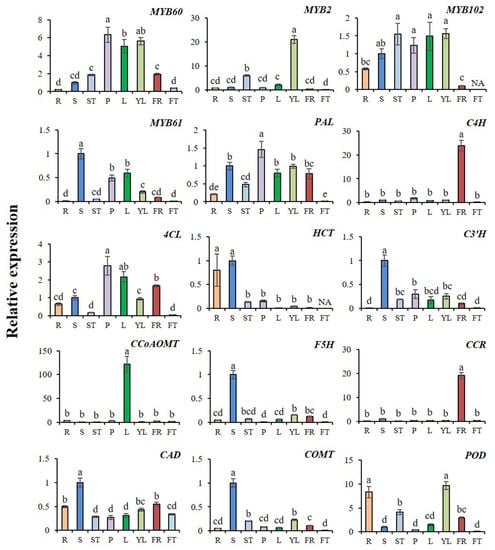
Figure 8.
Relative expression levels of AcMYBs and lignin-biosynthesis-related genes in different tissues of kiwifruit. The expression level of this gene in the stem was taken as 1. The relative expression level of genes was calculated by normalization method. R. Root, S. stem, ST. stem tip, P. petiole, L. leaf, YL. young leaf, FR. flower, FT. fruit. Different superscript letters indicate significant differences between different treatments at p < 0.05 level. NA indicates that the expression level is lower than the minimum value detectable by the instrument.
Correlation analysis showed (Table 2) that AcMYB102 and AcMYB61 had a very significant positive correlation with the initial response enzyme gene AcPAL of the common biosynthesis pathway of phenylpropane. AcMYB60 was closely related to the initial response enzyme gene AcPAL and the ligase gene Ac4CL. The expression levels of related genes, such as lignin-related hydroxylases AcC3′H and AcF5H, reductase gene AcCAD and methylase gene AcCOMT, also showed a significant positive correlation with AcMYB61. It should be noted that AcMYB2 was closely related to lignin biosynthesis and degradation, as well as to the related gene of suberized AcPOD. Based on these results, it could be speculated that there was co-expression between AcMYB60, AcMYB2, AcMYB102 and AcMYB61 or lignin-biosynthesis-related genes during plant growth and development.

Table 2.
Correlation between expression level of AcMYBs and lignin-biosynthesis-related genes in different tissues.
2.9. Determination of Lignin Autofluorescence in Different Tissues of Kiwifruit
Based on the autofluorescence characteristics of lignin under ultraviolet light, it was found that the deposition of lignin in different tissues of kiwifruit was different (Figure 9). It was mainly deposited in the phloem and conducting tissues of the stem, and it was also deposited in the veins, villi and inner wall of guard cells of the leaves. Secondly, a small amount of deposition was also observed in the phloem of stem tip and the conducting tissue of petiole. This result was consistent with the expression profiles of AcMYBs and lignin-biosynthesis-related genes in different tissues. Therefore, it was speculated that the screened AcMYB2, AcMYB60, AcMYB61 and AcMYB102 might be related to the existence of a certain function of lignin deposition.
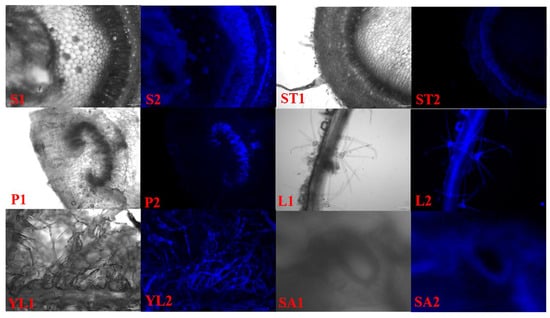
Figure 9.
Lignin sections in different tissues of kiwifruit. S. stem, ST. stem tip, P. petiole, L. leaf, YL. young leaf, SA. stomata; 1. bright field, 2. ultraviolet (UV).
