Figure 1.
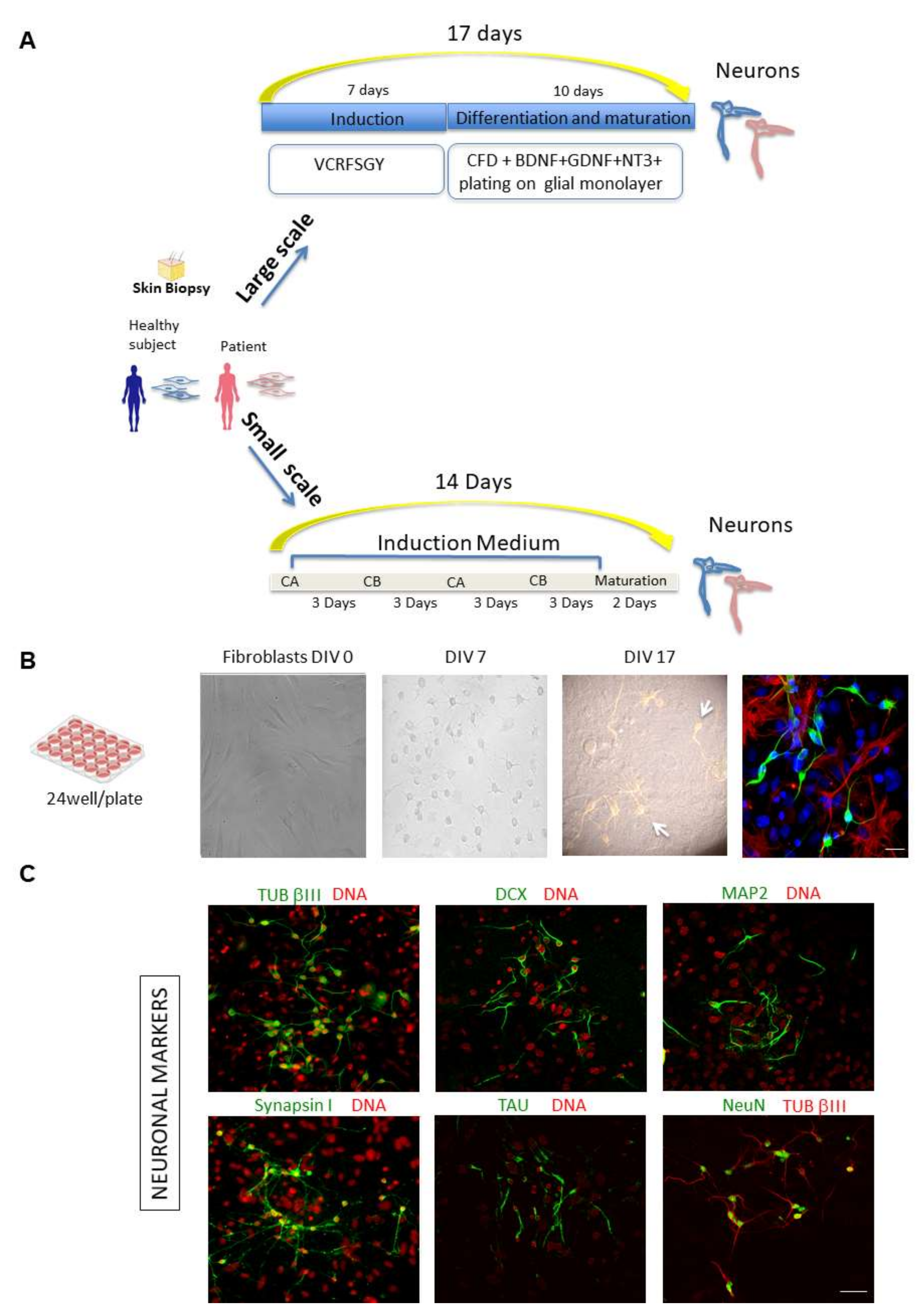
Chemical approaches to covert human fibroblasts into neurons: large-scale and small-scale experiments. (A) Schematic representation of the chemical approaches used to covert fibroblasts from healthy subjects and patients into neurons. Human fibroblasts isolated from skin biopsy (from Coriell BioBank, Camden, NJ, USA) were treated with specific chemical cocktails. Large scale: Fibroblasts were plated in DMEM in a 24-well plate (DIV 0). One day later, cells were transferred into induction medium with chemical compounds VCRFSGY (V, VPA; C, CHIR99021; R, Repsox; F, Forskolin; S, SP600625; G, GO6983; Y, Y-27632) and cultured for 7 days. Cells were then detached by enzymatic activity, further re-plated onto a layer of mouse glial cells and cultured in maturation medium with supplementary chemicals CFD (C, CHIR99021; R, Repsox; F, Forskolin; D, Dorsomorphin) and neurotrophic factors for 10 days. After fixation and permeabilization, cells were subjected to confocal microscopy analysis. Small scale: Schematic representation of the procedure for direct conversion of fibroblasts using chemical compounds on chip devises. A combination of medium CA, which mainly inhibits fibroblast signalling cascade and initiates neuronal fate transformation, in alternance with medium CB, which maintains cell survival and enhances reprogramming efficiency, is used to obtain the direct reprogramming towards a neuronal phenotype on chip. (CA: containing CHIR99021, LDN193189, RG108, dorsomorphin, P7C3-A20, A83-01, and ISX9); (CB: containing Forskolin, Y27632, DAPT, PD0325901, A83-01, purmorphamine, and P7C3-A20). (B) Representative phase-contrast images of a large-scale experiment to chemically convert human control fibroblasts showing a time course of cells at DIV 0, 7 and 17. At DIV 17, TUB βIII positive cells form a network on top of the glial monolayer. A representative composite confocal image of differentiating ciNs (DIV12) is shown by labelling with anti-TUB βIII antibody (green channel), laying on a layer of glial cells labelled with an anti-GFAP antibody (red channel). Nuclei are counterstained with Hoechst (blue channel). Scale Bar, 20 μm. (C) Representative double immunofluorescence images from a large-scale experiment show that ciNs express typical early and late neuronal markers, including TUB βIII, DCX, MAP2, Synapsin I, NeuN and TAU. Scale Bar, 50 μm.
Figure 1.
Chemical approaches to covert human fibroblasts into neurons: large-scale and small-scale experiments. (A) Schematic representation of the chemical approaches used to covert fibroblasts from healthy subjects and patients into neurons. Human fibroblasts isolated from skin biopsy (from Coriell BioBank, Camden, NJ, USA) were treated with specific chemical cocktails. Large scale: Fibroblasts were plated in DMEM in a 24-well plate (DIV 0). One day later, cells were transferred into induction medium with chemical compounds VCRFSGY (V, VPA; C, CHIR99021; R, Repsox; F, Forskolin; S, SP600625; G, GO6983; Y, Y-27632) and cultured for 7 days. Cells were then detached by enzymatic activity, further re-plated onto a layer of mouse glial cells and cultured in maturation medium with supplementary chemicals CFD (C, CHIR99021; R, Repsox; F, Forskolin; D, Dorsomorphin) and neurotrophic factors for 10 days. After fixation and permeabilization, cells were subjected to confocal microscopy analysis. Small scale: Schematic representation of the procedure for direct conversion of fibroblasts using chemical compounds on chip devises. A combination of medium CA, which mainly inhibits fibroblast signalling cascade and initiates neuronal fate transformation, in alternance with medium CB, which maintains cell survival and enhances reprogramming efficiency, is used to obtain the direct reprogramming towards a neuronal phenotype on chip. (CA: containing CHIR99021, LDN193189, RG108, dorsomorphin, P7C3-A20, A83-01, and ISX9); (CB: containing Forskolin, Y27632, DAPT, PD0325901, A83-01, purmorphamine, and P7C3-A20). (B) Representative phase-contrast images of a large-scale experiment to chemically convert human control fibroblasts showing a time course of cells at DIV 0, 7 and 17. At DIV 17, TUB βIII positive cells form a network on top of the glial monolayer. A representative composite confocal image of differentiating ciNs (DIV12) is shown by labelling with anti-TUB βIII antibody (green channel), laying on a layer of glial cells labelled with an anti-GFAP antibody (red channel). Nuclei are counterstained with Hoechst (blue channel). Scale Bar, 20 μm. (C) Representative double immunofluorescence images from a large-scale experiment show that ciNs express typical early and late neuronal markers, including TUB βIII, DCX, MAP2, Synapsin I, NeuN and TAU. Scale Bar, 50 μm.
