Micro-RNA and Proteomic Profiles of Plasma-Derived Exosomes from Irradiated Mice Reveal Molecular Changes Preventing Apoptosis in Neonatal Cerebellum
Abstract
1. Introduction
Experimental Design
2. Results
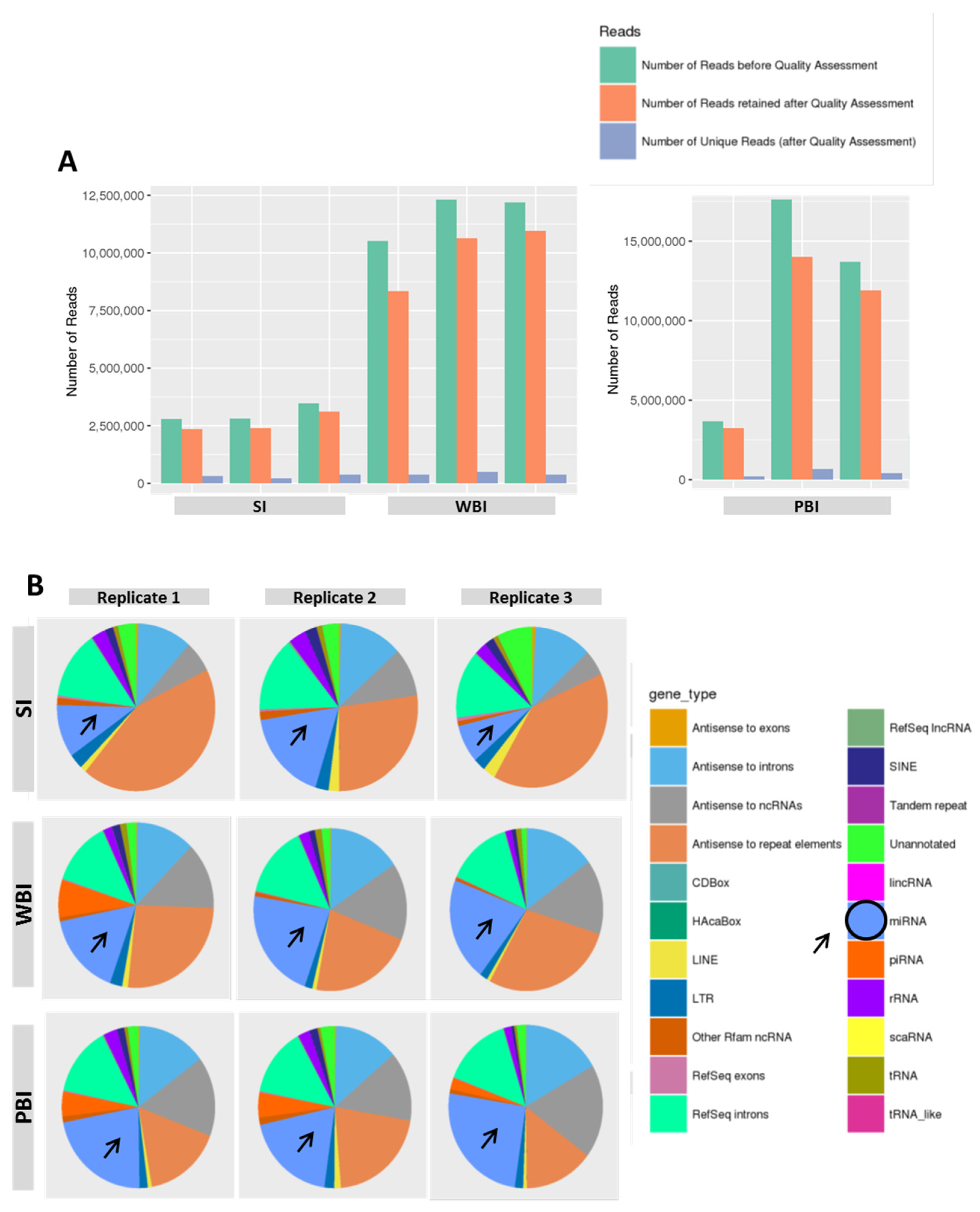
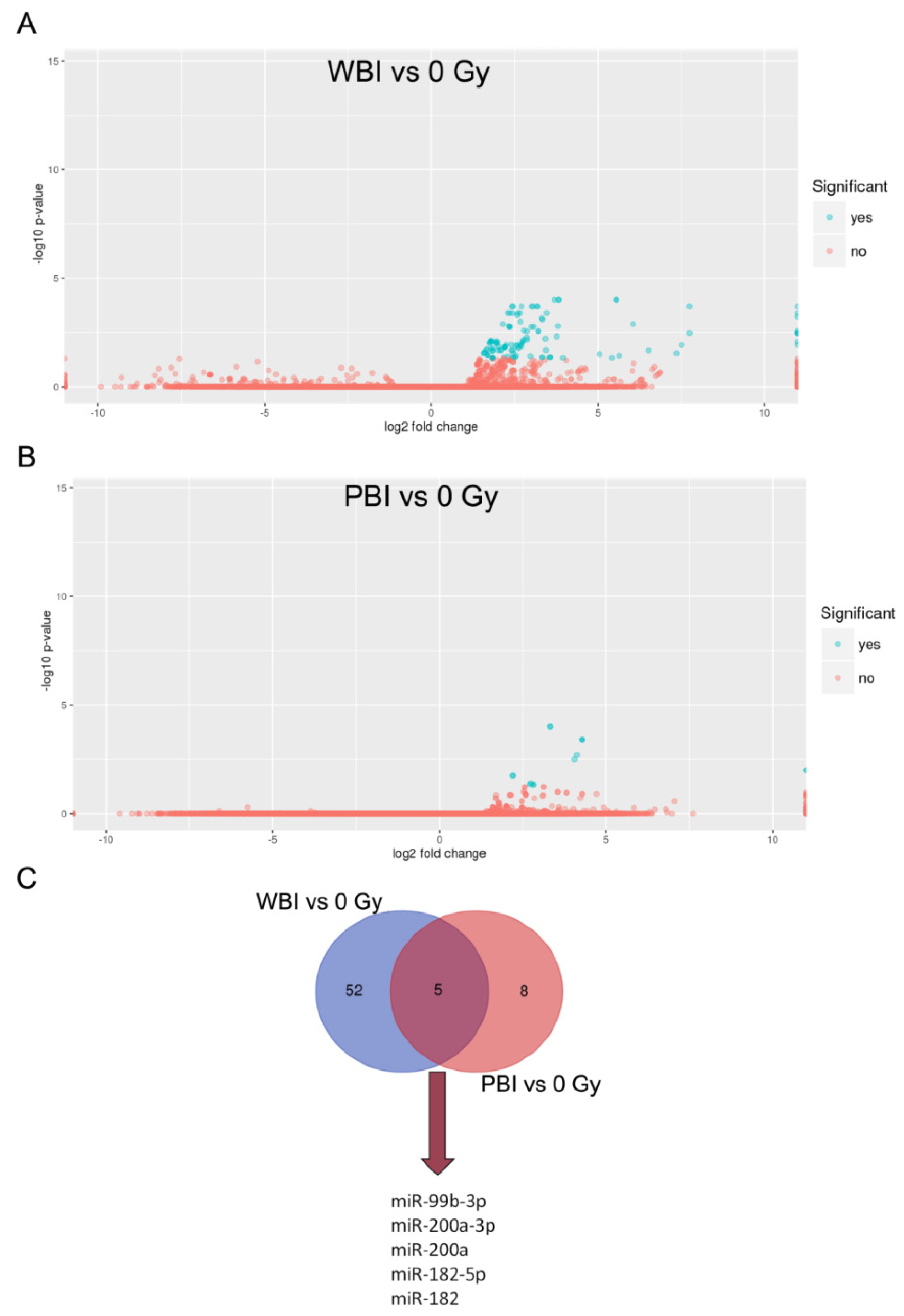
2.1. MicroRNA Profile of Circulating Plasma Exosomes from 2 Gy WBI and PBI Mice
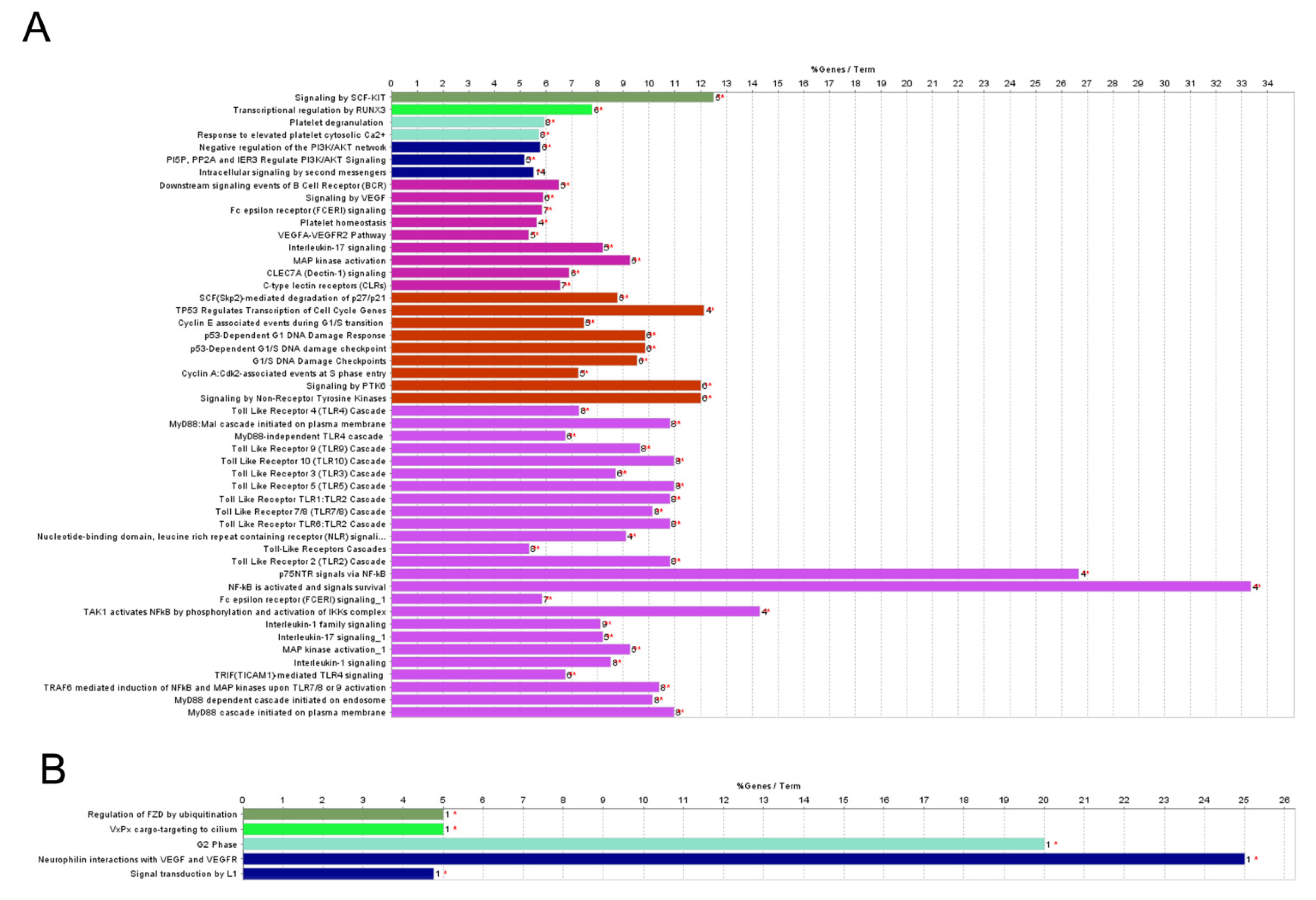
2.2. Pathway Analysis of Differentially Regulated miRNAs
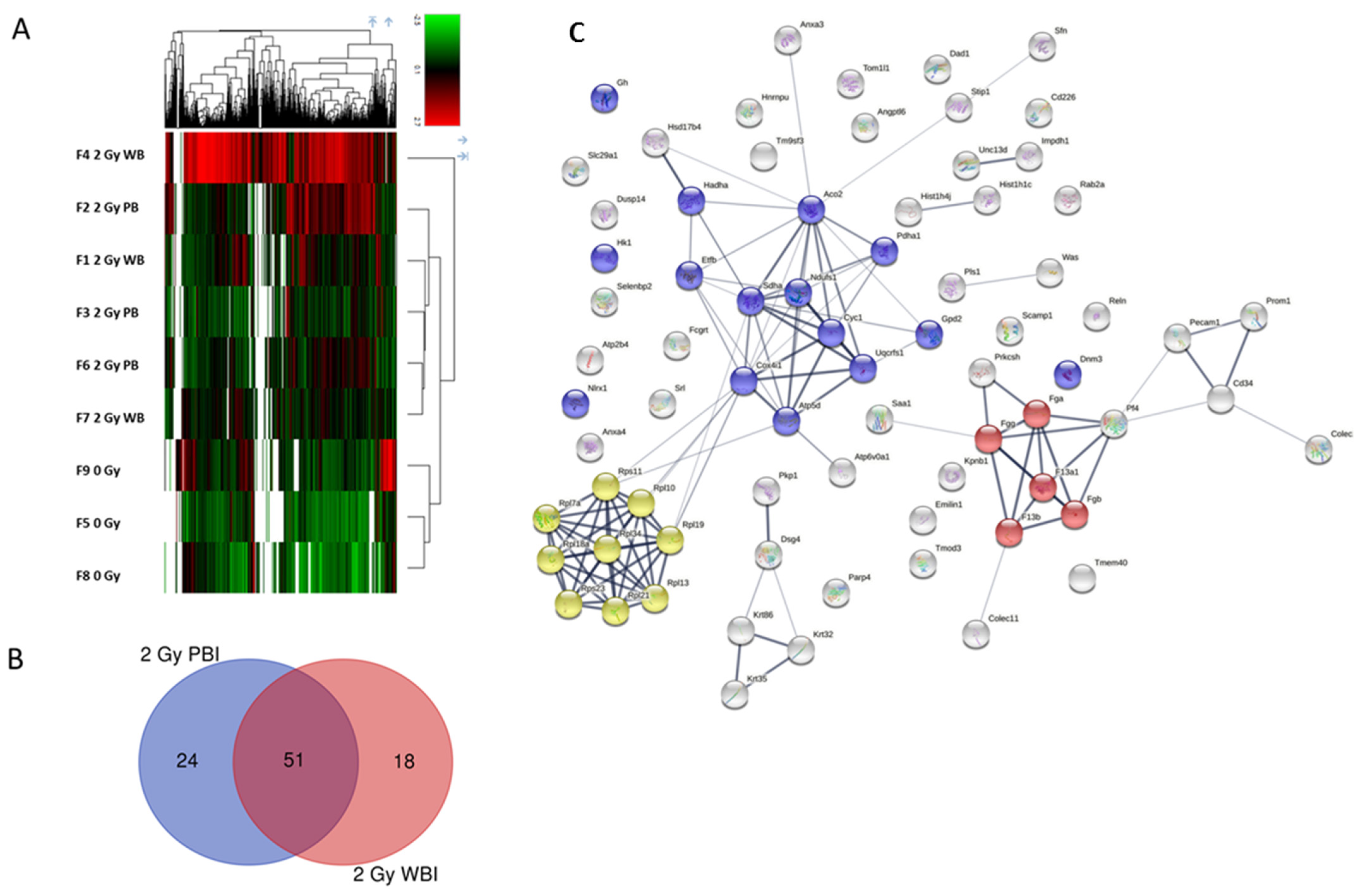
2.3. Proteomic Profiles of Circulating Plasma Exosomes from 2 Gy-Irradiated WBI and PBI Mice
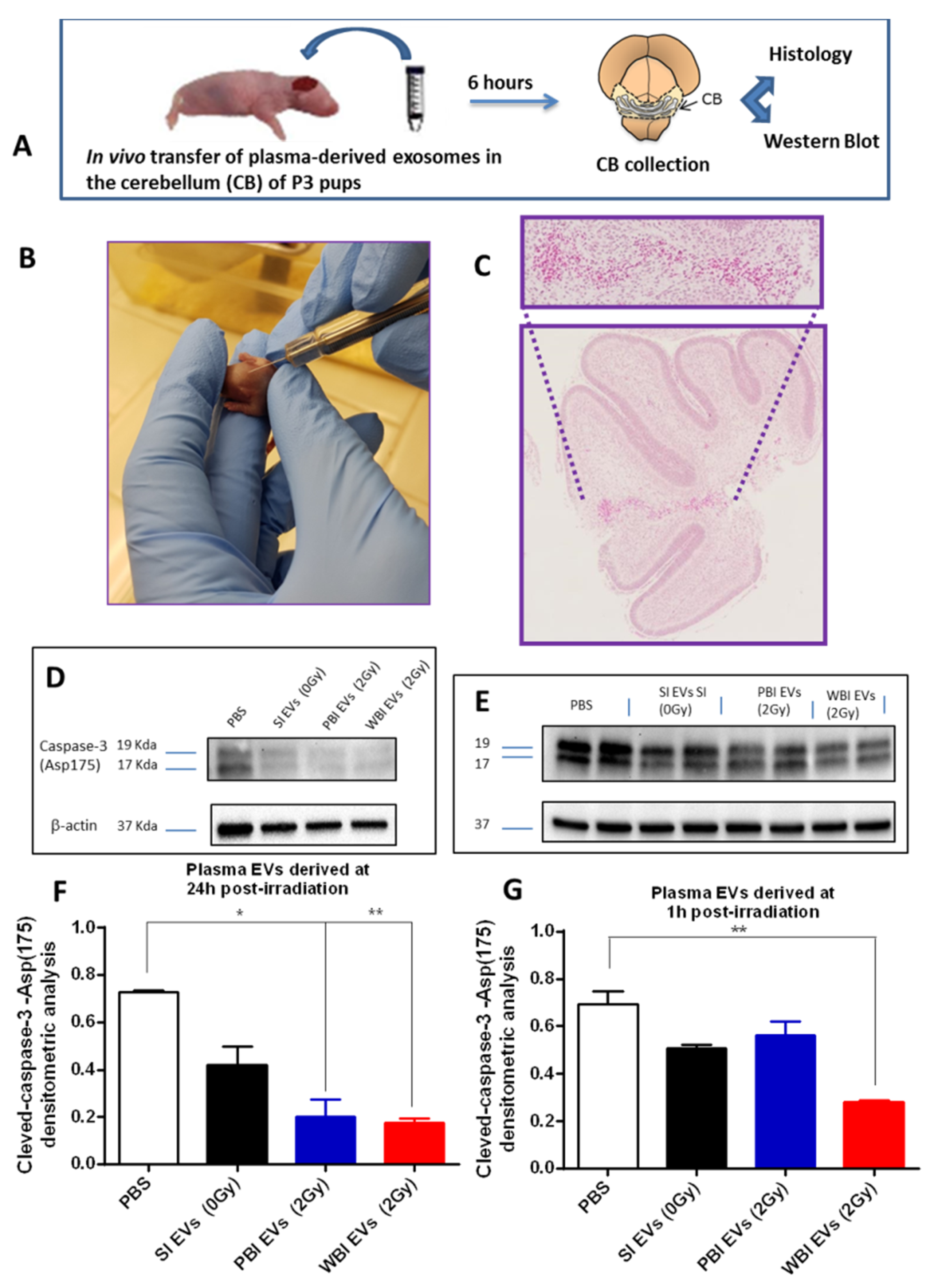
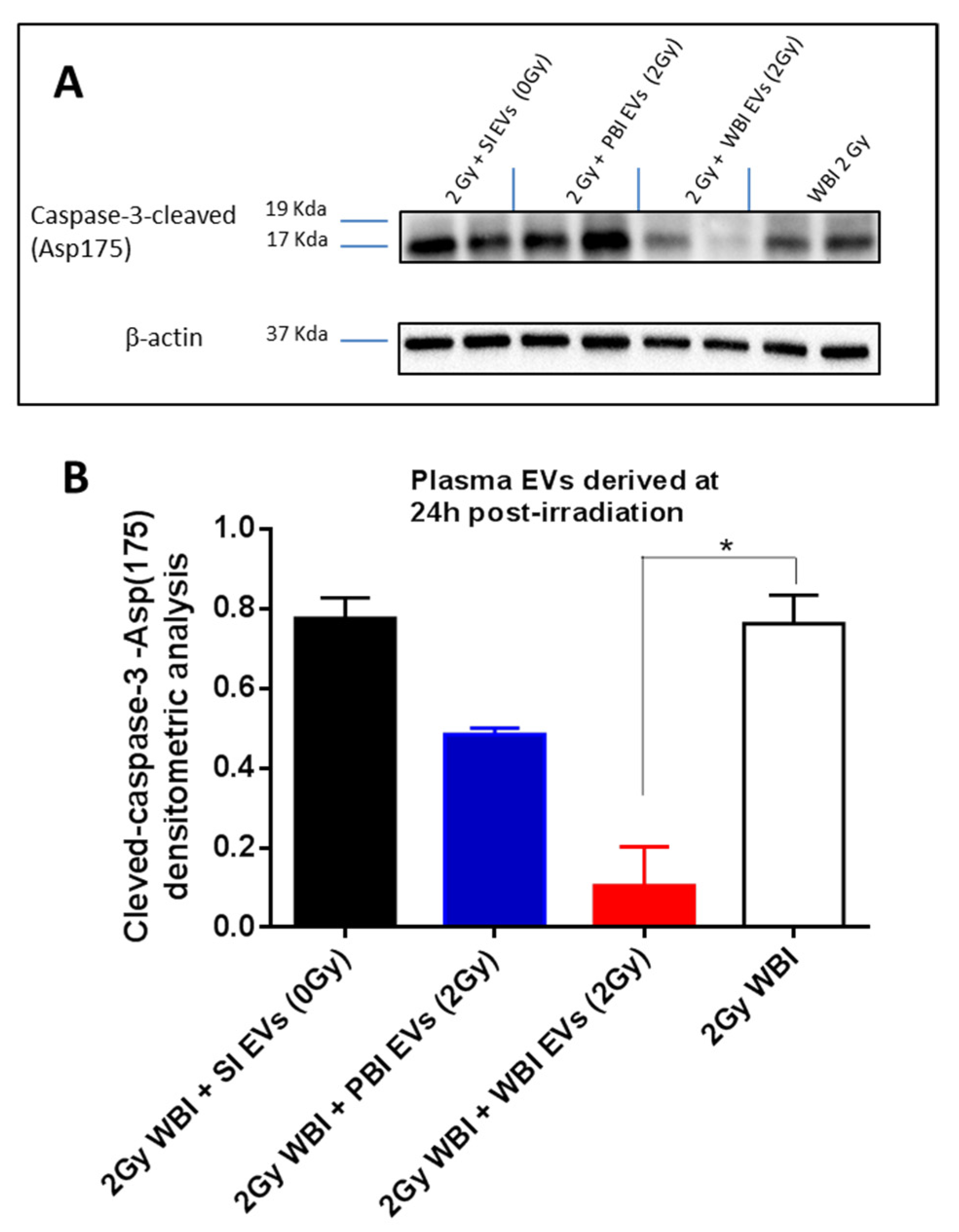
2.4. In Vivo Functional Testing of Plasma-Derived Exosomes on Neonatal Mouse Cerebellum
3. Discussion
4. Material and Methods
4.1. Mouse Irradiation
4.2. Isolation of Plasma-Derived Exosomes
4.3. MicroRNA Profiling from Plasma-Derived Exosomes
4.4. Pathway Analysis of Plasma-Derived Exosomal miRNA
4.5. Mass Spectrometry (MS) Sample Preparation and Measurement for Protein Profiling of Plasma-Derived Exosomes
4.6. Data Processing—Protein Identification
4.7. Data Processing—Label-Free Quantification
4.8. Data Availability
4.9. In Vivo Exosomes Transfer
4.10. Histological Analysis
4.11. Western Blot
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ward, J. New paradigms for low-dose radiation response in proceedings of the American statistical association conference on radiation and health. Radiat. Res. 1999, 151, 92–117. [Google Scholar]
- Iyer, R.; Lehnert, B.E. Effects of ionizing radiation in targeted and nontargeted cells. Biochem. Biophys. 2000, 376, 14–25. [Google Scholar] [CrossRef]
- Wright, E.G. Radiation-induced genomic instability in haemopoietic cells. Int. J. Radiat. Biol. 1998, 74, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.G. Inducible genomic instability: New insights into the biological effects of ionizing radiation. Med. Confl. Surv. 2000, 16, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Wolff, S. The adaptive response in radiobiology: Evolving insights and implications. Environ. Health Persp. 1998, 106, 277–283. [Google Scholar]
- Joiner, M.C.; Marples, B.; Lambin, P.; Short, S.C.; Turesson, I. Low-dose hypersensitivity: Current status and possible mechanisms. Int. J. Radiat. Oncol. Biol. Phys. 2001, 49, 379–389. [Google Scholar] [CrossRef]
- Amundson, S.A.; Bittner, M.; Meltzer, P.; Trent, J.; Fornace, A.J., Jr. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat. Res. 2001, 156, 657–661. [Google Scholar] [CrossRef]
- Budnik, V.; Ruiz-Cañada, C.; Wendler, F. Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 2016, 17, 160–172. [Google Scholar] [CrossRef]
- Coleman, B.M.; Hill, A.F. Extracellular vesicles—Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin. Cell Dev. Biol. 2015, 40, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Zheng, D.; Huo, M.; Li, B.; Wang, W.; Piao, H.; Wang, Y.; Zhu, Z.; Li, D.; Wang, T.; Liu, K. The Role of Exosomes and Exosomal MicroRNA in Cardiovascular Disease. Front. Cell Dev. Biol. 2021, 8, 616161. [Google Scholar] [CrossRef] [PubMed]
- Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef] [PubMed]
- Mallegol, J.; Van Niel, G.; Heyman, M. Phenotypic and functional characterization of intestinal epithelial exosomes. Blood Cells Mol. Dis. 2005, 35, 11–16. [Google Scholar] [CrossRef]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Mathivanan, S.; Lim, J.W.; Tauro, B.J.; Ji, H.; Moritz, R.L.; Simpson, R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell Proteom. 2010, 9, 197–208. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural. Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef]
