Progress in Methods for Copy Number Variation Profiling
Abstract
1. Introduction
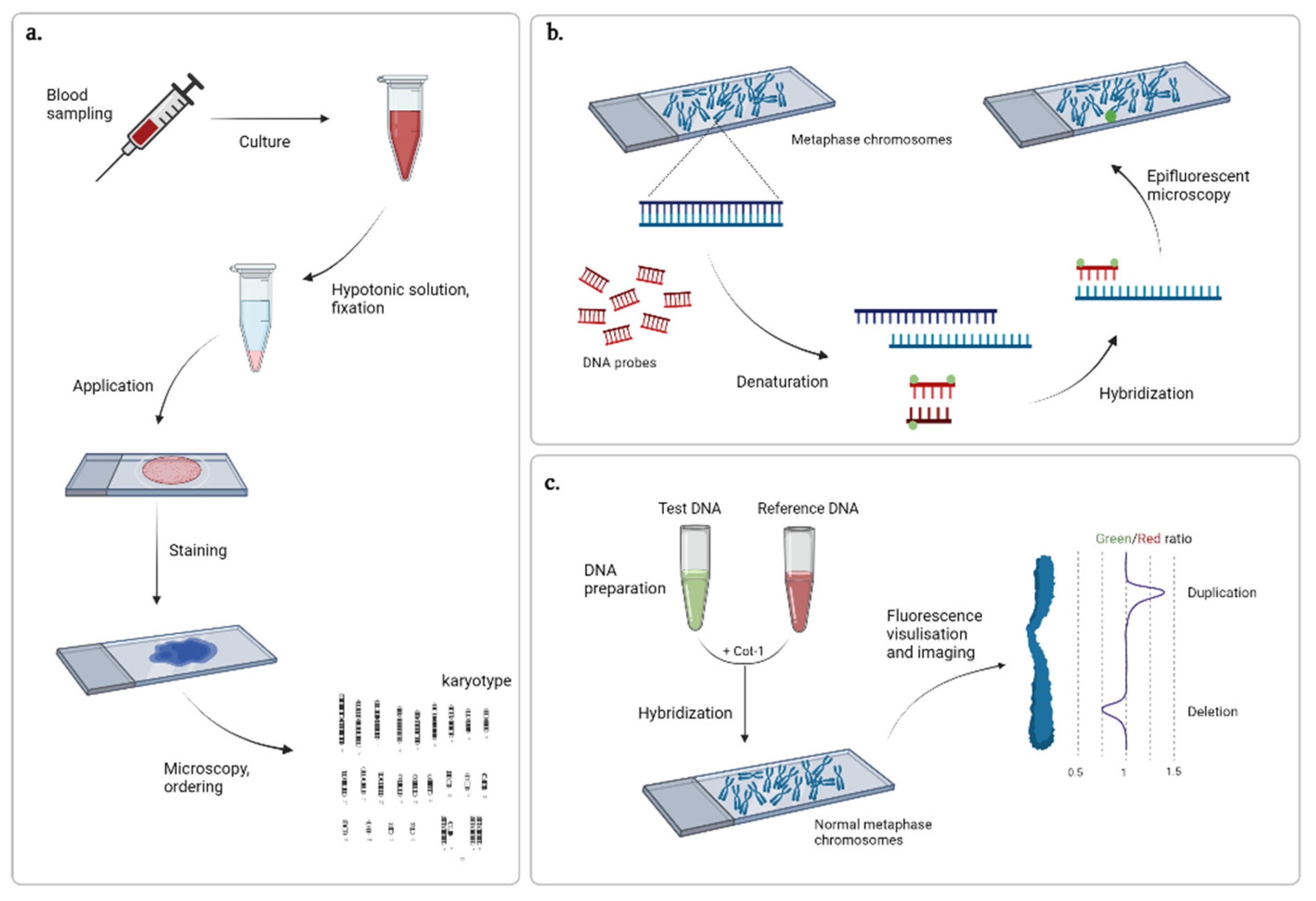
2. Methods of Cytogenetics
3. Chromosome Microarray Analysis (CMA)
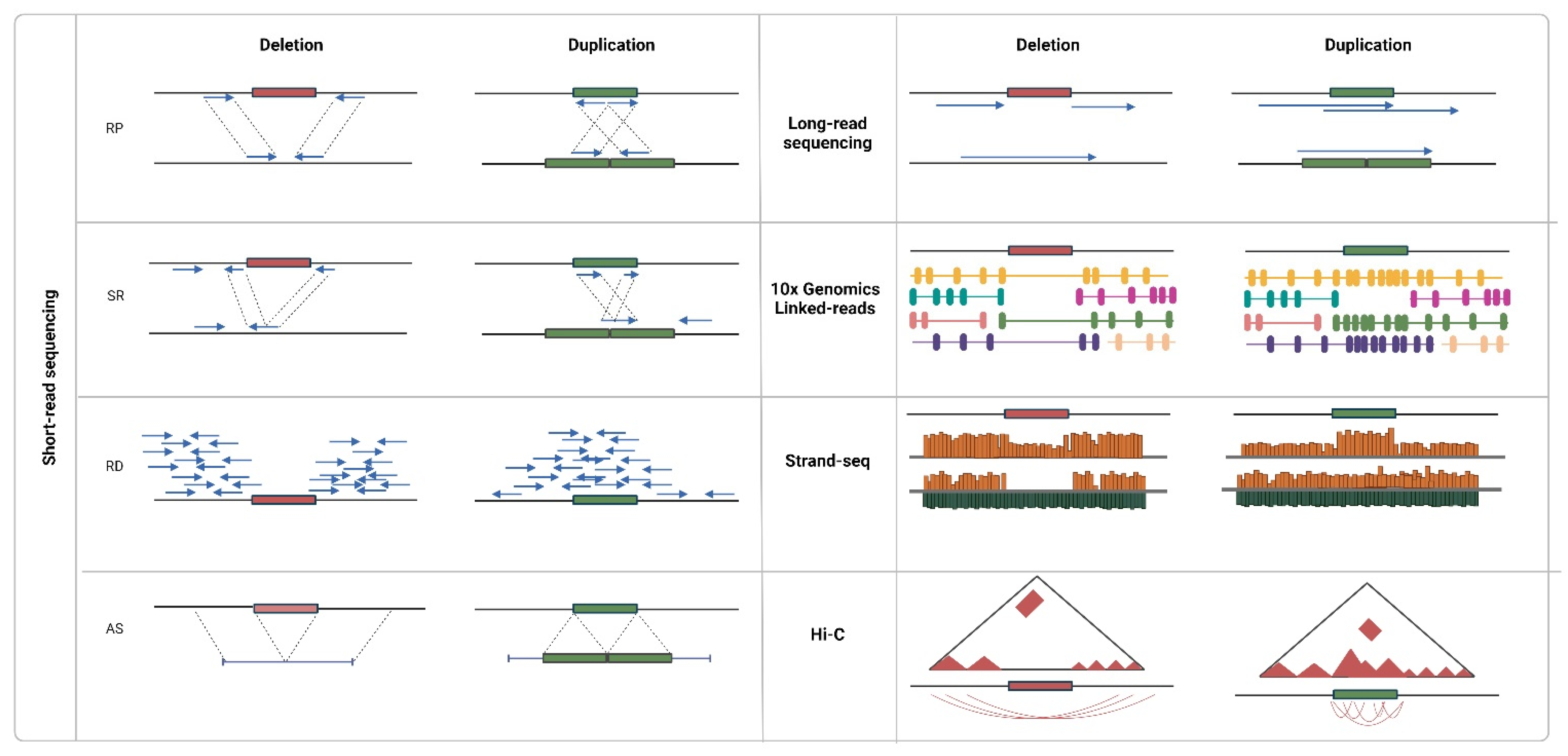
4. Sequencing Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hollox, E.J.; Zuccherato, L.W.; Tucci, S. Genome Structural Variation in Human Evolution. Trends Genet. 2022, 38, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Iafrate, A.J.; Feuk, L.; Rivera, M.N.; Listewnik, M.L.; Donahoe, P.K.; Qi, Y.; Scherer, S.W.; Lee, C. Detection of Large-Scale Variation in the Human Genome. Nat. Genet. 2004, 36, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global Variation in Copy Number in the Human Genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Zarrei, M.; MacDonald, J.R.; Merico, D.; Scherer, S.W. A Copy Number Variation Map of the Human Genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, T.H. Copy Number Variation Disorders. Curr. Genet. Med. Rep. 2017, 5, 183. [Google Scholar] [CrossRef]
- Roca, I.; González-Castro, L.; Fernández, H.; Couce, M.L.; Fernández-Marmiesse, A. Free-Access Copy-Number Variant Detection Tools for Targeted next-Generation Sequencing Data. Mutat. Res./Rev. Mutat. Res. 2019, 779, 114–125. [Google Scholar] [CrossRef]
- Seiser, E.L.; Innocenti, F. Hidden Markov Model-Based CNV Detection Algorithms for Illumina Genotyping Microarrays. Cancer Inform. 2015, 13, CIN-S16345. [Google Scholar] [CrossRef] [PubMed]
- Carter, N.P. Methods and Strategies for Analyzing Copy Number Variation Using DNA Microarrays. Nat. Genet. 2007, 39. [Google Scholar] [CrossRef]
- Mahmoud, M.; Gobet, N.; Cruz-Dávalos, D.I.; Mounier, N.; Dessimoz, C.; Sedlazeck, F.J. Structural Variant Calling: The Long and the Short of It. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Salgado, D.; Armean, I.M.; Baudis, M.; Beltran, S.; Capella-Gutierrez, S.; Carvalho-Silva, D.; Del Angel, V.D.; Dopazo, J.; Furlong, L.I.; Gao, B.; et al. The ELIXIR Human Copy Number Variations Community: Building Bioinformatics Infrastructure for Research. F1000Research 2020, 9, 1229. [Google Scholar] [CrossRef]
- Roizen, N.J.; Patterson, D. Down’s Syndrome. Lancet 2003, 361, 1281–1289. [Google Scholar] [CrossRef]
- Lanfranco, F.; Kamischke, A.; Zitzmann, M.; Nieschlag, E. Klinefelter’s Syndrome. Lancet 2004, 364, 273–283. [Google Scholar] [CrossRef]
- Cereda, A.; Carey, J.C. The Trisomy 18 Syndrome. Orphanet J. Rare Dis. 2012, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Nowell, P.C. The Minute Chromosome (Phl) in Chronic Granulocytic Leukemia. Blut 1962, 8, 65–66. [Google Scholar] [CrossRef]
- Bayani, J.; Squire, J.A. Traditional Banding of Chromosomes for Cytogenetic Analysis. Curr. Protoc. Cell Biol. 2004, 22, 22.3.1–22.3.7. [Google Scholar] [CrossRef]
- Swansbury, J. Introduction to the Analysis of the Human G-Banded Karyotype. Methods Mol. Biol. 2003, 220, 259–269. [Google Scholar] [PubMed]
- Gall, J.G.; Pardue, M.L. Formation and Detection of RNA-DNA Hybrid Molecules in Cytological Preparations. Proc. Natl. Acad. Sci. USA 1969, 63, 378–383. [Google Scholar] [CrossRef]
- Rudkin, G.T.; Stollar, B.D. High Resolution Detection of DNA-RNA Hybrids in Situ by Indirect Immunofluorescence. Nature 1977, 265, 472–473. [Google Scholar] [CrossRef]
- Bauman, J.G.; Wiegant, J.; Borst, P.; van Duijn, P. A New Method for Fluorescence Microscopical Localization of Specific DNA Sequences by in Situ Hybridization of Fluorochromelabelled RNA. Exp. Cell Res. 1980, 128. [Google Scholar] [CrossRef]
- Schrock, E.; du Manoir, S.; Veldman, T.; Schoell, B.; Wienberg, J.; Ferguson-Smith, M.A.; Ning, Y.; Ledbetter, D.H.; Bar-Am, I.; Soenksen, D.; et al. Multicolor Spectral Karyotyping of Human Chromosomes. Science 1996, 273, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Speicher, M.R.; Ballard, S.G.; Ward, D.C. Karyotyping Human Chromosomes by Combinatorial Multi-Fluor FISH. Nat. Genet. 1996, 12, 368–375. [Google Scholar] [CrossRef]
- Kallioniemi, A.; Kallioniemi, O.P.; Sudar, D.; Rutovitz, D.; Gray, J.W.; Waldman, F.; Pinkel, D. Comparative Genomic Hybridization for Molecular Cytogenetic Analysis of Solid Tumors. Science 1992, 258, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Kallioniemi, O.P.; Kallioniemi, A.; Piper, J.; Isola, J.; Waldman, F.M.; Gray, J.W.; Pinkel, D. Optimizing Comparative Genomic Hybridization for Analysis of DNA Sequence Copy Number Changes in Solid Tumors. Genes Chromosomes Cancer 1994, 10, 231–243. [Google Scholar] [CrossRef]