![Ijms 23 02147 g001 Ijms 23 02147 g001]()
Figure 2.
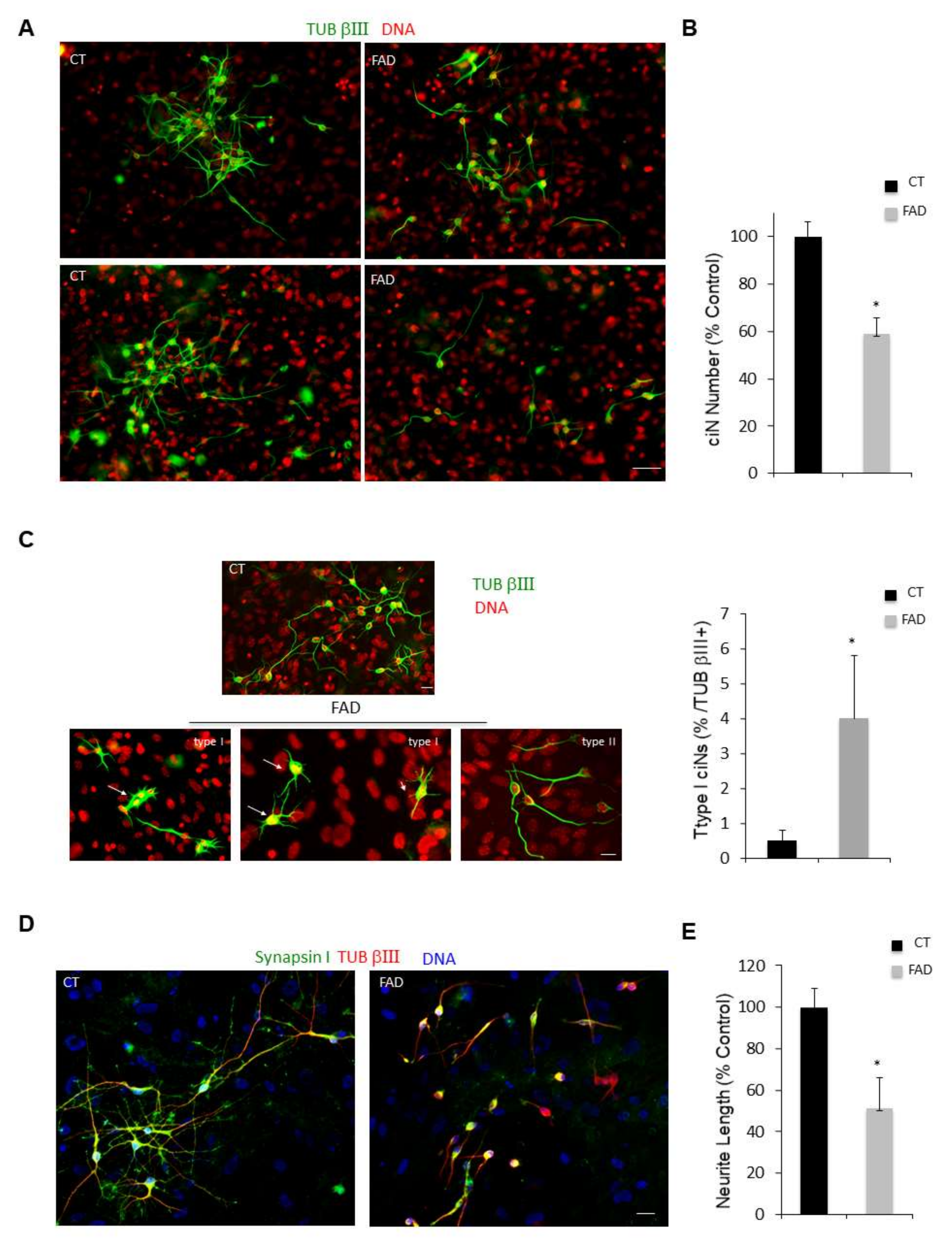
Patient fibroblasts have a reduced efficiency of reprogramming and patient ciNs present a less complex neuronal network. (A) Immunofluorescence images of ciNs obtained from direct conversions of healthy subject (CT) and Alzheimer’s Disease (FAD) patient fibroblasts, labelled with the neuronal marker TUB βIII (green channel) and DNA (red channel). Representative images show that FAD fibroblasts can be converted into neurons with a lower efficiency as compared with CT fibroblasts. Scale Bar, 50 μm. (B) Histogram data represent the percent change in the number of ciNs of FAD fibroblasts with respect to control fibroblasts. Bars in the plots represent means ± SD (n = 3, * p < 0.05 vs. control values). (C) Confocal fluorescence images show that following FAD fibroblast chemical conversion, two types of TUB βIII-positive ciNs are generated with different morphology. The so-called “type I” ciNs have flat body and a high number of short protrusions, whereas “type II” cells have a morphology similar to CT ciNs and present small cellular bodies with two or three neurites of variable length (TUB βIII, green channel; DNA red). Scale Bar, 10 μm. Histogram data show the percentage of Type I cells with respect to total TUB βIII + cells. (D) Analysis of neurite length of “type II” ciNs labelled with anti-Synapsin I antibody (a presynaptic marker, green channel), TUB βIII (red) and DNA (blue) show that FAD ciNs have a lower neurite complexity and a reduced neurite length as compared with CT cells. Scale Bar, 20 μm. (E) Histogram data represent the percent change in neurite length in FAD ciNs with respect to control neurons. Bars in the plots represent means ± SD (n = 3, * p < 0.05 vs. control values).
Figure 2.
Patient fibroblasts have a reduced efficiency of reprogramming and patient ciNs present a less complex neuronal network. (A) Immunofluorescence images of ciNs obtained from direct conversions of healthy subject (CT) and Alzheimer’s Disease (FAD) patient fibroblasts, labelled with the neuronal marker TUB βIII (green channel) and DNA (red channel). Representative images show that FAD fibroblasts can be converted into neurons with a lower efficiency as compared with CT fibroblasts. Scale Bar, 50 μm. (B) Histogram data represent the percent change in the number of ciNs of FAD fibroblasts with respect to control fibroblasts. Bars in the plots represent means ± SD (n = 3, * p < 0.05 vs. control values). (C) Confocal fluorescence images show that following FAD fibroblast chemical conversion, two types of TUB βIII-positive ciNs are generated with different morphology. The so-called “type I” ciNs have flat body and a high number of short protrusions, whereas “type II” cells have a morphology similar to CT ciNs and present small cellular bodies with two or three neurites of variable length (TUB βIII, green channel; DNA red). Scale Bar, 10 μm. Histogram data show the percentage of Type I cells with respect to total TUB βIII + cells. (D) Analysis of neurite length of “type II” ciNs labelled with anti-Synapsin I antibody (a presynaptic marker, green channel), TUB βIII (red) and DNA (blue) show that FAD ciNs have a lower neurite complexity and a reduced neurite length as compared with CT cells. Scale Bar, 20 μm. (E) Histogram data represent the percent change in neurite length in FAD ciNs with respect to control neurons. Bars in the plots represent means ± SD (n = 3, * p < 0.05 vs. control values).
