DNA Methylation Mediates EMT Gene Expression in Human Pancreatic Ductal Adenocarcinoma Cell Lines
Abstract
1. Introduction
2. Results
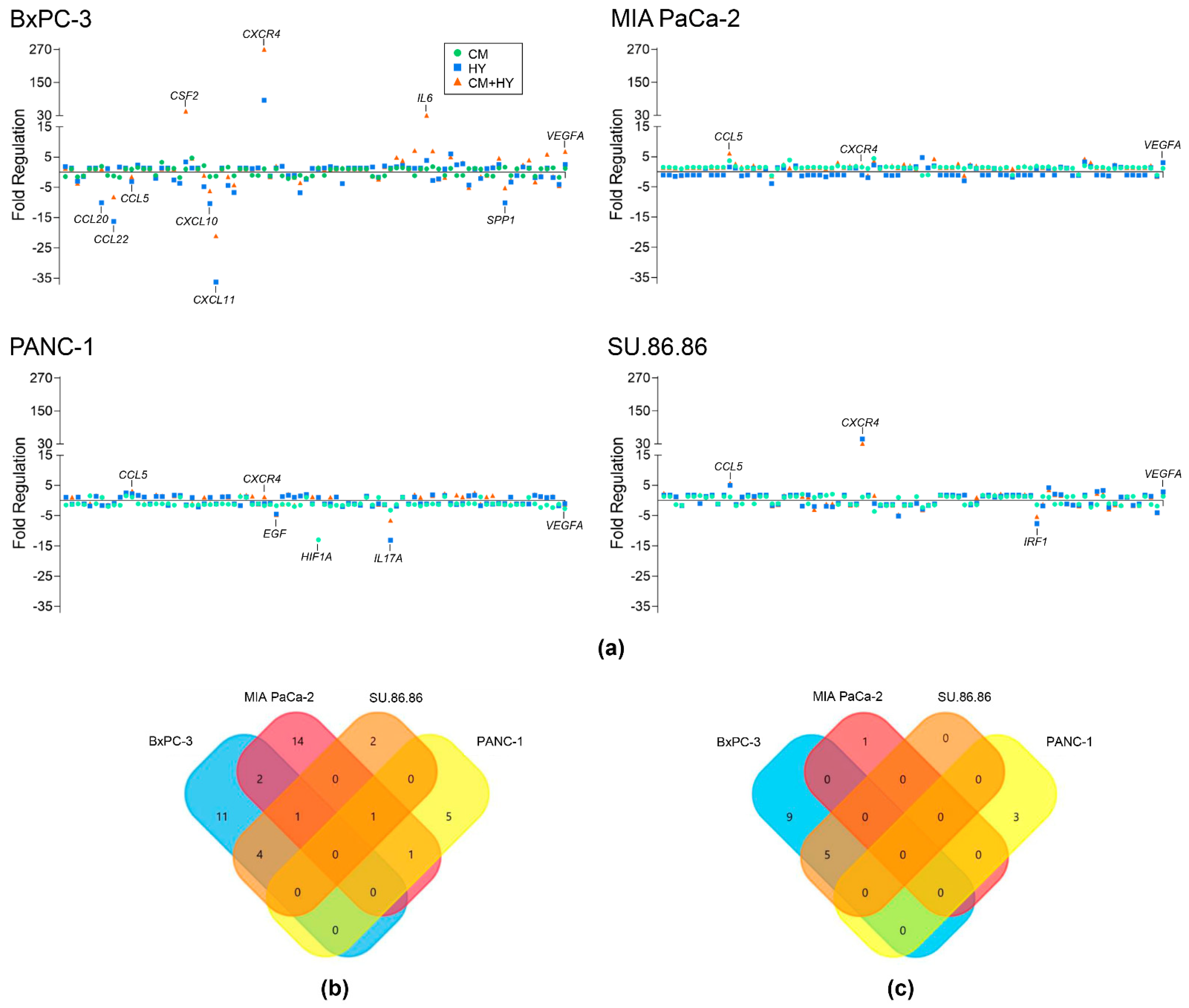
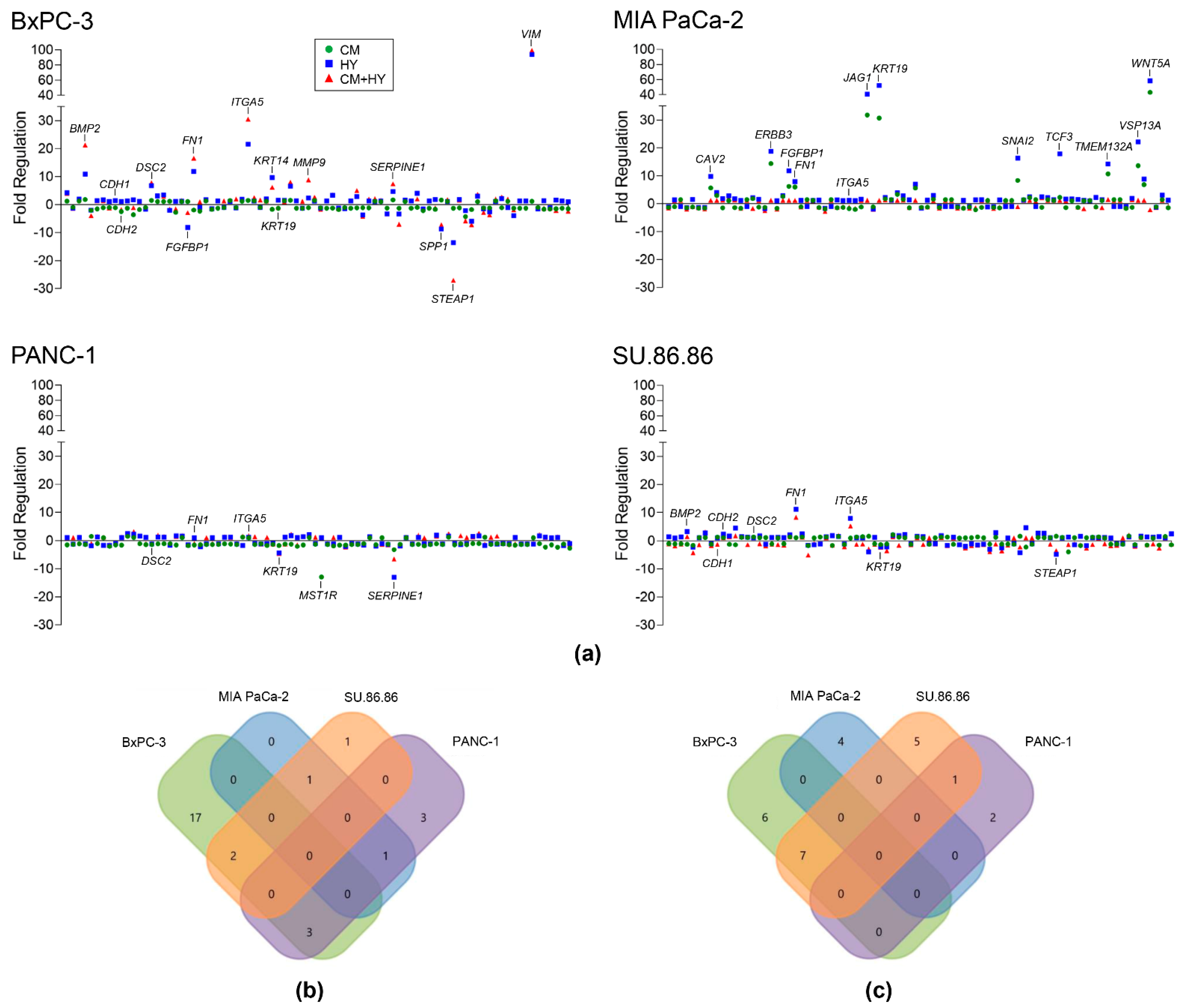
2.1. Inflammation and Hypoxia-Mediated Gene Expression