- Gebhart, E. Comparative Genomic Hybridization (CGH): Ten Years of Substantial Progress in Human Solid Tumor Molecular Cytogenetics. Cytogenet. Genome Res. 2004, 104, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Solinas-Toldo, S.; Lampel, S.; Stilgenbauer, S.; Nickolenko, J.; Benner, A.; Döhner, H.; Cremer, T.; Lichter, P. Matrix-Based Comparative Genomic Hybridization: Biochips to Screen for Genomic Imbalances. Genes Chromosomes Cancer 1997, 20, 399–407. [Google Scholar] [CrossRef]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.-L.; Chen, C.; Zhai, Y.; et al. High Resolution Analysis of DNA Copy Number Variation Using Comparative Genomic Hybridization to Microarrays. Nat. Genet. 1998, 20, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Osoegawa, K.; Mammoser, A.G.; Wu, C.; Frengen, E.; Zeng, C.; Catanese, J.J.; de Jong, P.J. A Bacterial Artificial Chromosome Library for Sequencing the Complete Human Genome. Genome Res. 2001, 11, 483. [Google Scholar] [CrossRef]
- Cowell, J.K.; Nowak, N.J. High-Resolution Analysis of Genetic Events in Cancer Cells Using Bacterial Artificial Chromosome Arrays and Comparative Genome Hybridization. Adv. Cancer Res. 2003, 90, 91–125. [Google Scholar] [CrossRef]
- Malan, V.; Chevallier, S.; Soler, G.; Coubes, C.; Lacombe, D.; Pasquier, L.; Soulier, J.; Morichon-Delvallez, N.; Turleau, C.; Munnich, A.; et al. Array-Based Comparative Genomic Hybridization Identifies a High Frequency of Copy Number Variations in Patients with Syndromic Overgrowth. Eur. J. Hum. Genet. 2009, 18, 227–232. [Google Scholar] [CrossRef]
- Pollack, J.R.; Perou, C.M.; Alizadeh, A.A.; Eisen, M.B.; Pergamenschikov, A.; Williams, C.F.; Jeffrey, S.S.; Botstein, D.; Brown, P.O. Genome-Wide Analysis of DNA Copy-Number Changes Using cDNA Microarrays. Nat. Genet. 1999, 23, 41–46. [Google Scholar] [CrossRef]
- Bashyam; Bair, R.; Kim, Y.H.; Wang, P.; Hernandez-Boussard, T.; Karikari, C.A.; Tibshirani, R.; Maitra, A.; Pollack, J.R. Array-Based Comparative Genomic Hybridization Identifies Localized DNA Amplifications and Homozygous Deletions in Pancreatic Cancer. Neoplasia 2005, 7, 556-IN16. [Google Scholar] [CrossRef] [PubMed]
- Dhami, P.; Coffey, A.J.; Abbs, S.; Vermeesch, J.R.; Dumanski, J.P.; Woodward, K.J.; Andrews, R.M.; Langford, C.; Vetrie, D. Exon Array CGH: Detection of Copy-Number Changes at the Resolution of Individual Exons in the Human Genome. Am. J. Hum. Genet. 2005, 76, 750. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Heller, R.; Chai, A.; Brown, P.O.; Davis, R.W. Parallel Human Genome Analysis: Microarray-Based Expression Monitoring of 1000 Genes. Proc. Natl. Acad. Sci. USA 1996, 93, 10614–10619. [Google Scholar] [CrossRef] [PubMed]
- DeRisi, J.; Penland, L.; Brown, P.O.; Bittner, M.L.; Meltzer, P.S.; Ray, M.; Chen, Y.; Su, Y.A.; Trent, J.M. Use of a cDNA Microarray to Analyse Gene Expression Patterns in Human Cancer. Nat. Genet. 1996, 14, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Lucito, R.; Healy, J.; Alexander, J.; Reiner, A.; Esposito, D.; Chi, M.; Rodgers, L.; Brady, A.; Sebat, J.; Troge, J.; et al. Representational Oligonucleotide Microarray Analysis: A High-Resolution Method to Detect Genome Copy Number Variation. Genome Res. 2003, 13, 2291–2305. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.T.; Scheffer, A.; Ben-Dor, A.; Sampas, N.; Lipson, D.; Kincaid, R.; Tsang, P.; Curry, B.; Baird, K.; Meltzer, P.S.; et al. Comparative Genomic Hybridization Using Oligonucleotide Microarrays and Total Genomic DNA. Proc. Natl. Acad. Sci. USA 2004, 101, 17765–17770. [Google Scholar] [CrossRef]
- Bignell, G.R.; Huang, J.; Greshock, J.; Watt, S.; Butler, A.; West, S.; Grigorova, M.; Jones, K.W.; Wei, W.; Stratton, M.R.; et al. High-Resolution Analysis of DNA Copy Number Using Oligonucleotide Microarrays. Genome Res. 2004, 14, 287–295. [Google Scholar] [CrossRef]
- Peiffer, D.A.; Le, J.M.; Steemers, F.J.; Chang, W.; Jenniges, T.; Garcia, F.; Haden, K.; Li, J.; Shaw, C.A.; Belmont, J.; et al. High-Resolution Genomic Profiling of Chromosomal Aberrations Using Infinium Whole-Genome Genotyping. Genome Res. 2006, 16, 1136–1148. [Google Scholar] [CrossRef]
- Shen, F.; Huang, J.; Fitch, K.R.; Truong, V.B.; Kirby, A.; Chen, W.; Zhang, J.; Liu, G.; McCarroll, S.A.; Jones, K.W.; et al. Improved Detection of Global Copy Number Variation Using High Density, Non-Polymorphic Oligonucleotide Probes. BMC Genet. 2008, 9, 27. [Google Scholar] [CrossRef]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am. J. Hum. Genet. 2010, 86, 749. [Google Scholar] [CrossRef] [PubMed]
- Haraksingh, R.R.; Abyzov, A.; Urban, A.E. Comprehensive Performance Comparison of High-Resolution Array Platforms for Genome-Wide Copy Number Variation (CNV) Analysis in Humans. BMC Genom. 2017, 18, 321. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Darvishi, K.; Shi, X.; Rajan, D.; Rigler, D.; Fitzgerald, T.; Lionel, A.C.; Thiruvahindrapuram, B.; MacDonald, J.R.; Mills, R.; et al. Comprehensive Assessment of Array-Based Platforms and Calling Algorithms for Detection of Copy Number Variants. Nat. Biotechnol. 2011, 29, 512–520. [Google Scholar] [CrossRef]