2.10. Expression Profiles of AcMYBs and Lignin-Biosynthesis-Related Genes in Kiwifruit under Different Stresses
Through cis-acting element analysis, it was found that the four screened AcMYBs members mostly contained stress-responsive elements, such as drought, low temperature and hypoxia, as well as light-responsive elements (Table S10). Therefore, their expression levels were determined by qRT-PCR under different abiotic stresses (dark, low temperature, PEG simulation of drought and salt) (Figure 10). From the results, we could find that both AcMYB2 and AcMYB102 could respond to salt stress, while AcMYB60 and AcMYB61 responded to low temperature and salt stress. As for the relative expression levels of lignin-biosynthesis-related genes, they were increased under low-temperature stress and salt stress. In addition, under dark conditions, AcMYBs and lignin-biosynthesis-related genes were mostly expressed at a relatively low level, suggesting that dark conditions might be unfavorable for lignin accumulation. Therefore, it was speculated that the four selected AcMYBs might affect lignin biosynthesis and respond to stress.
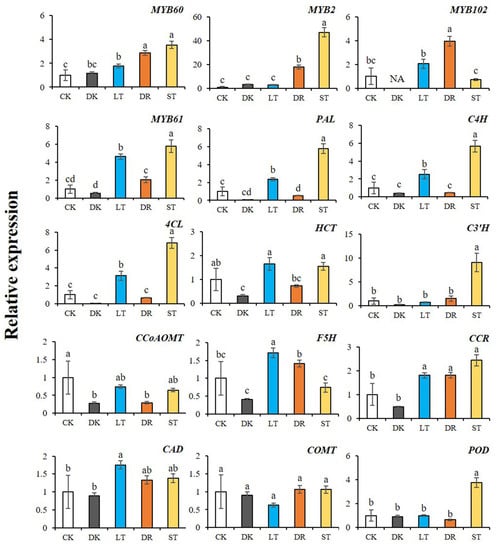
Figure 10.
Relative expression levels of AcMYBs and lignin-biosynthesis-related genes in kiwifruit under different stresses. CK. control, DK. dark, LT. low temperature, DR. PEG simulation of drought, ST. salt. Different superscript letters indicate significant differences between different treatments at p < 0.05 level. NA indicates that the expression level is lower than the minimum value detectable by the instrument.
Correlation analysis (Table 3) showed that AcMYB60, AcMYB2 and AcMYB61 had highly significant positive correlations with two initial response enzyme genes and one ligase gene AcPAL, AcC4H and Ac4CL of the common biosynthesis pathway of phenylpropane, respectively. Among the related genes, including lignin-related hydroxylases AcF5H, AcC3′H and methylase genes AcCOMT, AcCCoAOMT and reductase genes AcCCR and AcCAD, AcMYB61 was closely related to these genes, except for the methylase genes AcCCoAOMT and AcCOMT. As for AcMYB60 and AcMYB2, they were closely related to the AcC3′H and reductase gene AcCCR that determine the biosynthesis of G-and S-type lignin, as well as to the biosynthesis and degradation of lignin, and the related gene of AcPOD that is responsible for cork-rization. AcMYB102 was closely related to hydroxylase F5H. Based on these results, it could be speculated that AcMYB60, AcMYB2, AcMYB102 and AcMYB61 might co-express with lignin-biosynthesis-related genes in the process of plant stress response.

Table 3.
Correlation between expression level of AcMYBs and lignin-biosynthesis-related genes under different stresses.
3. Discussion
In recent years, a variety of plants have systematically identified the MYB TF family through genomic analysis [41,42]. However, no literature has been reported regarding the responses of members of the MYB family in kiwifruit to stress and their responses to lignin-biosynthesis-related genes. In this study, 181 AcMYB TFs were identified in the kiwifruit genome and subjected to chromosome distribution analysis, gene duplication event analysis, conservative domain identification, phylogenetic analysis, gene structure and protein motif analysis. Furthermore, AcMYBs participating in stress resistance was screened out based on transcriptomic data. Protein regulatory network prediction and promoter cis-acting element analysis were conducted to explore the responses of selected AcMYBs genes to lignin-biosynthesis-related genes and stress responses.
The results showed that the identified AcMYBs gene in kiwifruit was different in amino acid sequence length, physical and chemical properties (such as molecular weight and isoelectric point) and gene structure. It reflected the complexity and functional diversity of kiwifruit MYBs gene to a certain extent. At the same time, kiwifruit MYBs gene has tandem repeat events and has a closer collinearity with Arabidopsis thaliana. Notably, in this identification, we referred to the reported functions of members of AtMYBs in Arabidopsis thaliana and transcriptomic data of the previous research group, and we identified four MYBs that might participate in stress response. According to the functions, it was speculated that they might participate in the phenylpropanoid pathway, in which the lignin biosynthesis pathway was the central pathway [7]. It was suggested that it might affect lignin biosynthesis to a certain extent. Therefore, the prediction of protein regulatory network was performed for its interaction with lignin-biosynthesis-related genes. It was found that AcMYB61 could be directly associated with lignin-biosynthesis-related genes, while AcMYB2, AcMYB60 and AcMYB102 could indirectly affect lignin metabolism through anthocyanin-biosynthesis-related genes. At the same time, except for AcCCR and AcCAD, the promoter regions of other lignin-biosynthesis-related genes contained MYB-binding sites, such as AC-element, MRE, MBS and so on (Figure 7). Therefore, it was speculated that lignin-biosynthesis-related genes might be regulated by MYBs.