![Ijms 23 02147 g002 Ijms 23 02147 g002]()
Figure 3.
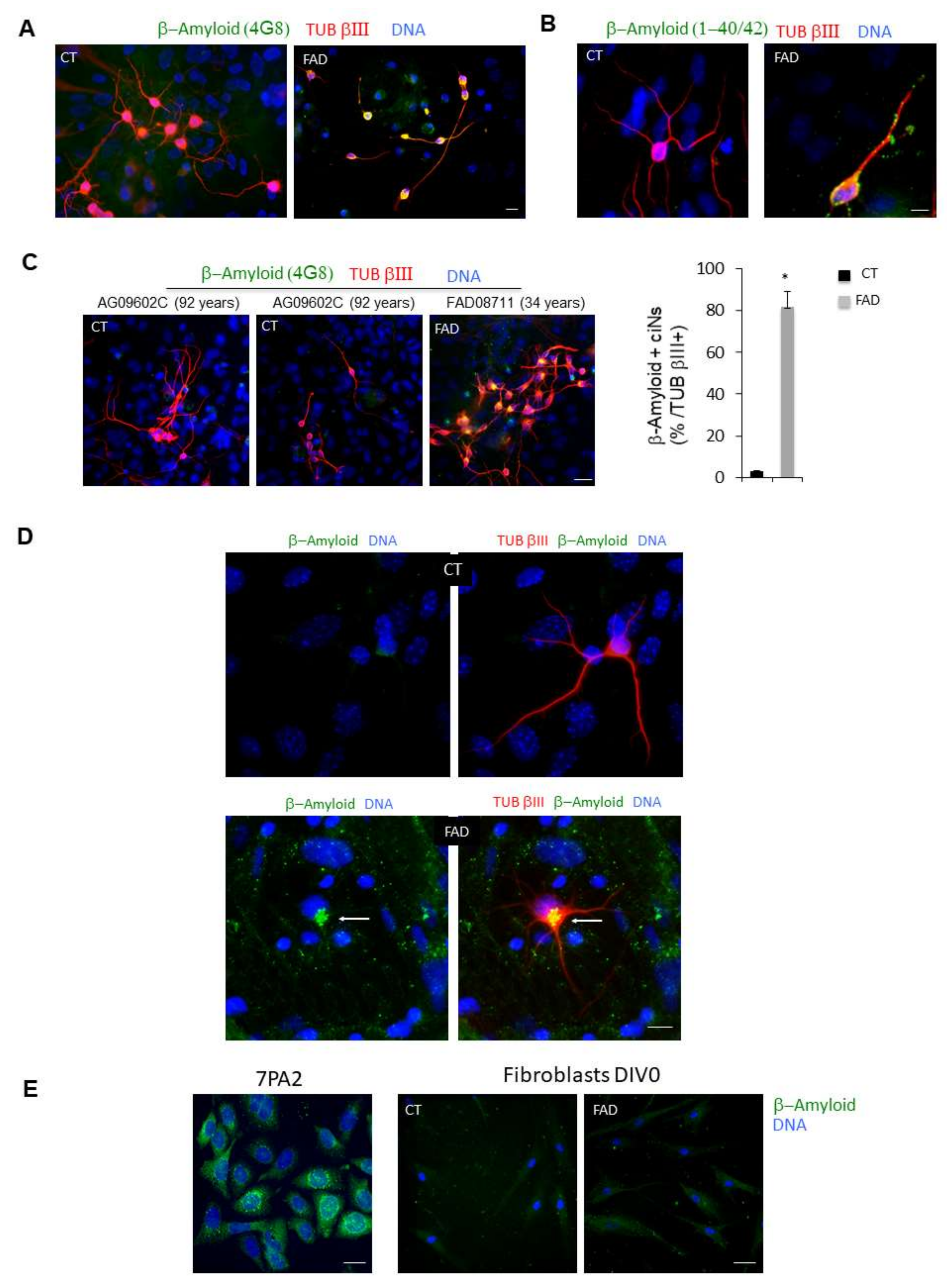
ciNs derived from FAD fibroblasts show intracellular β-Amyloid accumulation. (A) Confocal images show that, following chemical induction, fibroblasts acquire a neuronal phenotype (TUB βIII staining, red channel), and ciNs from FAD patients show a strong labelling of β-Amyloid, undetectable in CT ciNs. Nuclei were counterstained with Hoechst (blue channel). Scale Bar, 10 μm. (B) Close-up view images of ciNs obtained from CT and FAD converted fibroblasts (TUB βIII, red channel) showing the accumulation of β-Amyloid labelled with the antibody that recognises β-Amyloid 1-40/42 (green channel). Nuclei were counterstained with Hoechst (blue channel). Scale Bar, 10 μm. (C) Representative fluorescence images of TUB βIII positive ciNs (red channel) obtained from the direct conversion of fibroblasts from an old, healthy subject (92 years) showing the absence of aggregates of β-Amyloid (green channel), as compared with the FAD subject. Scale Bar, 20 μm. Histogram data represent the percentage of FAD ciNs showing β-Amyloid staining expressed as percentage of total number TUB βIII + cells (82 ± 7% β-Amyloid/ TUB βIII positive cells). Bars in the plots represent means ± SD (n = 3, * p < 0.05 vs. control values). (D) Triple confocal immunofluorescence images of ciNs showing the presence of β-Amyloid aggregates close to the nucleus in FAD cells. This aggregates have features similar to the so called “aggresomes” (white arrow) generally found in the cytoplasm as protein inclusions. β-Amyloid (green channel), TUB βIII (red channel), DNA (blue channel). No β-Amyloid aggregates are observed in CT cells. Double fluorescence images are also shown (green and blue channel) to better visualize the structure. Scale Bar, 10 μm. (E) 7PA2 CHO cells, over-expressing mutated Amyloid Precursor Protein (APP), were used as positive control for the staining with the anti β-Amyloid antibody (green channel). Differently, the same antibody failed to show any specific labelling in dermal fibroblasts at DIV 0 from both healthy subjects and FAD patients (Coriell Institute, Biobank, Camden, NJ, USA and NIH, Bethesda, MD, USA), thus indicating that direct chemical conversion of fibroblasts into neurons is crucial for detection of the pathological marker. Nuclei were counterstained with Hoechst (blue channel). Scale Bar, 20 μm.
