EMBER—Embedding Multiple Molecular Fingerprints for Virtual Screening
Abstract
:1. Introduction
- The EMBER (EMBedding multiplE molecular fingeRprints) embedding is proposed, which is made by multiple molecular fingerprints that have been generated using complementary methods to search for molecular substructures and are stacked as the spectra of a sort of “molecular image”; such an embedding aims at exploiting the ability of Convolutional Neural Networks (CNN) in learning the proper features, as they do for images;
- A multi classifier has been developed to prove the previous claim, which performs very well in screening ligands on twenty protein kinases presenting the closest binding sites to CDK1; moreover, our architectural design lowers the parameter numberl
- A curated data set made by nearly 90,000 ligands labeled as active/inactive against 20 Kinase target selected as the most similar to CDK1.
- An explainability analysis has been performed to assess the most relevant features for the classification task, and the results of this analysis confirm some very recent in vitro studies that outline the relevance of pharmacophore-like description fingerprints when addressing bioactivity classification for kinase inhibitors
1.1. Theoretical Remarks
1.1.1. Deep Neural Networks for Virtual Screening
1.1.2. Molecular Embeddings
2. Results
3. Discussion
4. Materials and Methods
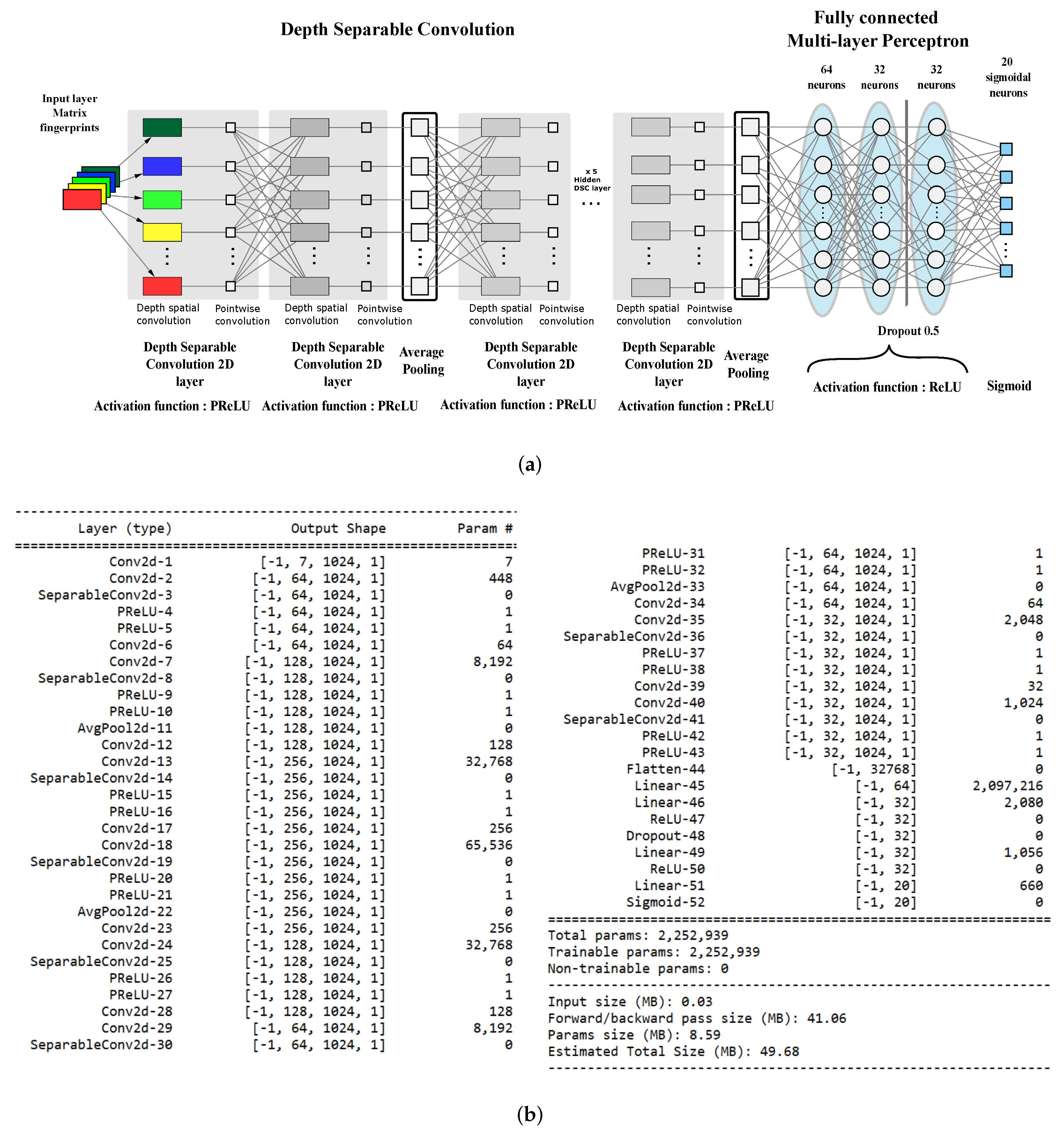
4.1. EMBER Multi-Fingerprint Embedding
4.2. Data Preparation
- Molecular weight > 100;
- Number of carbon atoms > 10;
- Number of nitrogen atoms > 2;
- Number of oxygen atoms > 2;
- At least one aromatic ring.