It should be noted that the selected AcMYB2, AcMYB60, AcMYB61 and AcMYB102 mostly expressed in stems or stem tips, petioles, leaves or young leaves with high expression levels, and the expression profiles of lignin-biosynthesis-related genes showed a similar trend. The results of correlation analysis also indicated that these four genes might co-express with lignin-biosynthesis-related genes in the process of plant growth and development. Moreover, the results of lignin autofluorescence measurement showed that lignin did deposit in guard cells, vein and villous parts of kiwifruit leaves, phloem and translocation tissue of stem, translocation tissue of petiole and phloem of stem tip.
In addition, the selected AcMYBs also showed responses to low-temperature stress, salt stress or drought stress, which were consistent with the results of their cis-acting element analysis. At the same time, the expression levels of structural genes for lignin metabolism were increased under low temperature or osmotic stress. The results of correlation analysis showed that the expression changes of AcMYB60, AcMYB2 and AcMYB61 under stress had a significant positive correlation with the lignin-biosynthesis-related genes. It indicated that there might be co-expression effect therebetween, so as to resist stress. Based on this speculation, the up-regulation of kiwifruit MYB61 expression under low temperature or osmotic stress caused the up-regulation of lignin-biosynthesis-related genes, such as AcPAL, AcC4H, Ac4CL, AcCCR, AcCAD and AcPOD, which caused more lignin deposition and helped to resist external stress. This conclusion coincides with the research conclusion that the degree of lignification of soybean root system and wheat under salt stress is relatively high [43,44]. However, AcMYB60, AcMYB2 and AcMYB102 could indirectly up-regulate the expression of AcCCR or AcCAD and then deposit lignin to help resist stress.
4. Materials and Methods
4.1. Plant Materials and Abiotic Stress Treatments
The kiwifruit tissue, including stem tip, young leaf, root, stem, leaf, flower and fruit (n = 3), were collected from ‘Jinyan’ at a kiwifruit orchard in Pujiang County, Chengdu, China (29°28′ N, 119°53′ E). Samples were frozen in liquid nitrogen and stored at −80 °C using the RNA extract.
The sterile tissue culture seedlings of ‘Jinyan’ kiwifruit with consistent growth were selected for stress treatment. The 60 aseptic seedlings were equally divided into five groups for treatment, including control treatment group, dark treatment group, low temperature treatment group, PEG simulation of drought treatment group and salt treatment group. Except for the LT treatment group, which grew at 4 ± 2 °C, the other four treatment groups were all grown in the culture room at light/dark 16 h/8 h at 25 ± 2 °C. The DK treatment group was shaded with black cloth; the DR treatment group was treated with 40% (w/v) PEG-6000 under simulated drought stress; and the ST treatment group was simulated with 100 mmol/L NaCl. Three strains were repeated four times. Samples were taken at 48 h of stress and stored at −80 °C immediately after freezing in liquid nitrogen.
4.2. Identification and Physicochemical Properties Analysis of AcMYB TFs
The kiwifruit genome database from Kiwifruit Genome Database (http://kiwifruitgenome.org/ (accessed on 20 October 2021)) was compared and integrated with the transcriptomic data of kiwifruit under stresses previously sequenced by the research team. Predictions were made using the Plant TFDB database (http://plantfdb.Gao-lab.org/ (accessed on 21 October 2021)). AtMYB in Arabidopsis thaliana dataset (downloaded by TAIR (https://www.arabidopsis.org/ (accessed on 21 October 2021)) as a probe) bidirectional blast was performed using TBtools and the NCBI conservative domain database. Then, the hidden Markov model file of MYB gene family (PF00249) was downloaded from PFAM (http://pfam.xfam.org/ (accessed on 21 October 2021)) website and retrieved using hmmer 3.0. Combined with the above three methods, the search results were integrated to finally determine the AcMYB-related TFs and AcMYB TFs. The physical and chemical properties analyses of AcMYB protein in kiwifruit were performed using the online tool ProtParam (http://web.expasy.org/protparam/ (accessed on 25 October 2021)).