Figure 3.
ciNs derived from FAD fibroblasts show intracellular β-Amyloid accumulation. (A) Confocal images show that, following chemical induction, fibroblasts acquire a neuronal phenotype (TUB βIII staining, red channel), and ciNs from FAD patients show a strong labelling of β-Amyloid, undetectable in CT ciNs. Nuclei were counterstained with Hoechst (blue channel). Scale Bar, 10 μm. (B) Close-up view images of ciNs obtained from CT and FAD converted fibroblasts (TUB βIII, red channel) showing the accumulation of β-Amyloid labelled with the antibody that recognises β-Amyloid 1-40/42 (green channel). Nuclei were counterstained with Hoechst (blue channel). Scale Bar, 10 μm. (C) Representative fluorescence images of TUB βIII positive ciNs (red channel) obtained from the direct conversion of fibroblasts from an old, healthy subject (92 years) showing the absence of aggregates of β-Amyloid (green channel), as compared with the FAD subject. Scale Bar, 20 μm. Histogram data represent the percentage of FAD ciNs showing β-Amyloid staining expressed as percentage of total number TUB βIII + cells (82 ± 7% β-Amyloid/ TUB βIII positive cells). Bars in the plots represent means ± SD (n = 3, * p < 0.05 vs. control values). (D) Triple confocal immunofluorescence images of ciNs showing the presence of β-Amyloid aggregates close to the nucleus in FAD cells. This aggregates have features similar to the so called “aggresomes” (white arrow) generally found in the cytoplasm as protein inclusions. β-Amyloid (green channel), TUB βIII (red channel), DNA (blue channel). No β-Amyloid aggregates are observed in CT cells. Double fluorescence images are also shown (green and blue channel) to better visualize the structure. Scale Bar, 10 μm. (E) 7PA2 CHO cells, over-expressing mutated Amyloid Precursor Protein (APP), were used as positive control for the staining with the anti β-Amyloid antibody (green channel). Differently, the same antibody failed to show any specific labelling in dermal fibroblasts at DIV 0 from both healthy subjects and FAD patients (Coriell Institute, Biobank, Camden, NJ, USA and NIH, Bethesda, MD, USA), thus indicating that direct chemical conversion of fibroblasts into neurons is crucial for detection of the pathological marker. Nuclei were counterstained with Hoechst (blue channel). Scale Bar, 20 μm.
![Ijms 23 02147 g003 Ijms 23 02147 g003]()
Figure 4.
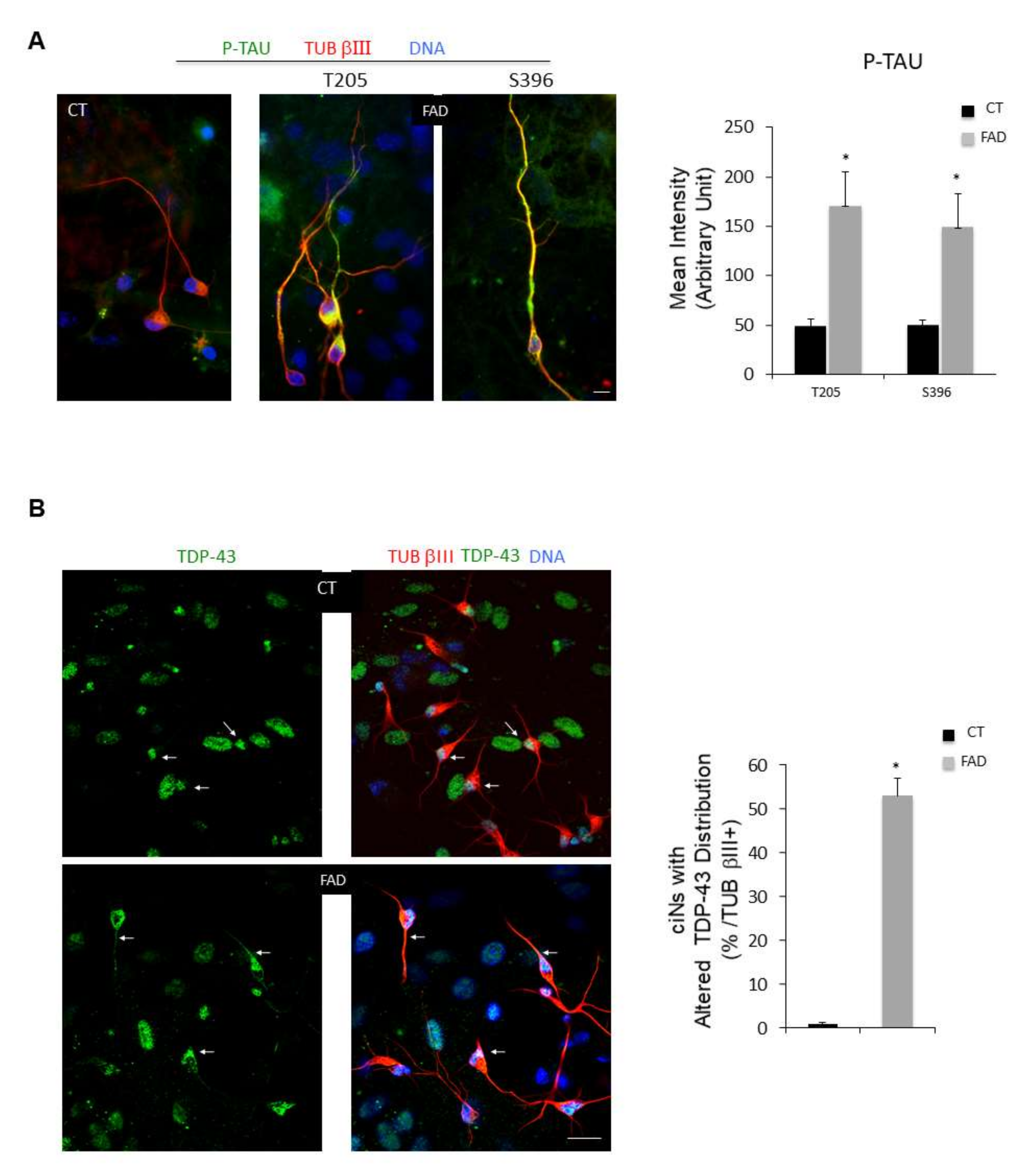
ciNs derived from FAD fibroblasts show an increase in P-TAU immunoreactivity and altered TDP-43 localization. (A) Representative triple fluorescence images of ciNs (TUB βIII labelling, red channel), obtained from chemically converted FAD fibroblasts, showing a strong labelling with two different anti P-TAU antibodies (green channel) (Thr205 and Ser396). Nuclei are counterstained with Hoechst (blue channel). Scale Bar, 10 μm. Histogram data represent the mean intensity values (arbitrary unit) of P-TAU immunolabelling in ciNs of CT and FAD by using two different phospho-antibodies (T205 and S396). Bars in the plots represent means ± SD from three independent experiments (* p < 0.05) (B) Fluorescence images of ciNs derived from CT and FAD chemically converted fibroblasts, triple labelled with TDP-43 (green), TUB βIII (red) and DNA (Blue). White arrows point to TDP-43 labelling distribution. In CT cells, all cells displayed TDP-43 exclusively accumulated in nuclei, whereas in FAD cells, TDP-43 labelling is decreased in the nucleus and redistributed along neurites. Scale Bar, 20 μm. Histogram represents the percentage of FAD ciNs showing altered TDP-43 distribution. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05).
Figure 4.
ciNs derived from FAD fibroblasts show an increase in P-TAU immunoreactivity and altered TDP-43 localization. (A) Representative triple fluorescence images of ciNs (TUB βIII labelling, red channel), obtained from chemically converted FAD fibroblasts, showing a strong labelling with two different anti P-TAU antibodies (green channel) (Thr205 and Ser396). Nuclei are counterstained with Hoechst (blue channel). Scale Bar, 10 μm. Histogram data represent the mean intensity values (arbitrary unit) of P-TAU immunolabelling in ciNs of CT and FAD by using two different phospho-antibodies (T205 and S396). Bars in the plots represent means ± SD from three independent experiments (* p < 0.05) (B) Fluorescence images of ciNs derived from CT and FAD chemically converted fibroblasts, triple labelled with TDP-43 (green), TUB βIII (red) and DNA (Blue). White arrows point to TDP-43 labelling distribution. In CT cells, all cells displayed TDP-43 exclusively accumulated in nuclei, whereas in FAD cells, TDP-43 labelling is decreased in the nucleus and redistributed along neurites. Scale Bar, 20 μm. Histogram represents the percentage of FAD ciNs showing altered TDP-43 distribution. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05).