4.3. The Proposed Architecture
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berdigaliyev, N.; Aljofan, M. An overview of drug discovery and development. Future Med. Chem. 2020, 12, 939–947. [Google Scholar] [CrossRef]
- Turner, J.R. New Drug Development: An Introduction to Clinical Trials, 2nd ed.; Springer: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- DiMasi, J.; Hansen, R.; Grabowski, H. The Price of Innovation: New Estimates of Drug Development Costs. J. Health Econ. 2003, 22, 151–185. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; MacKerell, A.D. Computer-Aided Drug Design Methods. Methods Mol. Biol. 2017, 1520, 85–106. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, I.J.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Muegge, I.; Mukherjee, P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug Discov. 2016, 11, 137–148. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Cicenas, J.; Tamosaitis, L.; Kvederaviciute, K.; Tarvydas, R.; Staniute, G.; Kalyan, K.; Meskinyte-Kausiliene, E.; Stankevicius, V.; Valius, M. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med. Oncol. 2017, 34, 26. [Google Scholar] [CrossRef]
- Diril, M.K.; Ratnacaram, C.K.; Padmakumar, V.; Du, T.; Wasser, M.; Coppola, V.; Tessarollo, L.; Kaldis, P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 2012, 109, 3826–3831. [Google Scholar] [CrossRef] [Green Version]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [Green Version]
- Jing, Y.; Bian, Y.; Hu, Z.; Wang, L.; Xie, X.Q.S. Deep Learning for Drug Design: An Artificial Intelligence Paradigm for Drug Discovery in the Big Data Era. AAPS J. 2018, 20, 1–10. [Google Scholar]
- Schneider, G. Mind and machine in drug design. Nat. Mach. Intell. 2019, 1, 128–130. [Google Scholar] [CrossRef]
- Kimber, T.B.; Chen, Y.; Volkamer, A. Deep Learning in Virtual Screening: Recent Applications and Developments. Int. J. Mol. Sci. 2021, 22, 4435. [Google Scholar] [CrossRef] [PubMed]
- Sydow, D.; Burggraaff, L.; Szengel, A.; van Vlijmen, H.W.T.; IJzerman, A.P.; van Westen, G.J.P.; Volkamer, A. Advances and Challenges in Computational Target Prediction. J. Chem. Inf. Model. 2019, 59, 1728–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahl, G.E.; Jaitly, N.; Salakhutdinov, R. Multi-task Neural Networks for QSAR Predictions. arXiv 2014, arXiv:1406.1231. [Google Scholar]
- Wallach, I.; Dzamba, M.; Heifets, A. AtomNet: A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery. arXiv 2015, arXiv:1510.02855. [Google Scholar]
- Duvenaud, D.K.; Maclaurin, D.; Iparraguirre, J.; Bombarell, R.; Hirzel, T.; Aspuru-Guzik, A.; Adams, R.P. Convolutional networks on graphs for learning molecular fingerprints. In Proceedings of the Advances in Neural Information Processing Systems 28 (NIPS 2015), Montreal, QC, USA, 7–12 December 2015; pp. 2224–2232. [Google Scholar]