- Sardar Sinha, M.; Ansell-Schultz, A.; Civitelli, L.; Hildesjö, C.; Larsson, M.; Lannfelt, L.; Ingelsson, M.; Hallbeck, M. Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018, 136, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Jelonek, K.; Widlak, P.; Pietrowska, M. The Influence of Ionizing Radiation on Exosome Composition, Secretion and Intercellular Communication. Protein Pept. Lett. 2016, 23, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.K.; Hinzman, C.P.; Mehta, K.Y.; Hanlon, B.K.; Garcia, M.; Fatanmi, O.O.; Singh, V.K. Plasma Derived Exosomal Biomarkers of Exposure to Ionizing Radiation in Nonhuman Primates. Int. J. Mol. Sci. 2018, 19, 3427. [Google Scholar] [CrossRef]
- Tuncay Cagatay, S.; Mayah, A.; Mancuso, M.; Giardullo, P.; Pazzaglia, S.; Saran, A.; Daniel, A.; Traynor, D.; Meade, A.D.; Lyng, F.; et al. Phenotypic and Functional Characteristics of Exosomes Derived from Irradiated Mouse Organs and Their Role in the Mechanisms Driving Non-Targeted Effects. Int. J. Mol. Sci. 2020, 21, 8389. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, R.; Chen, G. Exosome and Secretion: Action On? Adv. Exp. Med. Biol. 2020, 1248, 455–483. [Google Scholar]
- Rasini, V.; Dominici, M.; Kluba, T.; Siegel, G.; Lusenti, G.; Northoff, H.; Horwitz, E.M.; Schäfer, R. Mesenchymal stromal/stem cells markers in the human bone marrow. Cytotherapy 2013, 15, 292–306. [Google Scholar] [CrossRef]
- Pu, X.; Ma, S.; Gao, Y.; Xu, T.; Chang, P.; Dong, L. Mesenchymal Stem Cell-Derived Exosomes: Biological Function and Their Therapeutic Potential in Radiation Damage. Cells 2020, 10, 42. [Google Scholar] [CrossRef]
- Jacobs, S.A.; Roobrouck, V.D.; Verfaillie, C.M.; Van Gool, S.W. Immunological characteristics of human mesenchymal stem cells and multipotent adult progenitor cells. Immunol. Cell Biol. 2013, 91, 32–39. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Xue, J.; Li, X.; Lu, Y.; Gan, L.; Zhou, L.; Wang, Y.; Lan, J.; Liu, S.; Sun, L.; Jia, L.; et al. Gene-modified mesenchymal stem cells protect against radiation-induced lung injury. Mol. Ther. 2013, 21, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Al-Mayah, A.H.; Irons, S.L.; Pink, R.C.; Carter, D.R.; Kadhim, M.A. Possible role of exosomes containing RNA in mediating nontargeted effect of ionizing radiation. Radiat. Res. 2012, 177, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Arscott, W.T.; Tandle, A.T.; Zhao, S.; Shabason, J.E.; Gordon, I.K.; Schlaff, C.D.; Zhang, G.; Tofilon, P.J.; Camphausen, K.A. Ionizing radiation and glioblastoma exosomes: Implications in tumor biology and cell migration. Transl. Oncol. 2013, 6, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Al-Mayah, A.; Bright, S.; Chapman, K.; Irons, S.; Luo, P.; Carter, D.; Goodwin, E.; Kadhim, M. The non-targeted effects of radiation are perpetuated by exosomes. Mutat. Res. 2015, 772, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wang, J.; Ding, N.; Hu, W.; Zhang, X.; Wang, B.; Hua, J.; Wei, W.; Zhu, Q. Exosome-mediated microRNA transfer plays a role in radiation-induced bystander effect. RNA Biol. 2015, 12, 1355–1363. [Google Scholar] [CrossRef]