![Ijms 23 02147 g004 Ijms 23 02147 g004]()
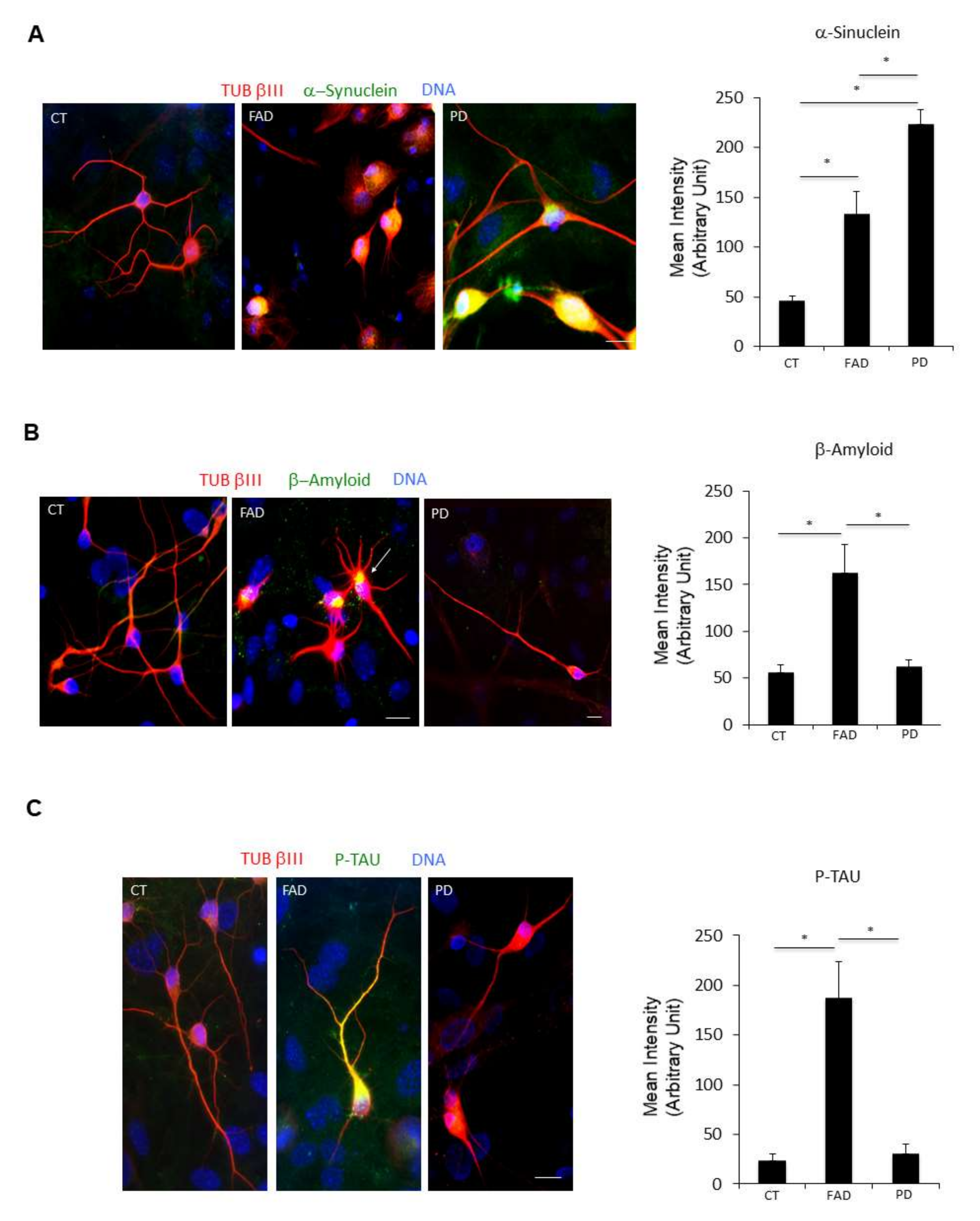
Figure 5.
ciNs from FAD and PD patients show neurodegenerative disease hallmarks. (A) Immunofluorescence images of ciNs labelled with an anti α-Synuclein antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). α-Synuclein antibody was able to strongly label PD ciNs, showing aggregates within the cell. Similarly, FAD ciNs show the presence of α-Synuclein labelling and aggregates, although with a weaker intensity. Scale Bar, 10 μm. Histogram data represent the mean intensity values (arbitrary unit) of α-Synuclein immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA). (B) Immunofluorescence images of ciNs obtained from direct conversions of CT, FAD and PD patient fibroblasts, labelled with an anti β-Amyloid antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). The images show that FAD ciNs have a strong labelling, with the anti β-Amyloid antibody consisting of aggregates localised close to the nucleus (white arrow), as compared with CT cells. On the contrary, PD ciNs do not show any evident aggregates of β-Amyloid. Scale Bar, 10 μm. Histogram data represent the mean intensity values (arbitrary unit) of β-Amyloid immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA). (C) Immunofluorescence images of ciNs labelled with an anti P-TAU antibody (Ser396, green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). P-TAU antibody was able to label the cell body and neurites of FAD ciNs, whereas it failed to stain PD ciNs. Scale Bar, 10 μm. Histogram shows the mean intensity values (arbitrary unit) of P-TAU immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA).
Figure 5.
ciNs from FAD and PD patients show neurodegenerative disease hallmarks. (A) Immunofluorescence images of ciNs labelled with an anti α-Synuclein antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). α-Synuclein antibody was able to strongly label PD ciNs, showing aggregates within the cell. Similarly, FAD ciNs show the presence of α-Synuclein labelling and aggregates, although with a weaker intensity. Scale Bar, 10 μm. Histogram data represent the mean intensity values (arbitrary unit) of α-Synuclein immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA). (B) Immunofluorescence images of ciNs obtained from direct conversions of CT, FAD and PD patient fibroblasts, labelled with an anti β-Amyloid antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). The images show that FAD ciNs have a strong labelling, with the anti β-Amyloid antibody consisting of aggregates localised close to the nucleus (white arrow), as compared with CT cells. On the contrary, PD ciNs do not show any evident aggregates of β-Amyloid. Scale Bar, 10 μm. Histogram data represent the mean intensity values (arbitrary unit) of β-Amyloid immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA). (C) Immunofluorescence images of ciNs labelled with an anti P-TAU antibody (Ser396, green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). P-TAU antibody was able to label the cell body and neurites of FAD ciNs, whereas it failed to stain PD ciNs. Scale Bar, 10 μm. Histogram shows the mean intensity values (arbitrary unit) of P-TAU immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA).