- Pereira, J.C.; Caffarena, E.R.; dos Santos, C.N. Boosting Docking-Based Virtual Screening with Deep Learning. J. Chem. Inf. Model. 2016, 56, 2495–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirohara, M.; Saito, Y.; Koda, Y.; Sato, K.; Sakakibara, Y. Convolutional neural network based on SMILES representation of compounds for detecting chemical motif. BMC Bioinform. 2018, 19, 526. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Luna, J.; Grisoni, F.; Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2020, 2, 573–584. [Google Scholar] [CrossRef]
- Grisoni, F.; Schneider, G. De novo Molecular Design with Generative Long Short-term Memory. CHIMIA Int. J. Chem. 2019, 73, 1006–1011. [Google Scholar] [CrossRef]
- Karpov, P.; Godin, G.; Tetko, I.V. Transformer-CNN: Swiss knife for QSAR modeling and interpretation. J. Cheminform. 2020, 12, 17. [Google Scholar] [CrossRef] [Green Version]
- Bjerrum, E.J. SMILES Enumeration as Data Augmentation for Neural Network Modeling of Molecules. arXiv 2017, arXiv:1703.07076. [Google Scholar]
- Mikolov, T.; Le, Q.V.; Sutskever, I. Exploiting Similarities among Languages for Machine Translation. arXiv 2013, arXiv:1309.4168. [Google Scholar]
- Kearnes, S.; McCloskey, K.; Berndl, M.; Pande, V.; Riley, P. Molecular Graph Convolutions: Moving Beyond Fingerprints. J. Comput.-Aided Mol. Des. 2016, 30, 595–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coley, C.W.; Barzilay, R.; Green, W.H.; Jaakkola, T.S.; Jensen, K.F. Convolutional Embedding of Attributed Molecular Graphs for Physical Property Prediction. J. Chem. Inf. Model. 2017, 57, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural Message Passing for Quantum Chemistry. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017. [Google Scholar]
- Torng, W.; Altman, R.B. Graph Convolutional Neural Networks for Predicting Drug-Target Interactions. J. Chem. Inf. Model. 2019, 59, 4131–4149. [Google Scholar] [CrossRef] [PubMed]
- Koge, D.; Ono, N.; Huang, M.; Altaf-Ul-Amin, M.; Kanaya, S. Embedding of Molecular Structure Using Molecular Hypergraph Variational Autoencoder with Metric Learning. Mol. Inform. 2021, 40, 2000203. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Oono, K.; Hayashi, K. Weisfeiler-Lehman Embedding for Molecular Graph Neural Networks. arXiv 2020, arXiv:2006.06909. [Google Scholar]
- Bender, A.; Glen, R.C. A Discussion of Measures of Enrichment in Virtual Screening: Comparing the Information Content of Descriptors with Increasing Levels of Sophistication. J. Chem. Inf. Model. 2005, 45, 1369–1375. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Lipovetsky, S.; Conklin, M. Analysis of regression in game theory approach. Appl. Stoch. Model. Bus. Ind. 2001, 17, 319–330. [Google Scholar] [CrossRef]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning Important Features Through Propagating Activation Differences. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017. [Google Scholar]
- Zhu, Y.; Alqahtani, S.; Hu, X. Aromatic Rings as Molecular Determinants for the Molecular Recognition of Protein Kinase Inhibitors. Molecules 2021, 26, 1776. [Google Scholar] [CrossRef] [PubMed]