- Veltman, J.A.; Fridlyand, J.; Pejavar, S.; Olshen, A.B.; Korkola, J.E.; DeVries, S.; Carroll, P.; Kuo, W.-L.; Pinkel, D.; Albertson, D.; et al. Array-Based Comparative Genomic Hybridization for Genome-Wide Screening of DNA Copy Number in Bladder Tumors. Cancer Res. 2003, 63, 2872–2880. [Google Scholar] [PubMed]
- Array-Based Comparative Genomic Hybridization for the Genomewide Detection of Submicroscopic Chromosomal Abnormalities. Am. J. Hum. Genet. 2003, 73, 1261–1270. [CrossRef] [PubMed]
- Comparative Genomic Hybridization–Array Analysis Enhances the Detection of Aneuploidies and Submicroscopic Imbalances in Spontaneous Miscarriages. Am. J. Hum. Genet. 2004, 74, 1168–1174. [CrossRef] [PubMed]
- Shaw-Smith, C.; Redon, R.; Rickman, L.; Rio, M.; Willatt, L.; Fiegler, H.; Firth, H.; Sanlaville, D.; Winter, R.; Colleaux, L.; et al. Microarray Based Comparative Genomic Hybridisation (array-CGH) Detects Submicroscopic Chromosomal Deletions and Duplications in Patients with Learning Disability/mental Retardation and Dysmorphic Features. J. Med. Genet. 2004, 41, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Schwaenen, C.; Nessling, M.; Wessendorf, S.; Salvi, T.; Wrobel, G.; Radlwimmer, B.; Kestler, H.A.; Haslinger, C.; Stilgenbauer, S.; Döhner, H.; et al. Automated Array-Based Genomic Profiling in Chronic Lymphocytic Leukemia: Development of a Clinical Tool and Discovery of Recurrent Genomic Alterations. Proc. Natl. Acad. Sci. USA 2004, 101, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- :Olshen, A.B.; Venkatraman, E.S.; Lucito, R.; Wigler, M. Circular Binary Segmentation for the Analysis of Array-Based DNA Copy Number Data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Jong, K.; Marchiori, E.; van der Vaart, A.; Ylstra, B.; Weiss, M.; Meijer, G. Chromosomal Breakpoint Detection in Human Cancer. In Proceedings of the Applications of Evolutionary Computing, Essex, UK, 14–16 April 2003; Springer: Berlin/Heidelberg, Germany, 2003; pp. 54–65. [Google Scholar]
- Hupé, P.; Stransky, N.; Thiery, J.P.; Radvanyi, F.; Barillot, E. Analysis of Array CGH Data: From Signal Ratio to Gain and Loss of DNA Regions. Bioinformatics 2004, 20, 3413–3422. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R.; Wang, P. Spatial Smoothing and Hot Spot Detection for CGH Data Using the Fused Lasso. Biostatistics 2008, 9, 18–29. [Google Scholar] [CrossRef]
- Jeng, X.J.; Cai, T.T.; Li, H. Optimal Sparse Segment Identification with Application in Copy Number Variation Analysis. J. Am. Stat. Assoc. 2010, 105, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.S.; Zhang, H. The screening and ranking algorithm to detect dna copy number variations. Ann. Appl. Stat. 2012, 6, 1306–1326. [Google Scholar] [CrossRef]
- de Vries, B.B.A.; Pfundt, R.; Leisink, M.; Koolen, D.A.; Vissers, L.E.L.; Janssen, I.M.; van Reijmersdal, S.; Nillesen, W.M.; Huys, E.H.L.P.; de Leeuw, N.; et al. Diagnostic Genome Profiling in Mental Retardation. Am. J. Hum. Genet. 2005, 77, 606. [Google Scholar] [CrossRef]
- Zhao, X.; Li, C.; Guillermo Paez, J.; Chin, K.; Jänne, P.A.; Chen, T.-H.; Girard, L.; Minna, J.; Christiani, D.; Leo, C.; et al. An Integrated View of Copy Number and Allelic Alterations in the Cancer Genome Using Single Nucleotide Polymorphism Arrays. Cancer Res. 2004, 64, 3060–3071. [Google Scholar] [CrossRef]
- Picard, F.; Robin, S.; Lavielle, M.; Vaisse, C.; Daudin, J.-J. A Statistical Approach for Array CGH Data Analysis. BMC Bioinform. 2005, 6, 27. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.A.; Hakonarson, H.; Bucan, M. PennCNV: An Integrated Hidden Markov Model Designed for High-Resolution Copy Number Variation Detection in Whole-Genome SNP Genotyping Data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Dellinger, A.E.; Saw, S.M.; Goh, L.K.; Seielstad, M.; Young, T.L.; Li, Y.J. Comparative Analyses of Seven Algorithms for Copy Number Variant Identification from Single Nucleotide Polymorphism Arrays. Nucleic Acids Res. 2010, 38, e105. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Motsinger, R.A. Evaluation of Calling Algorithms for Array-CGH. Front. Genet. 2013, 4, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Winchester, L.; Yau, C.; Ragoussis, J. Comparing CNV Detection Methods for SNP Arrays. Brief. Funct. Genom. 2009, 8, 353–366. [Google Scholar] [CrossRef]
- Korn, J.M.; Kuruvilla, F.G.; McCarroll, S.A.; Wysoker, A.; Nemesh, J.; Cawley, S.; Hubbell, E.; Veitch, J.; Collins, P.J.; Darvishi, K.; et al. Integrated Genotype Calling and Association Analysis of SNPs, Common Copy Number Polymorphisms and Rare CNVs. Nat. Genet. 2008, 40, 1253–1260. [Google Scholar] [CrossRef]