![Ijms 23 02147 g005 Ijms 23 02147 g005]()
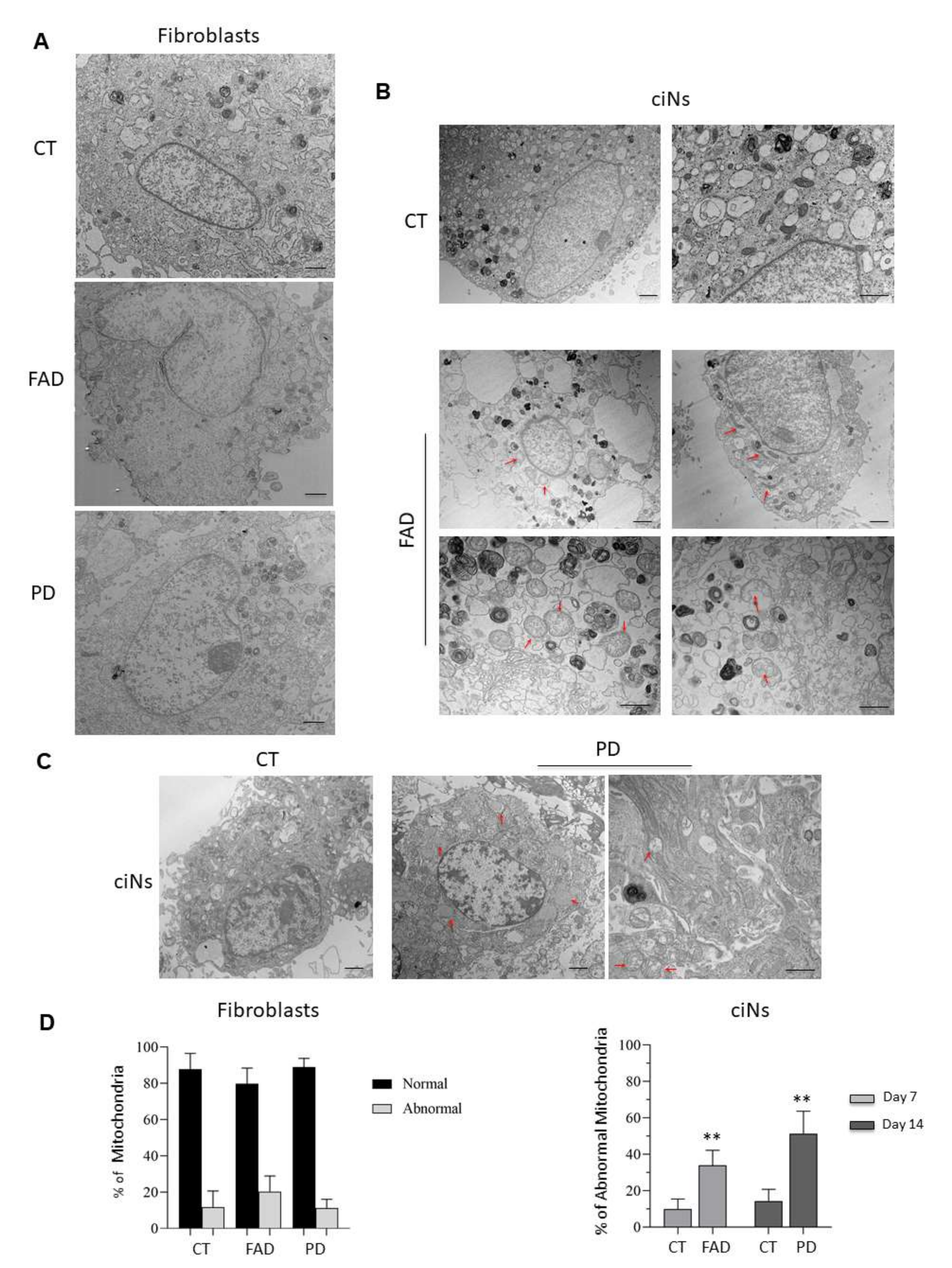
Figure 6.
Ultrastructural analysis and morphometric quantification of abnormal mitochondria. (A) Representative TEM images of healthy (CT), FAD and PD fibroblasts. Mitochondria with normal morphology appear with round shape and densely packed cristae in all three experimental groups. Scale Bar, 1 μm. (B) Morphometric analysis of TEM images at a 2500× (Scale Bar, 1 μm) and 6000× magnification (Scale bar 800 nm) of ciNs from CT and FAD patients. Images show mitochondrial abnormalities (red arrows) mainly consisting of elongated and distorted shape and swollen with a less dense matrix. (C) Morphometric analysis of TEM images at a 2500× (Scale Bar, 1 μm) and 6000× magnification (Scale bar 800 nm) of ciNs from CT and PD patients. Images show mitochondria swollen with a less dense matrix, and swollen with severe loss of cristae with an empty matrix (red arrows). (D) Quantitative assessment of mitochondrial morphology. The number of normal and abnormal mitochondria was measured in both fibroblasts and ciNs of CT, FAD and PD patients in 12 different fields of view (190 μm2). Percentage of abnormal mitochondria in ciNs of FAD and PD patients was calculated on the total number of counted mitochondria (approx. n = 500/experimental group). Data are depicted as mean ± SD, n = 3 independent cell cultures. ** p < 0.01 (two-way ANOVA).
Figure 6.
Ultrastructural analysis and morphometric quantification of abnormal mitochondria. (A) Representative TEM images of healthy (CT), FAD and PD fibroblasts. Mitochondria with normal morphology appear with round shape and densely packed cristae in all three experimental groups. Scale Bar, 1 μm. (B) Morphometric analysis of TEM images at a 2500× (Scale Bar, 1 μm) and 6000× magnification (Scale bar 800 nm) of ciNs from CT and FAD patients. Images show mitochondrial abnormalities (red arrows) mainly consisting of elongated and distorted shape and swollen with a less dense matrix. (C) Morphometric analysis of TEM images at a 2500× (Scale Bar, 1 μm) and 6000× magnification (Scale bar 800 nm) of ciNs from CT and PD patients. Images show mitochondria swollen with a less dense matrix, and swollen with severe loss of cristae with an empty matrix (red arrows). (D) Quantitative assessment of mitochondrial morphology. The number of normal and abnormal mitochondria was measured in both fibroblasts and ciNs of CT, FAD and PD patients in 12 different fields of view (190 μm2). Percentage of abnormal mitochondria in ciNs of FAD and PD patients was calculated on the total number of counted mitochondria (approx. n = 500/experimental group). Data are depicted as mean ± SD, n = 3 independent cell cultures. ** p < 0.01 (two-way ANOVA).