- Mendolia, I.; Contino, S.; Perricone, U.; Ardizzone, E.; Pirrone, R. Convolutional architectures for virtual screening. BMC Bioinform. 2020, 21, 310. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Inakollu, V.S.S.; Sherman, W. Boosting Virtual Screening Enrichments with Data Fusion: Coalescing Hits from Two-Dimensional Fingerprints, Shape, and Docking. J. Chem. Inf. Model. 2013, 53, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cruz, A.; Ramsey, S.; Dickson, C.J.; Duca, J.S.; Hornak, V.; Koes, D.R.; Kurtzman, T. Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLoS ONE 2019, 14, e0220113. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shen, C.; Huang, N. Predicting or Pretending: Artificial Intelligence for Protein-Ligand Interactions Lack of Sufficiently Large and Unbiased Datasets. Front. Pharmacol. 2020, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME—The Konstanz Information Miner: Version 2.0 and Beyond. SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Kooistra, A.J.; Kanev, G.K.; van Linden, O.P.; Leurs, R.; de Esch, I.J.; de Graaf, C. KLIFS: A structural kinase-ligand interaction database. Nucleic Acids Res. 2015, 44, D365–D371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Xia, J.; Tilahun, E.L.; Reid, T.E.; Zhang, L.; Wang, X.S. Benchmarking methods and data sets for ligand enrichment assessment in virtual screening. Methods 2015, 71, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]

| Target | Acc. | Loss | Sensitivity | MCC | AUC | F1-Score |
|---|---|---|---|---|---|---|
| ACK | 0.9957 | 0.0226 | 0.5000 | 0.6742 | 0.9834 | 0.6463 |
| ALK | 0.9930 | 0.0402 | 0.6575 | 0.7913 | 0.9904 | 0.7804 |
| CDK1 | 0.9910 | 0.0314 | 0.4537 | 0.6397 | 0.9850 | 0.6059 |
| CDK2 | 0.9859 | 0.0431 | 0.5281 | 0.6338 | 0.9845 | 0.6287 |
| CDK6 | 0.9966 | 0.0210 | 0.5865 | 0.7523 | 0.9895 | 0.7305 |
| INSR | 0.9893 | 0.0329 | 0.3779 | 0.5830 | 0.9858 | 0.5342 |
| ITK | 0.9945 | 0.0232 | 0.5886 | 0.7302 | 0.9905 | 0.7154 |
| JAK2 | 0.9898 | 0.0472 | 0.8474 | 0.9090 | 0.9950 | 0.9114 |
| JNK3 | 0.9967 | 0.0154 | 0.5905 | 0.7610 | 0.9901 | 0.7381 |
| MELK | 0.9957 | 0.0229 | 0.7081 | 0.8270 | 0.9897 | 0.8188 |
| CHK1 | 0.9895 | 0.0512 | 0.6385 | 0.7650 | 0.9846 | 0.7565 |
| CK2A1 | 0.9942 | 0.0253 | 0.5166 | 0.6944 | 0.9857 | 0.6667 |
| CLK2 | 0.9936 | 0.0259 | 0.2255 | 0.4137 | 0.9771 | 0.3485 |
| DYRK1A | 0.9916 | 0.0321 | 0.4080 | 0.5987 | 0.9776 | 0.5591 |
| EGFR | 0.9845 | 0.0604 | 0.7536 | 0.8331 | 0.9874 | 0.8357 |
| ERK2 | 0.9881 | 0.0563 | 0.7295 | 0.8292 | 0.9886 | 0.8272 |
| GSK3 | 0.9843 | 0.0554 | 0.5827 | 0.6892 | 0.9762 | 0.6856 |
| IRAK4 | 0.9936 | 0.0287 | 0.7611 | 0.8611 | 0.9938 | 0.8571 |
| MAP2K1 | 0.9931 | 0.0319 | 0.5497 | 0.7184 | 0.9795 | 0.6954 |
| PDK1 | 0.9945 | 0.0271 | 0.6310 | 0.7757 | 0.9875 | 0.7613 |
| Protein | TP/P 1% * | TP/P 2% * | TP/P 5% * | TP/P 10% * | EF 1% | EF 2% | EF 5% | EF 10% |
|---|---|---|---|---|---|---|---|---|
| ACK | 72/106 | 84/106 | 95/106 | 101/106 | 68 | 40 | 18 | 10 |
| ALK | 131/254 | 202/254 | 229/254 | 247/254 | 52 | 40 | 18 | 10 |
| CDK1 | 111/205 | 150/205 | 189/205 | 196/205 | 54 | 37 | 18 | 10 |
| CDK2 | 118/303 | 194/303 | 264/303 | 289/303 | 39 | 32 | 17 | 10 |
| CDK6 | 79/104 | 90/104 | 98/104 | 101/104 | 76 | 43 | 19 | 10 |