- Frenz, M.B.; Mee, A.S. Diagnostic radiation exposure and cancer risk. Gut 2005, 54, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Wang, X.; Teng, F.; Kong, L.; Yu, J. Abscopal effect of metastatic pancreatic cancer after local radiotherapy and granulocyte-macrophage colony-stimulating factor therapy. Cancer Biol. Ther. 2017, 18, 137–141. [Google Scholar] [CrossRef][Green Version]
- Wood, J.; Yasmin-Karim, S.; Mueller, R.; Viswanathan, A.N.; Ngwa, W. Single Radiotherapy Fraction with Local Anti-CD40 Therapy Generates Effective Abscopal Responses in Mouse Models of Cervical Cancer. Cancers 2020, 12, 1026. [Google Scholar] [CrossRef]
- Pouget, J.P.; Georgakilas, A.G.; Ravanat, J.L. Targeted and Off-Target (Bystander and Abscopal) Effects of Radiation Therapy: Redox Mechanisms and Risk/Benefit Analysis. Antioxid. Redox. Signal. 2018, 29, 1447–1487. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.X.; Huen, I.; Wu, Z.J.; Wang, H.; Duan, M.Y.; Guenther, I.; Bhanu Prakash, K.N.; Tang, F.R. Early postnatal irradiation-induced age-dependent changes in adult mouse brain: MRI based characterization. BMC Neurosci. 2021, 22, 28. [Google Scholar] [CrossRef]
- Mancuso, M.; Pasquali, E.; Leonardi, S.; Tanori, M.; Rebessi, S.; Di Majo, V.; Pazzaglia, S.; Toni, M.P.; Pimpinella, M.; Covelli, V.; et al. Oncogenic bystander radiation effects in Patched heterozygous mouse cerebellum. Proc. Natl. Acad. Sci. USA 2008, 105, 12445–12450. [Google Scholar] [CrossRef] [PubMed]
- STRING. Available online: http://string-db.org (accessed on 20 April 2021).
- Février, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Al-Mayah, A.H.; Bright, S.J.; Bowler, D.A.; Slijepcevic, P.; Goodwin, E.; Kadhim, M.A. Exosome-Mediated Telomere Instability in Human Breast Epithelial Cancer Cells after X Irradiation. Radiat. Res. 2017, 187, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.C.; Sun, D.; Dutta, A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013, 32, 1164–1172. [Google Scholar] [CrossRef]
- Fei, M.; Li, Z.; Cao, Y.; Jiang, C.; Lin, H.; Chen, Z. MicroRNA-182 improves spinal cord injury in mice by modulating apoptosis and the inflammatory response via IKKβ/NF-κB. Lab. Invest. 2021, 101, 1238–1253. [Google Scholar] [CrossRef]
- Li, W.; Wang, W.; Zuo, R.; Liu, C.; Shu, Q.; Ying, H.; Sun, K. 5 Induction of pro-inflammatory genes by serum amyloid A1 in human amnion fibroblasts. Sci. Rep. 2017, 7, 693. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Jin, H.; Liu, Q.; Sun, Q. miR-182-5p contributes to radioresistance in nasopharyngeal carcinoma by regulating BNIP3 expression. Mol. Med. Rep. 2021, 23, 130. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yu, L.; Dong, H.; Liu, Z.; Sun, Y. MiR-182 enhances radioresistance in non-small cell lung cancer cells by regulating FOXO3. Clin. Exp. Pharmacol. Physiol. 2019, 46, 137–143. [Google Scholar] [CrossRef]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kurie, J.M.; Ahn, Y.H. BMP4 depletion by miR-200 inhibits tumorigenesis and metastasis of lung adenocarcinoma cells. Mol Cancer. 2015, 14, 173. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tang, J.; Lei, H.; Cai, P.; Zhu, H.; Li, B.; Xu, X.; Xia, Y.; Tang, W. Decreased MiR-200a/141 suppress cell migration and proliferation by targeting PTEN in Hirschsprung’s disease. Cell Physiol. Biochem. 2014, 34, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Roslan, S.; Johnstone, C.N.; Wright, J.A.; Bracken, C.P.; Anderson, M.; Bert, A.G.; Selth, L.A.; Anderson, R.L.; Goodall, G.J.; et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene 2014, 33, 4077–4088. [Google Scholar] [CrossRef] [PubMed]