- Sun, W.; Wright, F.A.; Tang, Z.; Nordgard, S.H.; Van Loo, P.; Yu, T.; Kristensen, V.N.; Perou, C.M. Integrated Study of Copy Number States and Genotype Calls Using High-Density SNP Arrays. Nucleic Acids Res. 2009, 37, 5365–5377. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, K. Application of Nexus Copy Number Software for CNV Detection and Analysis. Curr. Protoc. Hum. Genet. 2010, 65, 4–14. [Google Scholar] [CrossRef]
- Colella, S.; Yau, C.; Taylor, J.M.; Mirza, G.; Butler, H.; Clouston, P.; Bassett, A.S.; Seller, A.; Holmes, C.C.; Ragoussis, J. QuantiSNP: An Objective Bayes Hidden-Markov Model to Detect and Accurately Map Copy Number Variation Using SNP Genotyping Data. Nucleic Acids Res. 2007, 35, 2013–2025. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Q.; Wang, Q.; Jia, P.; Zhao, Z. Computational Tools for Copy Number Variation (CNV) Detection Using next-Generation Sequencing Data: Features and Perspectives. BMC Bioinform. 2013, 14, S1. [Google Scholar] [CrossRef]
- Korbel, J.O.; Urban, A.E.; Affourtit, J.P.; Godwin, B.; Grubert, F.; Simons, J.F.; Kim, P.M.; Palejev, D.; Carriero, N.J.; Du, L.; et al. Paired-End Mapping Reveals Extensive Structural Variation in the Human Genome. Science 2007, 318, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wallis, J.W.; McLellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; McGrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An Algorithm for High-Resolution Mapping of Genomic Structural Variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Korbel, J.O.; Abyzov, A.; Mu, X.J.; Carriero, N.; Cayting, P.; Zhang, Z.; Snyder, M.; Gerstein, M.B. PEMer: A Computational Framework with Simulation-Based Error Models for Inferring Genomic Structural Variants from Massive Paired-End Sequencing Data. Genome Biol. 2009, 10, R23. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Hormozdiari, F.; Alkan, C.; Brudno, M. MoDIL: Detecting Small Indels from Clone-End Sequencing with Mixtures of Distributions. Nat. Methods 2009, 6, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.; Pyon, Y.S.; Li, J. A Model-Based Clustering Method for Genomic Structural Variant Prediction and Genotyping Using Paired-End Sequencing Data. PLoS ONE 2012, 7, e52881. [Google Scholar] [CrossRef][Green Version]
- Marschall, T.; Costa, I.G.; Canzar, S.; Bauer, M.; Klau, G.W.; Schliep, A.; Schönhuth, A. CLEVER: Clique-Enumerating Variant Finder. Bioinformatics 2012, 28, 2875–2882. [Google Scholar] [CrossRef] [PubMed]
- Trappe, K.; Emde, A.-K.; Ehrlich, H.-C.; Reinert, K. Gustaf: Detecting and Correctly Classifying SVs in the NGS Twilight Zone. Bioinformatics 2014, 30, 3484–3490. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A Pattern Growth Approach to Detect Break Points of Large Deletions and Medium Sized Insertions from Paired-End Short Reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Xuan, Z.; Makarov, V.; Ye, K.; Sebat, J. Sensitive and Accurate Detection of Copy Number Variants Using Read Depth of Coverage. Genome Res. 2009, 19, 1586–1592. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Tammi, M.T. CNV-Seq, a New Method to Detect Copy Number Variation Using High-Throughput Sequencing. BMC Bioinform. 2009, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Gusnanto, A.; Taylor, C.C.; Nafisah, I.; Wood, H.M.; Rabbitts, P.; Berri, S. Estimating Optimal Window Size for Analysis of Low-Coverage next-Generation Sequence Data. Bioinformatics 2014, 30, 1823–1829. [Google Scholar] [CrossRef]
- Benjamini, Y.; Speed, T.P. Summarizing and Correcting the GC Content Bias in High-Throughput Sequencing. Nucleic Acids Res. 2012, 40, e72. [Google Scholar] [CrossRef]
- Talevich, E.; Hunter Shain, A.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Dharanipragada, P.; Vogeti, S.; Parekh, N. iCopyDAV: Integrated Platform for Copy Number variations—Detection, Annotation and Visualization. PLoS ONE 2018, 13, e0195334. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.; Sun, W.; Crowley, J.J.; Szatkiewicz, J.P. Allele-Specific Copy-Number Discovery from Whole-Genome and Whole-Exome Sequencing. Nucleic Acids Res. 2015, 43, e90. [Google Scholar] [CrossRef][Green Version]
- Xi, R.; Lee, S.; Xia, Y.; Kim, T.-M.; Park, P.J. Copy Number Analysis of Whole-Genome Data Using BIC-seq2 and Its Application to Detection of Cancer Susceptibility Variants. Nucleic Acids Res. 2016, 44, 6274–6286. [Google Scholar] [CrossRef]
- Boeva, V.; Zinovyev, A.; Bleakley, K.; Vert, J.P.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-Free Calling of Copy Number Alterations in Deep-Sequencing Data Using GC-Content Normalization. Bioinformatics 2011, 27, 268–269. [Google Scholar] [CrossRef]
- Miller, C.A.; Hampton, O.; Coarfa, C.; Milosavljevic, A. ReadDepth: A Parallel R Package for Detecting Copy Number Alterations from Short Sequencing Reads. PLoS ONE 2011, 6, e16327. [Google Scholar] [CrossRef]