| INSR | 110/217 | 145/217 | 195/217 | 206/217 | 51 | 33 | 18 | 9 |
| ITK | 107/158 | 125/158 | 148/158 | 155/158 | 68 | 40 | 19 | 10 |
| JAK2 | 134/832 | 268/832 | 669/832 | 804/832 | 16 | 16 | 16 | 10 |
| JNK3 | 81/105 | 88/105 | 95/105 | 102/105 | 77 | 42 | 18 | 10 |
| MELK | 130/185 | 157/185 | 178/185 | 181/185 | 70 | 42 | 19 | 10 |
| CHK1 | 134/343 | 233/343 | 300/343 | 324/343 | 39 | 34 | 17 | 9 |
| CK2A1 | 100/151 | 117/151 | 141/151 | 146/151 | 66 | 39 | 19 | 10 |
| CLK2 | 59/102 | 73/102 | 87/102 | 96/102 | 58 | 36 | 17 | 9 |
| DYRK1A | 97/174 | 126/174 | 152/174 | 162/174 | 56 | 36 | 17 | 9 |
| EGFR | 134/702 | 268/702 | 586/702 | 664/702 | 19 | 19 | 17 | 9 |
| ERK2 | 133/525 | 267/525 | 471/525 | 505/525 | 25 | 25 | 18 | 10 |
| GSK3 | 132/393 | 226/393 | 327/393 | 353/393 | 34 | 29 | 17 | 9 |
| IRAK4 | 134/339 | 263/339 | 320/339 | 333/339 | 40 | 39 | 19 | 10 |
| MAP2K1 | 118/191 | 142/191 | 167/191 | 178/191 | 62 | 37 | 17 | 9 |
| PDK1 | 123/187 | 149/187 | 170/187 | 181/187 | 66 | 40 | 18 | 10 |
| Molecule ChEMBLID | Chemical Structure | |
|---|---|---|
| CHEMBL192216 |  | 2 nM |
| CHEMBL3644025 |  | 82 nM |
| CHEMBL445125 |  | 500 nM |
| CHEMBL2403087 |  | 183 nM |
| CHEMBL2403084 |  | 148 nM |
| Target | PDB ID | Ligand Code * | Actives | Inactives |
|---|---|---|---|---|
| ACK | 5ZXB | 9KO | 746 | 159,775 |
| ALK | 6E0R | HKJ | 1665 | 227,247 |
| CDK1 | 6GU2 | F9Z | 1241 | 124,473 |
| CDK2 | 6INL | AJR | 1924 | 225,087 |
| CDK6 | 5L2S | 6ZV | 646 | 256,561 |
| INSR | 5E1S | 5JA | 1423 | 195,990 |
| ITK | 4RFM | 3P6 | 1001 | 135,007 |
| JAK2 | 6M9H | J9D | 5526 | 577,409 |
| JNK3 | 2B1P | AIZ | 658 | 95,252 |
| MELK | 6GVX | TAK | 1215 | 246,662 |
| CHK1 | 6FC8 | D4Q | 2175 | 21,763 |
| CK2a1 | 6JWA | 5ID | 1053 | 10,534 |
| CLK2 | 6FYL | 3NG | 671 | 6800 |
| DYRK1A | 4YLK | 4E2 | 1126 | 11,274 |
| EGFR | 5GNK | 80U | 4757 | 47,541 |
| ERK2 | 6OPH | 6QB | 3525 | 35,237 |
| GSK3B | 5F94 | 3UO | 2578 | 25,768 |
| IRAK4 | 6EG9 | OLI | 2131 | 21,282 |
| MAPK2K1 | 4AN9 | ACP; 2P7 | 1254 | 12,508 |
| PDK1 | 3NAX | MP7 | 1117 | 11,166 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendolia, I.; Contino, S.; De Simone, G.; Perricone, U.; Pirrone, R. EMBER—Embedding Multiple Molecular Fingerprints for Virtual Screening. Int. J. Mol. Sci. 2022, 23, 2156. https://doi.org/10.3390/ijms23042156
Mendolia I, Contino S, De Simone G, Perricone U, Pirrone R. EMBER—Embedding Multiple Molecular Fingerprints for Virtual Screening. International Journal of Molecular Sciences. 2022; 23(4):2156. https://doi.org/10.3390/ijms23042156
Chicago/Turabian StyleMendolia, Isabella, Salvatore Contino, Giada De Simone, Ugo Perricone, and Roberto Pirrone. 2022. "EMBER—Embedding Multiple Molecular Fingerprints for Virtual Screening" International Journal of Molecular Sciences 23, no. 4: 2156. https://doi.org/10.3390/ijms23042156
APA StyleMendolia, I., Contino, S., De Simone, G., Perricone, U., & Pirrone, R. (2022). EMBER—Embedding Multiple Molecular Fingerprints for Virtual Screening. International Journal of Molecular Sciences, 23(4), 2156. https://doi.org/10.3390/ijms23042156