- Magenta, A.; Cencioni, C.; Fasanaro, P.; Zaccagnini, G.; Greco, S.; Sarra-Ferraris, G.; Antonini, A.; Martelli, F.; Capogrossi, M.C. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011, 18, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Kozak, J.; Jonak, K.; Maciejewski, R. The function of miR-200 family in oxidative stress response evoked in cancer chemotherapy and radiotherapy. Biomed. Pharmacother. 2020, 125, 110037. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Jang, S.C.; Crescitelli, R.; Cvjetkovic, A.; Belgrano, V.; Olofsson Bagge, R.; Sundfeldt, K.; Ochiya, T.; Kalluri, R.; Lötvall, J. Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. J. Extracell. Vesicles 2019, 8, 1635420. [Google Scholar] [CrossRef]
- Todkar, K.; Chikhi, L.; Desjardins, V.; El-Mortada, F.; Pépin, G.; Germain, M. Selective packaging of mitochondrial proteins into extracellular vesicles prevents the release of mitochondrial DAMPs. Nat. Commun. 2021, 12, 1971. [Google Scholar] [CrossRef]
- Panfoli, I.; Ravera, S.; Podestà, M.; Cossu, C.; Santucci, L.; Bartolucci, M.; Bruschi, M.; Calzia, D.; Sabatini, F.; Bruschettini, M.; et al. Exosomes from human mesenchymal stem cells conduct aerobic metabolism in term and preterm newborn infants. FASEB J. 2016, 30, 1416–1424. [Google Scholar] [CrossRef]
- Montay-Gruel, P.; Zhu, Y.; Petit, B.; Leavitt, R.; Warn, M.; Giedzinski, E.; Ollivier, J.; Sinclair, D.A.M.; Vozenin, M.C.; Limoli, C.L. Extracellular Vesicles for the Treatment of Radiation-Induced Normal Tissue Toxicity in the Lung. Front. Oncol. 2021, 10, 602763. [Google Scholar] [CrossRef]
- Hosseinkhani, B.; Kuypers, S.; van den Akker, N.M.S.; Molin, D.G.M.; and Michiels, L. Extracellular Vesicles Work as a Functional Inflammatory Mediator Between Vascular Endothelial Cells and Immune Cells. Front. Immunol. 2018, 9, 1789. [Google Scholar] [CrossRef] [PubMed]
- Puhm, F.; Afonyushkin, T.; Resch, U.; Obermayer, G.; Rohde, M.; Penz, T.; Schuster, M.; Wagner, G.; Rendeiro, A.F.; Melki, I.; et al. Mitochondria Are a Subset of Extracellular Vesicles Released by Activated Monocytes and Induce Type I IFN and TNF Responses in Endothelial Cells. Circ. Res. 2019, 125, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Privratsky, J.R.; Newman, D.K.; and Newman, P.J. PECAM-1: Conflicts of interest in inflammation. Life Sci. 2010, 87, 69–82. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Tanori, M.; Mancuso, M.; Rebessi, S.; Leonardi, S.; Di Majo, V.; Covelli, V.; Atkinson, M.J.; Hahn, H.; Saran, A. Linking DNA damage to medulloblastoma tumorigenesis in patched heterozygous knockout mice. Oncogene 2006, 25, 1165–1173. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Tanno, B.; Antonelli, F.; Giardullo, P.; Babini, G.; Subedi, P.; Azimzadeh, O.; Khan, Z.N.; Oleksenko, K.; Metzger, F.; et al. Out-of-Field Hippocampus from Partial-Body Irradiated Mice Displays Changes in Multi-Omics Profile and Defects in Neurogenesis. Int. J. Mol. Sci. 2021, 20, 4290. [Google Scholar] [CrossRef] [PubMed]
- Makale, M.T.; McDonald, C.R.; Hattangadi-Gluth, J.A.; Kesari, S. Mechanisms of radiotherapy-associated cognitive disability in patients with brain tumours. Nat. Rev. Neurol. 2017, 13, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Piccinini, A.M.; Midwood, K.S. DAMPening inflammation by modulating TLR Signalling. Mediat. Inflamm. 2010, 2010, 672395. [Google Scholar] [CrossRef] [PubMed]