![Ijms 23 02147 g006 Ijms 23 02147 g006]()
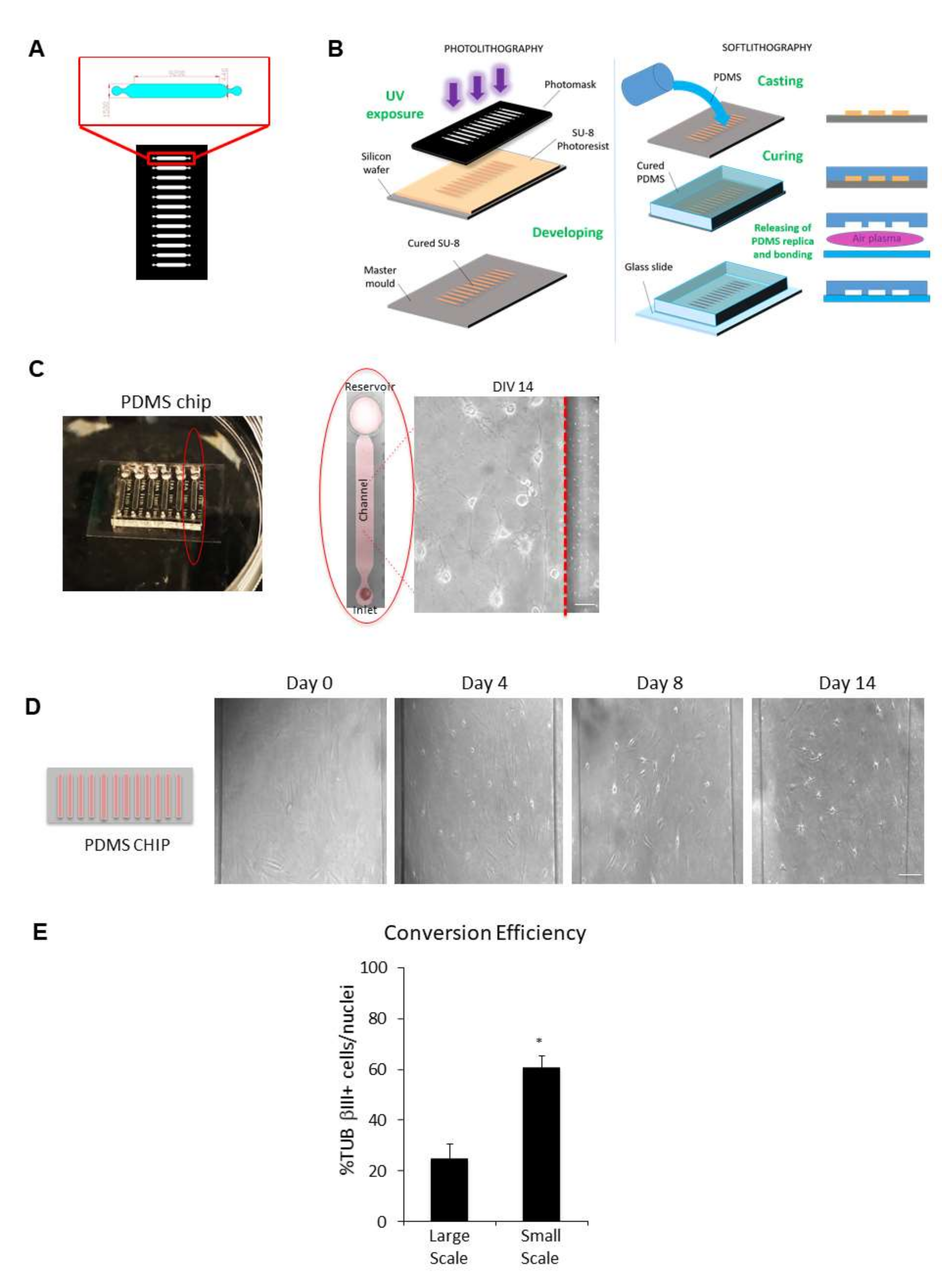
Figure 7.
Microfluidic technology combined with chemical reprogramming of fibroblasts enable neuronal differentiation. (A) Microfluidic channel design: the high resolution photomask is a glass layer directly printed from a CAD file by a laser-writer. Microchannel areas are transparent, while complementary area is black and opaque. The photomask includes a series of microchannels with the shape shown in the red zoom box (dimension in micrometres). (B) Photolithography (left panel): the SU-8 negative photoresist layer is spun with a 200 μm of thickness on a silicon wafer and prepared with a pre-baking (15 min at 95 °C) for UV light etching. After light exposure through the photomask, crosslinked areas with the microchannel shape are obtained on the photoresist layer. A post-baking (7 min at 95 °C) treatment and a final chemical bath (developing) remove the uncrosslinked areas, and after a supplementary hard baking, the final mould is obtained. Soft lithography (right panel): the replica of the mould is obtained through a casting process of liquid 1:10 PDMS (Polydimethylsiloxane). The transparent polymer is thermally cured (30 min at 80 °C) and removed from the mould. A plasma treatment is used to bond the PDMS structure to a glass slide. (C) Representative image of the chip device used in this study. A single channel is outlined by a red ellipse. Cells were chemically converted directly into the channel, as shown in the phase-contrast image of ciNs at DIV 14. Scale Bar, 10 μm. (D) (Left panel) A schematic drawing of a typical chip device with 12 microchannels in which cells are directly converted. (Right panel) Phase-contrast images showing the direct conversion of human fibroblasts from a healthy subject on chip with small molecules. The time-course of chemical conversion starts from fibroblasts (DIV 0) with the typical flat and elongated morphology. Soon after the addition of CA medium, followed by CB medium addition (DIV 1 up to DIV 14), an increasing number of fibroblasts on chip change their morphology and acquire small rounded nuclei with cytoplasmic protrusions. Particularly, morphological changes appear slowly in the first days of chemical conversion and then enhance during DIV 4 up to DIV 8. Scale Bar, 50 μm. (E) Histogram showing a comparison between the efficiency of transdifferentiation in large-scale (24-well plate) and in small-scale (chip) chemical conversion procedures. Data are shown as percentage of TUB βIII positive cells/nuclei. A strong increase in TUB βIII positive cells with a neuronal morphology is observed upon chemical conversion directly on chip. Bars in the plots represent means ± SD (n = 3, * p < 0.05).
Figure 7.
Microfluidic technology combined with chemical reprogramming of fibroblasts enable neuronal differentiation. (A) Microfluidic channel design: the high resolution photomask is a glass layer directly printed from a CAD file by a laser-writer. Microchannel areas are transparent, while complementary area is black and opaque. The photomask includes a series of microchannels with the shape shown in the red zoom box (dimension in micrometres). (B) Photolithography (left panel): the SU-8 negative photoresist layer is spun with a 200 μm of thickness on a silicon wafer and prepared with a pre-baking (15 min at 95 °C) for UV light etching. After light exposure through the photomask, crosslinked areas with the microchannel shape are obtained on the photoresist layer. A post-baking (7 min at 95 °C) treatment and a final chemical bath (developing) remove the uncrosslinked areas, and after a supplementary hard baking, the final mould is obtained. Soft lithography (right panel): the replica of the mould is obtained through a casting process of liquid 1:10 PDMS (Polydimethylsiloxane). The transparent polymer is thermally cured (30 min at 80 °C) and removed from the mould. A plasma treatment is used to bond the PDMS structure to a glass slide. (C) Representative image of the chip device used in this study. A single channel is outlined by a red ellipse. Cells were chemically converted directly into the channel, as shown in the phase-contrast image of ciNs at DIV 14. Scale Bar, 10 μm. (D) (Left panel) A schematic drawing of a typical chip device with 12 microchannels in which cells are directly converted. (Right panel) Phase-contrast images showing the direct conversion of human fibroblasts from a healthy subject on chip with small molecules. The time-course of chemical conversion starts from fibroblasts (DIV 0) with the typical flat and elongated morphology. Soon after the addition of CA medium, followed by CB medium addition (DIV 1 up to DIV 14), an increasing number of fibroblasts on chip change their morphology and acquire small rounded nuclei with cytoplasmic protrusions. Particularly, morphological changes appear slowly in the first days of chemical conversion and then enhance during DIV 4 up to DIV 8. Scale Bar, 50 μm. (E) Histogram showing a comparison between the efficiency of transdifferentiation in large-scale (24-well plate) and in small-scale (chip) chemical conversion procedures. Data are shown as percentage of TUB βIII positive cells/nuclei. A strong increase in TUB βIII positive cells with a neuronal morphology is observed upon chemical conversion directly on chip. Bars in the plots represent means ± SD (n = 3, * p < 0.05).