4.3. Chromosomal Distribution, Gene Duplication Events and Collinearity Analysis of AcMYBs
The chromosomal distribution of AcMYB gene was retrieved from the Kiwifruit Genome Database gff3 file and visualized by Map Inspect. Gene duplication events in the AcMYB TFs were detected using MC Scan X with default parameters. Additionally, the homology analysis atlas of kiwifruit, Arabidopsis thaliana and rice was constructed by using TBtools software.
4.4. Conservative Motif, Gene Structure Analysis and Phylogenetic Analysis of AcMYBs
The conserved motifs of AcMYBs were revealed using MEME (https://meme-suite.org/ (accessed on 21 October 2021)), and the gene structure was extracted and visualized using TBtools. For phylogenetic analysis, the sequences of AtMYBs protein in Arabidopsis thaliana downloaded by TAIR were used as reference, and the conservative domains of MYB protein in kiwifruit and Arabidopsis thaliana were preserved. After Clustal W alignment, a neighbor-joining (NJ) phylogenetic tree was constructed using Mega X, and the bootstrap value was set to 1000 repetitions.
4.5. Expression Analysis Based on Transcriptomic Data
Based on the transcriptomic data of kiwifruit under drought and high-temperature stresses sequenced by our research group at the early stage, the low-quality reading (padj > 0.05) was removed, and the transcriptional abundance of the gene was expressed as FPKM (number of fragments per million kilobases). log2(FPKM) was used for hierarchical clustering, and the results were visualized by TBtools.
4.6. Prediction of AcMYBs Protein Regulatory Network
The String website (http://string-db.org/ (accessed on 21 November 2021)) was used to analyze the protein regulatory network of AcMYBs protein in kiwifruit, with the model plant Arabidopsis thaliana as the species parameter.
4.7. MYB-Binding Site Analysis
The genomic DNA sequences of 2 kb upstream of the initiation codon of phenylpropanoid pathway-related candidate gene was extracted using TBtools software and submitted to PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 22 November 2021)) for the cis-acting element and functional sites analysis in the promoter regions.
4.8. Extraction of Total RNA and qRT-PCR Analysis
The extraction of total RNA in kiwifruit referred to the modified CTAB-LiCl method [45]. The cDNA prepared by PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) was used as the template. Quantitative PCR was performed using TB Green® Premix Ex TaqTM II (TaKaRa, Dalian, China). Gene-specific primers were designed using Primer 5.0 (Table S1). The 2−ΔΔCQ method was used to quantify the relative expression level of the gene. AcActin (GenBank Accession No.EF063572) gene was used as the endogenous internal reference gene. Three biological and technical repetitions were completed for each sample based on qRT-PCR.
4.9. Determination of Lignin Autofluorescence in Different Tissues of Kiwifruit
Different tissues, such as leaves, petioles and stems of ‘Jinyan’ kiwifruit seedlings, were sectioned by hand, and the blue fluorescence of lignified tissues under ultraviolet (UV) fluorescence was observed under the fluorescence phase contrast micro-imaging system, according to the methods described by Jia [46] and Han [47].
4.10. Data Analysis
The experimental data were all expressed as mean standard error (M ± SE). The statistical analysis and chart drawing were performed using Excel 2010 software, while the significance analysis was performed using SPSS 20.0 software using one-way ANOVA or Tukey test (p < 0.05). The correlation analysis was a two-variable linear correlation analysis, expressed as Pearson correlation coefficient (R).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23042323/s1.