- Burdelya, L.G.; Krivokrysenko, V.I.; Tallant, T.C.; Strom, E.; Gleiberman, A.S.; Gupta, D.; Kurnasov, O.V.; Fort, F.L.; Osterman, A.L.; Didonato, J.A.; et al. An agonist of toll-like receptor 5 has radioprotective activity in mouse and primate models. Science 2008, 320, 226–230. [Google Scholar] [CrossRef]
- Shakhov, A.N.; Singh, V.K.; Bone, F.; Cheney, A.; Kononov, Y.; Krasnov, P.; Bratanova-Toshkova, T.K.; Shakhova, V.V.; Young, J.; Weil, M.M.; et al. Prevention and mitigation of acute radiation syndrome in mice by synthetic lipopeptide agonists of Toll-like receptor 2 (TLR2). PLoS ONE 2012, 7, e33044. [Google Scholar] [CrossRef]
- Takemura, N.; Kawasaki, T.; Kunisawa, J.; Sato, S.; Lamichhane, A.; Kobiyama, K.; Aoshi, T.; Ito, J.; Mizuguchi, K.; Karuppuchamy, T.; et al. Blockade of TLR3 protects mice from lethal radiation-induced gastrointestinal syndrome. Nat. Commun. 2014, 5, 3492. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, C.; Mitchel, R.E.; Cui, J.; Lin, J.; Yang, Y.; Liu, X.; Cai, J. A critical role of toll-like receptor 4 (TLR4) and its’ in vivo ligands in basal radio-resistance. Cell Death Dis. 2013, 4, e649. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Bhanja, P.; Liu, L.; Alfieri, A.A.; Yu, D.; Kandimalla, E.R.; Agrawal, S.; Guha, C. TLR9 agonist protects mice from radiation-induced gastrointestinal syndrome. PLoS ONE 2012, 7, e29357. [Google Scholar] [CrossRef]
- Lam, R.K.; Fung, Y.K.; Han, W.; Li, L.; Chiu, S.K.; Cheng, S.H.; Yu, K.N. Modulation of NF-κB in rescued irradiated cells. Radiat. Prot. Dosim. 2015, 167, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Tanno, B.; Babini, G.; Leonardi, S.; De Stefano, I.; Merla, C.; Novelli, F.; Antonelli, F.; Casciati, A.; Tanori, M.; Pasquali, E.; et al. miRNA-Signature of Irradiated Ptch1+/− Mouse Lens is Dependent on Genetic Background. Radiat. Res. 2021, 197, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Mutschelknaus, L.; Peters, C.; Winkler, K.; Yentrapalli, R.; Heider, T.; Atkinson, M.J.; Moertl, S. Exosomes Derived from Squamous Head and Neck Cancer Promote Cell Survival after Ionizing Radiation. PLoS ONE 2016, 11, e0152213. [Google Scholar] [CrossRef] [PubMed]
- Moertl, S.; Buschmann, D.; Azimzadeh, O.; Schneider, M.; Kell, R.; Winkler, K.; Tapio, S.; Hornhardt, S.; Merl-Pham, J.; Pfaffl, M.W.; et al. Radiation Exposure of Peripheral Mononuclear Blood Cells Alters the Composition and Function of Secreted Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 2336. [Google Scholar] [CrossRef] [PubMed]
- Gan, F.; Wang, R.; Lyu, P.; Li, Y.; Fu, R.; Du, Y.; Gong, P.; Yao, Y. Plasma-Derived Exosomes Boost the Healing of Irradiated Wound by Regulating Cell Proliferation and Ferroptosis. J. Biomed. Nanotechnol. 2021, 17, 100–114. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, Y.; Zhang, Y.; Gao, L.; Zhang, N.; Feng, H. Therapeutic Potential of Umbilical Cord Mesenchymal Stem Cells for Inhibiting Myofibroblastic Differentiation of Irradiated Human Lung Fibroblasts. Tohoku J. Exp. Med. 2015, 236, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Kong, F.; Yuan, Y.; Seth, P.; Xu, W.; Wang, H.; Xiao, F.; Wang, L.; Zhang, Q.; Yang, Y.; et al. Decorin-Modified Umbilical Cord Mesenchymal Stem Cells (MSCs) Attenuate Radiation-Induced Lung Injuries via Regulating Inflammation, Fibrotic Factors, and Immune Responses. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 945–956. [Google Scholar] [CrossRef]