- Gordeeva, V.; Sharova, E.; Babalyan, K.; Sultanov, R.; Govorun, V.M.; Arapidi, G. Benchmarking Germline CNV Calling Tools from Exome Sequencing Data. Sci. Rep. 2021, 11, 14416. [Google Scholar] [CrossRef]
- Fromer, M.; Moran, J.L.; Chambert, K.; Banks, E.; Bergen, S.E.; Ruderfer, D.M.; Handsaker, R.E.; McCarroll, S.A.; O’Donovan, M.C.; Owen, M.J.; et al. Discovery and Statistical Genotyping of Copy-Number Variation from Whole-Exome Sequencing Depth. Am. J. Hum. Genet. 2012, 91, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Oldridge, D.A.; Diskin, S.J.; Zhang, N.R. CODEX: A Normalization and Copy Number Variation Detection Method for Whole Exome Sequencing. Nucleic Acids Res. 2015, 43, e39. [Google Scholar] [CrossRef]
- Plagnol, V.; Curtis, J.; Epstein, M.; Mok, K.Y.; Stebbings, E.; Grigoriadou, S.; Wood, N.W.; Hambleton, S.; Burns, S.O.; Thrasher, A.J.; et al. A Robust Model for Read Count Data in Exome Sequencing Experiments and Implications for Copy Number Variant Calling. Bioinformatics 2012, 28, 2747–2754. [Google Scholar] [CrossRef]
- Love, M.I.; Myšičková, A.; Sun, R.; Kalscheuer, V.; Vingron, M.; Haas, S.A. Modeling Read Counts for CNV Detection in Exome Sequencing Data. Stat. Appl. Genet. Mol. Biol. 2011, 10. [Google Scholar] [CrossRef]
- D’Aurizio, R.; Pippucci, T.; Tattini, L.; Giusti, B.; Pellegrini, M.; Magi, A. Enhanced Copy Number Variants Detection from Whole-Exome Sequencing Data Using EXCAVATOR2. Nucleic Acids Res. 2016, 44, e154. [Google Scholar] [CrossRef] [PubMed]
- Kuśmirek, W.; Szmurło, A.; Wiewiórka, M.; Nowak, R.; Gambin, T. Comparison of kNN and K-Means Optimization Methods of Reference Set Selection for Improved CNV Callers Performance. BMC Bioinform. 2019, 20, 266. [Google Scholar] [CrossRef]
- Klambauer, G.; Schwarzbauer, K.; Mayr, A.; Clevert, D.-A.; Mitterecker, A.; Bodenhofer, U.; Hochreiter, S. cn.MOPS: Mixture of Poissons for Discovering Copy Number Variations in next-Generation Sequencing Data with a Low False Discovery Rate. Nucleic Acids Res. 2012, 40, e69. [Google Scholar] [CrossRef] [PubMed]
- Krumm, N.; Sudmant, P.H.; Ko, A.; O’Roak, B.J.; Malig, M.; Coe, B.P.; Quinlan, A.R.; Nickerson, D.A.; Eichler, E.E. Copy Number Variation Detection and Genotyping from Exome Sequence Data. Genome Res. 2012, 22, 1525–1532. [Google Scholar] [CrossRef]
- Johansson, L.F.; van Dijk, F.; de Boer, E.N.; van Dijk-Bos, K.K.; Jongbloed, J.D.; van der Hout, A.H.; Westers, H.; Sinke, R.J.; Swertz, M.A.; Sijmons, R.H.; et al. CoNVaDING: Single Exon Variation Detection in Targeted NGS Data. Hum. Mutat. 2016, 37, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Fowler, A.; Mahamdallie, S.; Ruark, E.; Seal, S.; Ramsay, E.; Clarke, M.; Uddin, I.; Wylie, H.; Strydom, A.; Lunter, G.; et al. Accurate Clinical Detection of Exon Copy Number Variants in a Targeted NGS Panel Using DECoN. Wellcome Open Res. 2016, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Pevzner, P.A.; Tang, H.; Waterman, M.S. An Eulerian Path Approach to DNA Fragment Assembly. Proc. Natl. Acad. Sci. USA 2001, 98, 9748–9753. [Google Scholar] [CrossRef] [PubMed]
- Nijkamp, J.F.; van den Broek, M.A.; Geertman, J.-M.A.; Reinders, M.J.T.; Daran, J.-M.G.; de Ridder, D. De Novo Detection of Copy Number Variation by Co-Assembly. Bioinformatics 2012, 28, 3195–3202. [Google Scholar] [CrossRef]
- Iqbal, Z.; Caccamo, M.; Turner, I.; Flicek, P.; McVean, G. De Novo Assembly and Genotyping of Variants Using Colored de Bruijn Graphs. Nat. Genet. 2012, 44, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.L.; Schröder, J.; Penington, J.S.; Do, H.; Molania, R.; Dobrovic, A.; Speed, T.P.; Papenfuss, A.T. GRIDSS: Sensitive and Specific Genomic Rearrangement Detection Using Positional de Bruijn Graph Assembly. Genome Res. 2017, 27, 2050–2060. [Google Scholar] [CrossRef]
- Wang, J.; Mullighan, C.G.; Easton, J.; Roberts, S.; Heatley, S.L.; Ma, J.; Rusch, M.C.; Chen, K.; Harris, C.C.; Ding, L.; et al. CREST Maps Somatic Structural Variation in Cancer Genomes with Base-Pair Resolution. Nat. Methods 2011, 8, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Using ERDS to Infer Copy-Number Variants in High-Coverage Genomes. Am. J. Hum. Genet. 2012, 91, 408–421. [CrossRef] [PubMed]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, e1004572. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2015, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Handsaker, R.E.; Korn, J.M.; Nemesh, J.; McCarroll, S.A. Discovery and Genotyping of Genome Structural Polymorphism by Sequencing on a Population Scale. Nat. Genet. 2011, 43, 269–276. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Clark, R.A.; Sokolova, S.; Leibowitz, M.L.; Zhang, Y.; Hurles, M.E.; Mell, J.C.; Hall, I.M. Genome-Wide Mapping and Assembly of Structural Variant Breakpoints in the Mouse Genome. Genome Res. 2010, 20, 623–635. [Google Scholar] [CrossRef]