Author Contributions
H.X. and D.L. conceived and designed the experiments; X.L. (Xinling Liu), Z.L., X.Z., M.W., T.W. performed the experiments, with assistance from H.D., L.L., X.L. (Xiulan Lv), J.W., Q.D. and K.X.; X.L. (Xinling Liu), H.X. and D.L. analyzed the data and constructed the figures; X.L. (Xinling Liu) wrote the draft; H.X. and D.L. provided financial support; H.X. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Sichuan Science and Technology Department Projects (2021YFYZ0010, SCCXTD-2021) and Leshan Normal University Project (RC202001) for funding this research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are reported in this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shi, M.Z.; Xie, D.Y. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Griffiths, A.G.; Cousins, G.R.; Verry, I.M.; Williams, W.M. Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 2015, 205, 882–893. [Google Scholar] [CrossRef]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Gen. Genet. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, Y.L.; Yang, L.; Song, J.B.; Yang, Z.M. AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant Soil 2014, 376, 433–443. [Google Scholar] [CrossRef]
- Rowan, D.D.; Cao, M.; Lin-Wang, K.; Cooney, J.; Jensen, D.J.; Austin, P.T.; Hunt, M.B.; Norling, C.; Hellens, R.; Schaffer, R.; et al. Environmental regulation of leaf colour in red 35S: PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Taiz, L.; Zeiger, E. Plant Physiology; Sinauer Associates: New York, NY, USA, 2006. [Google Scholar]
- Dichio, B.; Montanaro, G.; Sofo, A.; Xiloyannis, C. Stem and whole-plant hydraulics in olive (Olea europaea) and kiwifruit (Actinidia deliciosa). Trees 2012, 27, 183–191. [Google Scholar] [CrossRef]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.H.; Espley, R.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2018, 221, 309–325. [Google Scholar] [CrossRef] [Green Version]
- Legay, S.; Sivadon, P.; Blervacq, A.-S.; Pavy, N.; Baghdady, A.; Tremblay, L.; Levasseur, C.; Ladouce, N.; Lapierre, C.; Séguin, A.; et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 2010, 188, 774–786. [Google Scholar] [CrossRef]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamidha Kumarihami, H.M.P.; Gook, K.J.; YunHee, K.; Mockhee, L.; YoungSuk, L.; YongBum, K.; Joonyup, K. Preharvest Application of Chitosan Improves the Postharvest Life of ‘Garmrok’ Kiwifruit through the Modulation of Genes Related to Ethylene Biosynthesis, Cell Wall Modification and Lignin Metabolism. Foods 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Ryoma, K.; Rina, Y.; Yumi, F.; Shiduku, T.; Yuya, U.; Go, S.; Keita, K.; Gan, K.; Katsuhiko, S.; Mamoru, O.; et al. Expression analysis of defense-related genes in wild kiwifruit (Actinidia rufa) tolerant to bacterial canker. J. Gen. Plant Pathol. 2021, 87, 361–365. [Google Scholar]
- Srivastava, S.; Vishwakarma, R.K.; Arafat, Y.A.; Gupta, S.K.; Khan, B.M. Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in developing seedlings of Leucaena leucocephala. Physiol. Mol. Biol. Plants 2015, 21, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A Guard-Cell-Specific MYB Transcription Factor Regulates Stomatal Movements and Plant Drought Tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Kirik, V.; Kölle, K.; Wohlfarth, T.; Misera, S.; Bäumlein, H. Ectopic expression of a novel MYB gene modifies the architecture of the Arabidopsis inflorescence. Plant J. 2010, 13, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Denekamp, M.; Smeekens, S.C. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol. 2003, 132, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2007, 146, 323–324. [Google Scholar] [CrossRef] [Green Version]
- Jianli, Z.; Chanhui, L.; Ruiqin, Z.; Zheng-Hua, Y. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar]
- Bhargava, A.; Mansfield, S.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 Functions in Regulation of Secondary Cell Wall Formation in the Arabidopsis inflorescence Stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuluaga, D.L.; Gonzali, S.; Loreti, E.; Pucciariello, C.; Degl’Innocenti, E.; Guidi, L.; Alpi, A.; Perata, P. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct. Plant Biol. 2008, 35, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of Production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef] [PubMed]
- Kelle, T.; Abbott, J.; Moritz, T.; Doerner, P. Arabidopsis Regulator of Axillary Meristems1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 2006, 18, 598–611. [Google Scholar] [CrossRef] [Green Version]