- Gong, W.; Guo, M.; Han, Z.; Wang, Y.; Yang, P.; Xu, C.; Wang, Q.; Du, L.; Li, Q.; Zhao, H.; et al. Mesenchymal stem cells stimulate intestinal stem cells to repair radiation-induced intestinal injury. Cell Death Dis. 2016, 7, e2387. [Google Scholar] [CrossRef]
- Nguyen, V.V.T.; Witwer, K.W.; Verhaar, M.C.; Strunk, D.; van Balkom, B.W.M. Functional assays to assess the therapeutic potential of extracellular vesicles. J. Extracell. Vesicles 2020, 10, e12033. [Google Scholar] [CrossRef] [PubMed]
- Dickens, A.M.; Tovar-Y.-Romo, L.B.; Yoo, S.W.; Trout, A.L.; Bae, M.; Kanmogne, M.; Megra, B.; Williams, D.W.; Witwer, K.W.; Gacias, M.; et al. Astrocyte-shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions. Sci. Signal. 2017, 10, eaai7696. [Google Scholar] [CrossRef]
- Couch, Y.; Akbar, N.; Roodselaar, J.; Evans, M.C.; Gardiner, C.; Sargent, I.; Romero, I.A.; Bristow, A.; Buchan, A.M.; Haughey, N.; et al. Circulating endothelial cell-derived extracellular vesicles mediate the acute phase response and sickness behaviour associated with CNS inflammation. Sci. Rep. 2017, 7, 9574. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Stoica, B.A.; Loane, D.J.; Yang, M.; Abulwerdi, G.; Khan, N.; Kumar, A.; Thom, S.R.; Faden, A.I. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J. Neuroinflamm. 2017, 14, 47. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.A.; et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, D.; Kuo, W.P.; Frühbeis, C.; Sun, J.J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Krämer-Albers, E.M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130510. [Google Scholar] [CrossRef]
- Guitart, K.; Loers, G.; Buck, F.; Bork, U.; Schachner, M.; Kleene, R. Improvement of neuronal cell survival by astrocyte-derived exosomes under hypoxic and ischemic conditions depends on prion protein. Glia 2016, 64, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Giedzinski, E.; Angulo, M.C.; Lui, T.; Lu, C.; Park, A.L.; Tang, S.; Martirosian, V.; Ru, N.; Chmielewski, N.N.; et al. Functional equivalence of stem cell and stem cell-derived extracellular vesicle transplantation to repair the irradiated brain. Stem Cells Transl. Med. 2020, 9, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, M.; Pasquali, E.; Leonardi, S.; Rebessi, S.; Tanori, M.; Giardullo, P.; Borra, F.; Pazzaglia, S.; Naus, C.C.; Di Majo, V.; et al. Role of connexin43 and ATP in long-range bystander radiation damage and oncogenesis in vivo. Oncogene 2011, 30, 4601–4608. [Google Scholar] [CrossRef]
- Mancuso, M.; Pasquali, E.; Giardullo, P.; Leonardi, S.; Tanori, M.; Di Majo, V.; Pazzaglia, S.; Saran, A. The radiation bystander effect and its potential implications for human health. Curr. Mol. Med. 2012, 12, 613–624. [Google Scholar] [CrossRef]
- Mancuso, M.; Giardullo, P.; Leonardi, S.; Pasquali, E.; Casciati, A.; De Stefano, I.; Tanori, M.; Pazzaglia, S.; Saran, A. Dose and spatial effects in long-distance radiation signaling in vivo: Implications for abscopal tumorigenesis. Int. J. Radiat. Oncol. Biol. Phys. 2013, 85, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Lyng, F.M.; Maguire, P.; McClean, B.; Seymour, C.; Mothersill, C. The involvement of calcium and MAP kinase signaling pathways in the production of radiation-induced bystander effects. Radiat. Res. 2006, 165, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Folkard, M.; Michael, B.D.; Prise, K.M. Targeted cytoplasmic irradiation induces bystander responses. Proc. Natl. Acad. Sci. USA 2004, 101, 13495–13500. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The proteome of native adult muller glial cells from murine retina. Mol. Cell Proteom. 2016, 15, 462–480. [Google Scholar] [CrossRef]