- Mohiyuddin, M.; Mu, J.C.; Li, J.; Bani Asadi, N.; Gerstein, M.B.; Abyzov, A.; Wong, W.H.; Lam, H.Y.K. MetaSV: An Accurate and Integrative Structural-Variant Caller for next Generation Sequencing. Bioinformatics 2015, 31, 2741–2744. [Google Scholar] [CrossRef]
- Michaelson, J.J.; Sebat, J. forestSV: Structural Variant Discovery through Statistical Learning. Nat. Methods 2012, 9, 819–821. [Google Scholar] [CrossRef]
- Parikh, H.; Mohiyuddin, M.; Lam, H.Y.K.; Iyer, H.; Chen, D.; Pratt, M.; Bartha, G.; Spies, N.; Losert, W.; Zook, J.M.; et al. Svclassify: A Method to Establish Benchmark Structural Variant Calls. BMC Genom. 2016, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Wu, Y.; Gao, J. DeepSV: Accurate Calling of Genomic Deletions from High-Throughput Sequencing Data Using Deep Convolutional Neural Network. BMC Bioinform. 2019, 20, 665. [Google Scholar] [CrossRef]
- Mills, R.E.; Walter, K.; Stewart, C.; Handsaker, R.E.; Chen, K.; Alkan, C.; Abyzov, A.; Yoon, S.C.; Ye, K.; Cheetham, R.K.; et al. Mapping Copy Number Variation by Population-Scale Genome Sequencing. Nature 2011, 470, 59–65. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Fritz, M.H.-Y.; et al. An Integrated Map of Structural Variation in 2504 Human Genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef]
- Collins, R.L.; Brand, H.; Karczewski, K.J.; Zhao, X.; Alföldi, J.; Francioli, L.C.; Khera, A.V.; Lowther, C.; Gauthier, L.D.; Wang, H.; et al. A Structural Variation Reference for Medical and Population Genetics. Nature 2020, 581, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Cretu Stancu, M.; van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; de Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Espejo Valle-Inclan, J.; et al. Mapping and Phasing of Structural Variation in Patient Genomes Using Nanopore Sequencing. Nat. Commun. 2017, 8, 1326. [Google Scholar] [CrossRef] [PubMed]
- English, A.C.; Salerno, W.J.; Reid, J.G. PBHoney: Identifying Genomic Variants via Long-Read Discordance and Interrupted Mapping. BMC Bioinform. 2014, 15, 180. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, S.; Cao, S.; Liu, Y.; Cui, Z.; Wang, Y.; Guo, H. Long-Read Sequencing Settings for Efficient Structural Variation Detection Based on Comprehensive Evaluation. BMC Bioinform. 2021, 22, 552. [Google Scholar] [CrossRef] [PubMed]
- Spies, N.; Weng, Z.; Bishara, A.; McDaniel, J.; Catoe, D.; Zook, J.M.; Salit, M.; West, R.B.; Batzoglou, S.; Sidow, A. Genome-Wide Reconstruction of Complex Structural Variants Using Read Clouds. Nat. Methods 2017, 14, 915–920. [Google Scholar] [CrossRef]
- Elyanow, R.; Wu, H.T.; Raphael, B.J. Identifying Structural Variants Using Linked-Read Sequencing Data. Bioinformatics 2018, 34, 353–360. [Google Scholar] [CrossRef]
- Hills, M.; O’Neill, K.; Falconer, E.; Brinkman, R.; Lansdorp, P.M. BAIT: Organizing Genomes and Mapping Rearrangements in Single Cells. Genome Med. 2013, 5, 82. [Google Scholar] [CrossRef]
- Wang, S.; Lee, S.; Chu, C.; Jain, D.; Kerpedjiev, P.; Nelson, G.M.; Walsh, J.M.; Alver, B.H.; Park, P.J. HiNT: A Computational Method for Detecting Copy Number Variations and Translocations from Hi-C Data. Genome Biol. 2020, 21, 73. [Google Scholar] [CrossRef]
- Schwartz, D.C.; Li, X.; Hernandez, L.I.; Ramnarain, S.P.; Huff, E.J.; Wang, Y.K. Ordered Restriction Maps of Saccharomyces Cerevisiae Chromosomes Constructed by Optical Mapping. Science 1993, 262, 110–114. [Google Scholar] [CrossRef]
- Latreille, P.; Norton, S.; Goldman, B.S.; Henkhaus, J.; Miller, N.; Barbazuk, B.; Bode, H.B.; Darby, C.; Du, Z.; Forst, S.; et al. Optical Mapping as a Routine Tool for Bacterial Genome Sequence Finishing. BMC Genom. 2007, 8, 321. [Google Scholar] [CrossRef]
- Pendleton, M.; Sebra, R.; Pang, A.W.C.; Ummat, A.; Franzen, O.; Rausch, T.; Stütz, A.M.; Stedman, W.; Anantharaman, T.; Hastie, A.; et al. Assembly and Diploid Architecture of an Individual Human Genome via Single-Molecule Technologies. Nat. Methods 2015, 12, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Optical Genome Mapping Enables Constitutional Chromosomal Aberration Detection. Am. J. Hum. Genet. 2021, 108, 1409–1422. [CrossRef]
- Li, L.; Leung, A.K.-Y.; Kwok, T.-P.; Lai, Y.Y.Y.; Pang, I.K.; Chung, G.T.-Y.; Mak, A.C.Y.; Poon, A.; Chu, C.; Li, M.; et al. OMSV Enables Accurate and Comprehensive Identification of Large Structural Variations from Nanochannel-Based Single-Molecule Optical Maps. Genome Biol. 2017, 18, 230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cheng, H.; Hong, X.; Di Narzo, A.F.; Franzen, O.; Peng, S.; Ruusalepp, A.; Kovacic, J.C.; Bjorkegren, J.L.M.; Wang, X.; et al. EnsembleCNV: An Ensemble Machine Learning Algorithm to Identify and Genotype Copy Number Variation Using SNP Array Data. Nucleic Acids Res. 2019, 47, e39. [Google Scholar] [CrossRef] [PubMed]