- Kirik, V.; Lee, M.M.; Wester, K.; Herrmann, U.; Zheng, Z.; Oppenheimer, D.; Schiefelbein, J.; Hulskamp, M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development (Camb. Engl.) 2005, 132, 1477–1485. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.H.; Kirik, V.; Hulskamp, M.; Nam, K.H.; Hagely, K.; Lee, M.M.; Schiefelbein, J. The MYB23 Gene Provides a Positive Feedback Loop for Cell Fate Specification in the Arabidopsis Root Epidermis. Plant Cell 2009, 21, 1080–1094. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.M.; Schiefelbein, J. WEREWOLF, a MYB-Related Protein in Arabidopsis, Is a Position-Dependent Regulator of Epidermal Cell Patterning. Cell 1999, 99, 473–483. [Google Scholar] [CrossRef] [Green Version]
- Mandaokar, A.; Browse, J. MYB108 Acts Together with MYB24 to Regulate Jasmonate-Mediated Stamen Maturation in Arabidopsis. Plant Physiol. 2008, 149, 851–862. [Google Scholar] [CrossRef] [Green Version]
- Mandaokar, A.; Thines, B.; Shin, B.; Lange, B.M.; Choi, G.; Koo, Y.J.; Yoo, Y.J.; Choi, Y.D.; Choi, G.; Browse, J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006, 46, 984–1008. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, X.; Zhang, Y.; Miao, Z.; Xie, C.; Meng, X.; Deng, J.; Wen, J.; Mysore, K.; Frugier, F.; et al. Opposing control by transcription factors MYB61 and MYB3 Increases Freezing Tolerance by relieving C-repeat Binding Factor suppression. Plant Physiol. 2016, 172, 1306–1323. [Google Scholar] [CrossRef] [Green Version]
- Xia, H.; Ni, Z.; Hu, R.; Lin, L.; Deng, H.; Wang, J.; Tang, Y.; Sun, G.; Wang, X.; Li, H.; et al. Melatonin Alleviates Drought Stress by a Non-Enzymatic and Enzymatic Antioxidative System in Kiwifruit Seedlings. Int. J. Mol. Sci. 2020, 21, 852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, H.; Zhou, Y.; Deng, H.; Lin, L.; Deng, Q.; Wang, J.; Lv, X.; Zhang, X.; Liang, D. Melatonin improves heat tolerance in Actinidia deliciosa via carotenoid biosynthesis and heat shock proteins expression. Physiol. Plant. 2021, 172, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.-K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.; Campbell, M. AtMYB61, an R2R3-MYB Transcription Factor Controlling Stomatal Aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef] [Green Version]
- Prouse, M.B.; Campbell, M.M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 2012, 1819, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Patzlaff, A.; Newman, L.J.; Dubos, C.; Whetten, R.W.; Smith, C.; McInnis, S.; Bevan, M.W.; Sederoff, R.R.; Campbell, M.M. Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003, 53, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Morikawa, S.; Nakamura, H.; Hojo, H.; Yoshimura, S.; Zhang, R.; Aimoto, S.; Ametani, Y.; Hirata, Z.; Sarai, A.; et al. Comparison of the free and DNA-complexed forms of the DMA-binding domain from c-Myb. Nat. Struct. Mol. Biol. 1995, 2, 309–320. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, S.; Li, H.; Wang, L.; Liu, Y.; Niu, L.; Yang, Q.; Meng, D.; Fu, Y. Genome-wide analysis and characterization of R2R3-MYB family in pigeon pea (Cajanus cajan) and their functional identification in phenylpropanoids biosynthesis. Planta 2021, 254, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kaur, L.; Zhawar, V.K. Phenolic parameters under exogenous ABA, water stress, salt stress in two wheat cultivars varying in drought tolerance. Indian J. Plant Physiol. 2015, 20, 151–156. [Google Scholar] [CrossRef]
- Neves, G.Y.S.; Marchiosi, R.; Ferrarese, M.L.L.; Siqueira-Soares, R.C.; Ferrarese-Filho, O. Root Growth Inhibition and Lignification Induced by Salt Stress in Soybean. J. Agron. Crop Sci. 2010, 196, 467–473. [Google Scholar] [CrossRef]
- Gasic, K.; Hernandez, A.; Korban, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 2004, 22, 437–438. [Google Scholar] [CrossRef]
- Jia, X.-L.; Wang, G.-L.; Xiong, F.; Yu, X.-R.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. De novo assembly, transcriptome characterization, lignin accumulation and anatomic characteristics: Novel insights into lignin biosynthesis during celery leaf development. Sci. Rep. 2015, 5, srep08259. [Google Scholar] [CrossRef] [Green Version]
- Han, M.-H.; Yang, N.; Wan, Q.-W.; Teng, R.-M.; Duan, A.-Q.; Wang, Y.-H.; Zhuang, J. Exogenous melatonin positively regulates lignin biosynthesis in Camellia sinensis. Int. J. Biol. Macromol. 2021, 179, 485–499. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).