![Ijms 23 02147 g007 Ijms 23 02147 g007]()
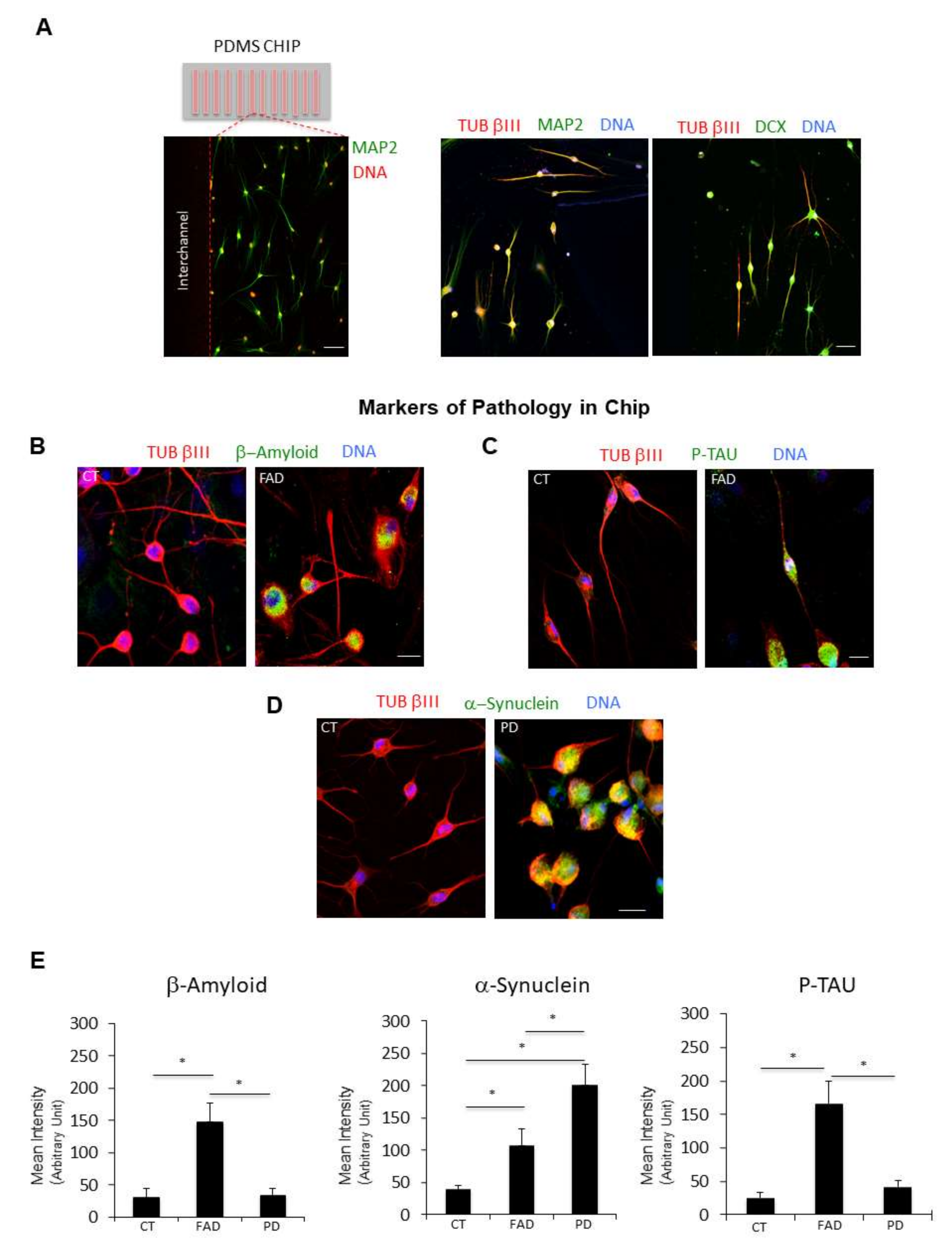
Figure 8.
Microfluidic technology combined with chemical reprogramming of fibroblasts enable detection of neuropathological markers. (A) (Left panel) A low magnification confocal immunofluorescence image shows the differentiation of cells expressing the late neuronal marker MAP2 (green channel). Nuclei are counterstained with Hoechst (red channel). (Right panel) Two representative confocal immunofluorescence images at higher magnification show ciNs expressing the neuronal markers TUB βIII, MAP2 and DCX. Scale Bar 20 μm. (B) Close-up view of ciNs, directly converted on chip, from CT and FAD patients, labelled with β-Amyloid antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). ciNs from FAD fibroblasts show accumulation of the pathological marker β-Amyloid. Scale Bar, 10 μm. (C) Close-up view of ciNs, directly converted on chip, from CT and FAD patients, labelled with P-TAU (Thr205) antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). ciNs from FAD fibroblasts show a strong labelling with the anti-P-TAU antibody. Scale Bar, 10 μm. (D) ciNs of CT and PD subjects were labelled with α-Synuclein antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). Chemically converted cells on chip confirm the presence of α-Synuclein aggregates only in patient cells as compared with CT cells. Scale Bar, 10 μm. (E) Histogram data represent the mean intensity values (arbitrary unit) of β-Amyloid, α-Synuclein and P-TAU immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA).
Figure 8.
Microfluidic technology combined with chemical reprogramming of fibroblasts enable detection of neuropathological markers. (A) (Left panel) A low magnification confocal immunofluorescence image shows the differentiation of cells expressing the late neuronal marker MAP2 (green channel). Nuclei are counterstained with Hoechst (red channel). (Right panel) Two representative confocal immunofluorescence images at higher magnification show ciNs expressing the neuronal markers TUB βIII, MAP2 and DCX. Scale Bar 20 μm. (B) Close-up view of ciNs, directly converted on chip, from CT and FAD patients, labelled with β-Amyloid antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). ciNs from FAD fibroblasts show accumulation of the pathological marker β-Amyloid. Scale Bar, 10 μm. (C) Close-up view of ciNs, directly converted on chip, from CT and FAD patients, labelled with P-TAU (Thr205) antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). ciNs from FAD fibroblasts show a strong labelling with the anti-P-TAU antibody. Scale Bar, 10 μm. (D) ciNs of CT and PD subjects were labelled with α-Synuclein antibody (green channel) and the neuronal marker TUB βIII (red channel). Nuclei were counterstained with Hoechst (blue channel). Chemically converted cells on chip confirm the presence of α-Synuclein aggregates only in patient cells as compared with CT cells. Scale Bar, 10 μm. (E) Histogram data represent the mean intensity values (arbitrary unit) of β-Amyloid, α-Synuclein and P-TAU immunolabelling in ciNs of CT, FAD and PD. Bars in the plots represent means ± SD from three independent experiments (* p < 0.05, one-way ANOVA).
![Ijms 23 02147 g008 Ijms 23 02147 g008]()
Table 1.
Human dermal fibroblasts used in the study.
Table 1.
Human dermal fibroblasts used in the study.
| | Fibroblasts | Origin |
|---|
| Healthy | AG11732 | Female 17 years |
| AG04062A | Male 31 years |
| Subject | AG04060 | Male 44 years |
| (CT) | AG04148 | Male 56 years |
| | AG07141 | Female 66 years |
| | AG07801 | Male 70 years |
| | AG09602C | Female 92 years |
| Familiar | | |
| Alzheimer’s | AG08711 | Female 34 years |
| Disease | AG06848 | Male 56 years |
| (FAD) | AG06840 | Female 56 years |
| (A246E) | | |
| in PSEN1 | | |
| Parkinson’s | AG20442 | Male 53 years |
| Disease | AG20446 | Male 57 years |
| (PD) | AG20443 | Male 71 years |
| Idiopathic | AG08395B | Female 85 years |