- Hladik, D.; Dalke, C.; von Toerne, C.; Hauck, S.M.; Azimzadeh, O.; Philipp, J.; Ung, M.C.; Schlattl, H.; Rößler, U.; Graw, J.; et al. CREB Signaling Mediates Dose-Dependent Radiation Response in the Murine Hippocampus Two Years after Total Body Exposure. J. Proteome Res. 2020, 19, 337–345. [Google Scholar] [CrossRef]
- Navarro, P.; Trevisan-Herraz, M.; Bonzon-Kulichenko, E.; Núñez, E.; Martínez-Acedo, P.; Pérez-Hernández, D.; Jorge, I.; Mesa, R.; Calvo, E.; Carrascal, M.; et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014, 13, 1234–1247. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]

| Accession | Description | Gene Symbol | Abundance Ratio (2 Gy, PBI)/0 Gy) |
|---|---|---|---|
| Q9D0M3 | Cytochrome c1, heme protein, mitochondrial | Cyc1 | 0.010 |
| O54990 | Prominin-1 | Prom1 | 0.010 |
| Q8JZM8 | Mucin-4 | Muc4 | 0.010 |
| Q62168 | Keratin, type I cuticular Ha2 | Krt32 | 0.010 |
| Q497I4 | Keratin, type I cuticular Ha5 | Krt35 | 0.010 |
| O70456 | 14-3-3 protein sigma | Sfn | 0.010 |
| P97350 | Plakophilin-1 | Pkp1 | 0.010 |
| Q8BZ98 | Dynamin-3 | Dnm3 | 0.010 |
| P70315 | Wiskott–Aldrich syndrome protein homolog | Was | 0.010 |
| Q923U0 | TOM1-like protein 1 | Tom1l1 | 0.010 |
| P47963 | 60S ribosomal protein L13 | Rpl13 | 0.010 |
| P12970 | 60S ribosomal protein L7a | Rpl7a | 0.010 |
| P62281 | 40S ribosomal protein S11 | Rps11 | 0.010 |
| O09167 | 60S ribosomal protein L21 | Rpl21 | 0.010 |
| P62717 | 60S ribosomal protein L18a | Rpl18a | 0.010 |
| Q8VCM7 | Fibrinogen gamma chain | Fgg | 0.031 |
| Q8K0E8 | Fibrinogen beta chain | Fgb | 0.038 |
| E9PV24 | Fibrinogen alpha chain | Fga | 0.040 |
| Q9D3D9 | ATP synthase subunit delta, mitochondrial | Atp5d | 0.040 |
| Q7TMD7 | Desmoglein-4 | Dsg4 | 0.094 |
| Q99K41 | EMILIN-1 | Emilin1 | 0.096 |
| P97861 | Keratin, type II cuticular Hb6 | Krt86 | 0.106 |
| Q60841 | Reelin | Reln | 0.139 |
| Q9JHJ0 | Tropomodulin-3 | Tmod3 | 0.143 |
| Q3SXB8 | Collectin-11 | Colec11 | 0.143 |
| Q8BH61 | Coagulation factor XIII A chain | F13a1 | 0.151 |
| Q8R0Z6 | Angiopoietin-related protein 6 | Angptl6 | 0.154 |
| P15864 | Histone H1.2 | Hist1h1c | 0.168 |
| Q9JLY7 | Dual specificity protein phosphatase 14 | Dusp14 | 0.180 |
| Q63836 | Selenium-binding protein 2 | Selenbp2 | 0.184 |
| Q8CF98 | Collectin-10 | Colec10 | 0.184 |
| P62806 | Histone H4 | Hist1h4j | 0.187 |
| Q9D1R9 | 60S ribosomal protein L34 | Rpl34 | 0.189 |
| P84099 | 60S ribosomal protein L19 | Rpl19 | 0.201 |
| Q07968 | Coagulation factor XIII B chain | F13b | 0.204 |
| Q6ZWV3 | 60S ribosomal protein L10 | Rpl10 | 0.210 |
| P62267 | 40S ribosomal protein S23 | Rps23 | 0.244 |
| P17710 | Hexokinase-1 | Hk1 | 5.598 |
| P19783 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | Cox4i1 | 6.534 |
| Q64521 | Glycerol-3-phosphate dehydrogenase, mitochondrial | Gpd2 | 6.570 |
| Q64314 | Hematopoietic progenitor cell antigen CD34 | Cd34 | 8.780 |
| Q9Z126 | Platelet factor 4 | Pf4 | 14.195 |
| Q9DCW4 | Electron transfer flavoprotein subunit beta | Etfb | 100.00 |
| Q8K021 | Secretory carrier-associated membrane protein 1 | Scamp1 | 100.00 |
| Q4FJU9 | Transmembrane protein 40 | Tmem40 | 100.00 |
| P70168 | Importin subunit beta-1 | Kpnb1 | 100.00 |
| Q61559 | IgG receptor FcRn large subunit p51 | Fcgrt | 100.00 |
| Q8VEK3 | Heterogeneous nuclear ribonucleoprotein U | Hnrnpu | 100.00 |