- Pounraja, V.K.; Jayakar, G.; Jensen, M.; Kelkar, N.; Girirajan, S. A Machine-Learning Approach for Accurate Detection of Copy-Number Variants from Exome Sequencing. Genome Res. 2019, 29, 1134–1143. [Google Scholar] [CrossRef]
- Akbarinejad, S.; Hadadian Nejad Yousefi, M.; Goudarzi, M. SVNN: An Efficient PacBio-Specific Pipeline for Structural Variations Calling Using Neural Networks. BMC Bioinform. 2021, 22, 335. [Google Scholar] [CrossRef]
- Kosugi, S.; Momozawa, Y.; Liu, X.; Terao, C.; Kubo, M.; Kamatani, Y. Comprehensive Evaluation of Structural Variation Detection Algorithms for Whole Genome Sequencing. Genome Biol. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Zook, J.M.; Hansen, N.F.; Olson, N.D.; Chapman, L.; Mullikin, J.C.; Xiao, C.; Sherry, S.; Koren, S.; Phillippy, A.M.; Boutros, P.C.; et al. A Robust Benchmark for Detection of Germline Large Deletions and Insertions. Nat. Biotechnol. 2020, 38, 1347–1355. [Google Scholar] [CrossRef]

| Array Platform | Specification * | Resolution ** | Description |
|---|---|---|---|
| Agilent SurePrint G3 Human CGH | 1 × 1 M | 2.1 kb | enhanced coverage on known genes, promoters, miRNAs, PAR, and telomeric regions |
| 2 × 400 K | 5.3 kb | ||
| 4 × 180 K | 13 kb | ||
| 8 × 60 K | 41 kb | ||
| Agilent Human Genome CGH | 2 × 105 | 35 kb | |
| 4 × 44 K | 43 kb | ||
| Agilent SurePrint G3 Human Genome CGH + SNP | 2 × 400 K | 7.2 Kb | |
| 4 × 180 K | 25.3 kb | ||
| Agilent SurePrint G3 Unrestricted CGH ISCA v2 | 4 × 180 K | 25 kb | enhanced coverage on ISCA (International Standards for Cytogenomic Arrays) regions |
| 8 × 60 K | 60 kb | ||
| 4 × 44 K | 75 kb | ||
| Agilent SurePrint G3 ISCA v2 CGH + SNP | 4 × 180 K | 25.3 kb | |
| Agilent SurePrint G3 Human High-Resolution Discovery | 1 × 1 M | 2.6 kb | association studies |
| Agilent SurePrint G3 Human CNV | 2 × 400 K | 1 kb | |
| Agilent Human CNV Association | 2 × 105 K | 232 b | |
| Agilent SurePrint G3 CGH Postnatal Research | 4 × 180 K | 2.4 kb | regions identified by Baylor College of Medicine experts |
| 8 × 60 K | 3.7 kb | ||
| Agilent GenetiSure Postnatal Research CGH + SNP | 2 × 400 K | 9.8 kb | disease-associated regions (The Clinical Genome/ISCA database) |
| Agilent GenetiSure Pre-Screen | 4 × 180 K | 31 kb | CNV identification from embryo biopsies and single-cell samples; increased density on chromosomes 13, 18, 20, 21, 22, and X |
| 8 × 60 K | 50 kb | ||
| Agilent GenetiSure Cyto CGH | 4 × 180 K | 3.5 kb | disease-associated regions linked to developmental delay, intellectual disability, neuropsychiatric disorders, congenital anomalies, or dysmorphic features |
| 8 × 60 K | 7.1 kb | ||
| Agilent GenetiSure Cyto CGH + SNP | 4 × 180 K | 7.3 kb | |
| Agilent GenetiSure Cancer Research CGH + SNP | 2 × 400 K | 9.8 kb | cancer regions of the genome COSMIC (Catalogue of Somatic Mutation in Cancer) CGC (Cancer Genetics Consortium) databases |
| Illumina HumanCytoSNP | 12 × 300 K | 6.2 kb | enhanced coverage of ~250 disease regions, including subtelomeric regions, pericentromeric regions, and sex chromosomes |
| Illumina Infinium CytoSNP-850 K | 8 × 850 K | 1.8 kb | comprehensive coverage of cytogenetically relevant genes for congenital disorders and cancer research ICCG (International Collaboration for Clinical Genomics) and CCMC (Cancer Cytogenomics Microarray Consortium) |
| Illumina Infinium Core | 24 × 300 K | 5.8 kb | genome-wide tag SNPs found across diverse world populations |
| Illumina Infinium Exome | 24 × 300 K | 0.21 kb | comprehensive coverage of putative functional exonic variants (including markers representing a range of common conditions, such as type 2 diabetes, cancer, and metabolic, and psychiatric disorders) |
| Illumina Infinium CoreExome | 24 × 600 K | 1.82 kb | all of the markers from the Infinium Core-24 BeadChip and the Infinium Exome-24 BeadChip |
| Illumina Infinium Global Diversity Array | 8 × 2 M | 0.63 kb | common and low frequency variants in global populations, curated clinical research variants |
| Illumina Infinium Global Screening Array | 24 × 700 K | 2.3 kb | multiethnic genome-wide content, curated clinical research variants |
| Illumina Infinium Omni2.5 | 8 × 2.4 M | 0.65 kb | common and rare SNP content from the 1000 Genomes Project (MAF > 2.5%) |
| Illumina Infinium Omni2.5Exome | 8 × 2.7 M | 0.56 kb | combined Infinum Omni2.5 and Infinium Exome-24 markers |
| Illumina Infinium Omni5 | 4 × 4.3 M | 0.36 kb | comprehensive coverage of the genome including common, intermediate, and rare SNPs |
| Illumina Infinium Omni5 Exome | 4 × 4.6 M | 0.33 kb | comprehensive genome-wide backbone combined with putative functional exonic variants |
| Illumina Infinium OmniExpress | 24 × 700 K | 2.23 kb | high coverage of common variants for genome-wide association studies |