| B2RUP2 | Protein unc-13 homolog D | Unc13d | 100.00 |
| Q7TQ48 | Sarcalumenin | Srl | 100.00 |
| P51660 | Peroxisomal multifunctional enzyme type 2 | Hsd17b4 | 100.00 |
| P53994 | Ras-related protein Rab-2A | Rab2a | 100.00 |
| Q91VD9 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Ndufs1 | 100.00 |
| Q8BMS1 | Trifunctional enzyme subunit alpha, mitochondrial | Hadha | 100.00 |
| P06880 | Somatotropin | Gh | 100.00 |
| Q6Q477 | Plasma membrane calcium-transporting ATPase 4 | Atp2b4 | 100.00 |
| O35639 | Annexin A3 | Anxa3 | 100.00 |
| Q3TL44 | NLR family member X1 | Nlrx1 | 100.00 |
| Q9JIM1 | Equilibrative nucleoside transporter 1 | Slc29a1 | 100.00 |
| P35486 | Pyruvate dehydrogenase E1 component subunit alpha, mitochondrial | Pdha1 | 100.00 |
| O08795 | Glucosidase 2 subunit beta | Prkcsh | 100.00 |
| Q9ET30 | Transmembrane 9 superfamily member 3 | Tm9sf3 | 100.00 |
| Q8K2B3 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | Sdha | 100.00 |
| P50096 | Inosine-5’-monophosphate dehydrogenase 1 | Impdh1 | 100.00 |
| Q99KI0 | Aconitate hydratase, mitochondrial | Aco2 | 100.00 |
| P97429 | Annexin A4 | Anxa4 | 100.00 |
| Q9Z1G4 | V-type proton ATPase 116 kDa subunit a isoform 1 | Atp6v0a1 | 100.00 |
| Q08481 | Platelet endothelial cell adhesion molecule | Pecam1 | 100.00 |
| Q3V0K9 | Plastin-1 | Pls1 | 100.00 |
| E9PYK3 | Protein mono-ADP-ribosyltransferase PARP4 | Parp4 | 100.00 |
| Q9CR68 | Cytochrome b-c1 complex subunit Rieske, mitochondrial | Uqcrfs1 | 100.00 |
| P61804 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit DAD1 | Dad1 | 100.00 |
| P05366 | Serum amyloid A-1 protein | Saa1 | 100.00 |
| Q60864 | Stress-induced-phosphoprotein 1 | Stip1 | 100.00 |
| Q8K4F0 | CD226 antigen | Cd226 | 100.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pazzaglia, S.; Tanno, B.; De Stefano, I.; Giardullo, P.; Leonardi, S.; Merla, C.; Babini, G.; Tuncay Cagatay, S.; Mayah, A.; Kadhim, M.; et al. Micro-RNA and Proteomic Profiles of Plasma-Derived Exosomes from Irradiated Mice Reveal Molecular Changes Preventing Apoptosis in Neonatal Cerebellum. Int. J. Mol. Sci. 2022, 23, 2169. https://doi.org/10.3390/ijms23042169
Pazzaglia S, Tanno B, De Stefano I, Giardullo P, Leonardi S, Merla C, Babini G, Tuncay Cagatay S, Mayah A, Kadhim M, et al. Micro-RNA and Proteomic Profiles of Plasma-Derived Exosomes from Irradiated Mice Reveal Molecular Changes Preventing Apoptosis in Neonatal Cerebellum. International Journal of Molecular Sciences. 2022; 23(4):2169. https://doi.org/10.3390/ijms23042169
Chicago/Turabian StylePazzaglia, Simonetta, Barbara Tanno, Ilaria De Stefano, Paola Giardullo, Simona Leonardi, Caterina Merla, Gabriele Babini, Seda Tuncay Cagatay, Ammar Mayah, Munira Kadhim, and et al. 2022. "Micro-RNA and Proteomic Profiles of Plasma-Derived Exosomes from Irradiated Mice Reveal Molecular Changes Preventing Apoptosis in Neonatal Cerebellum" International Journal of Molecular Sciences 23, no. 4: 2169. https://doi.org/10.3390/ijms23042169
APA StylePazzaglia, S., Tanno, B., De Stefano, I., Giardullo, P., Leonardi, S., Merla, C., Babini, G., Tuncay Cagatay, S., Mayah, A., Kadhim, M., Lyng, F. M., von Toerne, C., Khan, Z. N., Subedi, P., Tapio, S., Saran, A., & Mancuso, M. (2022). Micro-RNA and Proteomic Profiles of Plasma-Derived Exosomes from Irradiated Mice Reveal Molecular Changes Preventing Apoptosis in Neonatal Cerebellum. International Journal of Molecular Sciences, 23(4), 2169. https://doi.org/10.3390/ijms23042169