| Illumina Infinium OmniExpressExome | 8 × 1 M | 1.36 kb | tag SNPs and functional exonic content |
| Illumina Infinium OncoArray | 24 × 500 K | 5.4 kb | genetic variants associated with five common cancers |
| Illumina Infinium PsychArray | 24 × 700 K | 1.74 kb | genetic variants associated with common psychiatric disorders |
| Affymetrix Genome-Wide Human SNP Array 6.0 | 1 × 1.8 M | 0.68 kb | comprehensive coverage of the genome |
| Affymetrix CytoScan XON Suite | 24 × 6.85 M | 0.5 kb | enhanced coverage in 7000 clinically relevant gene, exon-level copy number changes |
| Affymetrix CytoScan HD | 24 × 2.7 M | 1.3 kb | enhanced coverage on cytogenetic relevant region |
| Tool | Description | aCGH | SNP-Array | Reference | |
|---|---|---|---|---|---|
| Affymetrix | Illumina | ||||
| ADM-2 | search for intervals in which a Z-score based on the average weighted log ratio exceeds a user-specified threshold | ✓ | technical documentation (Agilent) | ||
| Birdsuite | integration of common CNP genotypes and CNVs discovered using HMM | ✓ | [61] | ||
| ChAS | HMM on the log2 ratios processed through a Bayes wavelet shrinkage estimator | ✓ | technical documentation (Affymetrix) | ||
| cnvPartition | recursive partitioning approach based on preliminary copy number estimates | ✓ | technical documentation (Illumina) | ||
| DNAcopy | circular binary segmentation | ✓ | [48] | ||
| GenoCN | estimation of HMM, parameters from data, germline, and somatic modes | ✓ | ✓ | [62] | |
| iPattern | normalization of the total intensities across individuals, Gaussian mixture model fitting | ✓ | ✓ | [42] | |
| Nexus | the probe’s log-ratio rank segmentation | ✓ | ✓ | ✓ | [63] |
| PennCN | HMM, also counted for the population frequency of the B allele | ✓ | ✓ | [57] | |
| QuantiSNP | objective Bayes-HMM, fixed rate of heterozygosity for each SNP | ✓ | ✓ | [64] | |
| Tool | Description | Data | Mode | Reference | ||
|---|---|---|---|---|---|---|
| WES | Targeted | Germline | Somatic | |||
| cn.MOPS | mixture Poisson model and Bayes approach | ✓ | ✓ | ✓ | ✓ | [92] |
| CNVkit | in- and off-target regions, rolling median bias correction, CBS | ✓ | ✓ | ✓ | [78] | |
| CODEX | log-linear decomposition-based normalization, Poisson likelihood-based segmentation | ✓ | ✓ | ✓ | ✓ | [87] |
| CoNIFER | singular value decomposition-based normalization, ± 1.5 SVD-ZRPKM threshold | ✓ | ✓ | [93] | ||
| CoNVaDING | ratio scores and Z-scores of the sample of interest compared to the selected control | ✓ | ✓ | [94] | ||
| DECoN | ExomeDepth modification (the distance between exons is taken into account) | ✓ | ✓ | [95] | ||
| ExomeDepth | beta-binomial distribution, optimized reference set, HMM | ✓ | ✓ | ✓ | [88] | |
| XHMM | principal component analysis normalization, HMM | ✓ | ✓ | [86] | ||
| Approach | Tool | Description | Reference |
|---|---|---|---|
| RP | BreakDancer | search for regions that include more anomalous read pairs than expected | [67] |
| SR | Pindel | pattern growth approach for breakpoint identification | [73] |
| RD | CNVnator | mean-shift technique, multiple-bandwidth partitioning, and GC correction | [79] |
| AS | Cortex | bubble-calling in the colored de Bruijn graph | [98] |
| RP + RD | GenomeSTRiP | connected components algorithm for read pair clustering, Gaussian mixture model for read depth genotyping | [104] |
| RP + SR | DELLY | graph-based paired-end clustering, breakpoints refinement using split-read alignment | [105] |
| RP + AS | Hydra | assembly of discordant mate pairs and aligned to the reference genome with MEGABLAST | [106] |
| RP + SR + AS | Manta | breakend graph construction, independent for each edge variation hypothesis refinement and scoring with diploid model | [103] |
| RP + SR + RD | Lumpy | probabilistic representation of an SV breakpoint | [102] |
| Ensemble | MetaSV | merging calls from tools (BreakDancer, CNVnator, BreakSeq, Pindel), breakpoint refinement by aligning the assembled CNV regions | [107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gordeeva, V.; Sharova, E.; Arapidi, G. Progress in Methods for Copy Number Variation Profiling. Int. J. Mol. Sci. 2022, 23, 2143. https://doi.org/10.3390/ijms23042143
Gordeeva V, Sharova E, Arapidi G. Progress in Methods for Copy Number Variation Profiling. International Journal of Molecular Sciences. 2022; 23(4):2143. https://doi.org/10.3390/ijms23042143
Chicago/Turabian StyleGordeeva, Veronika, Elena Sharova, and Georgij Arapidi. 2022. "Progress in Methods for Copy Number Variation Profiling" International Journal of Molecular Sciences 23, no. 4: 2143. https://doi.org/10.3390/ijms23042143
APA StyleGordeeva, V., Sharova, E., & Arapidi, G. (2022). Progress in Methods for Copy Number Variation Profiling. International Journal of Molecular Sciences, 23(4), 2143. https://doi.org/10.3390/ijms23042143