Changes after 2-Day Exposure
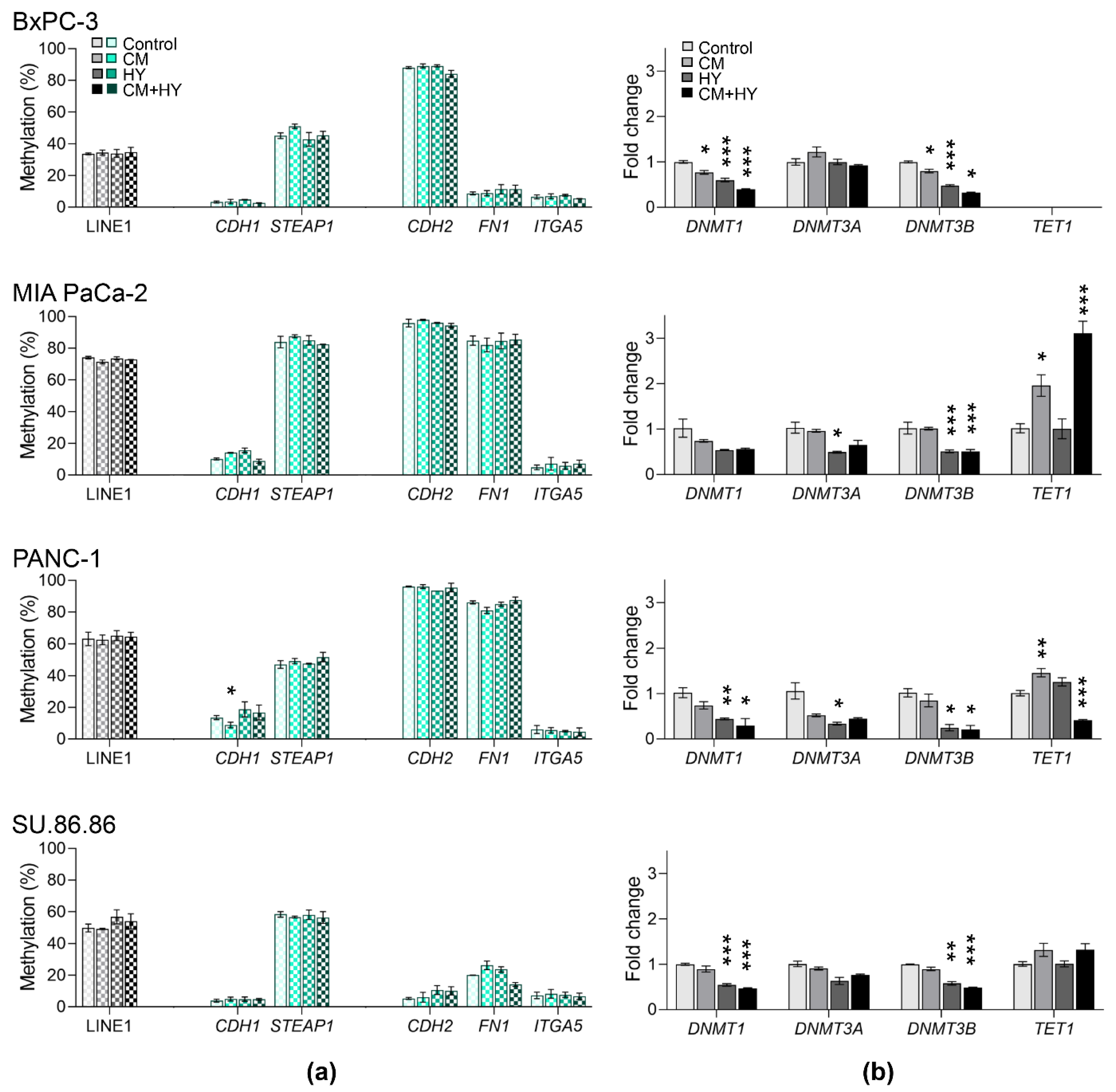
2.2. Global and Gene-Specific DNA Methylation in Individual Cell Lines
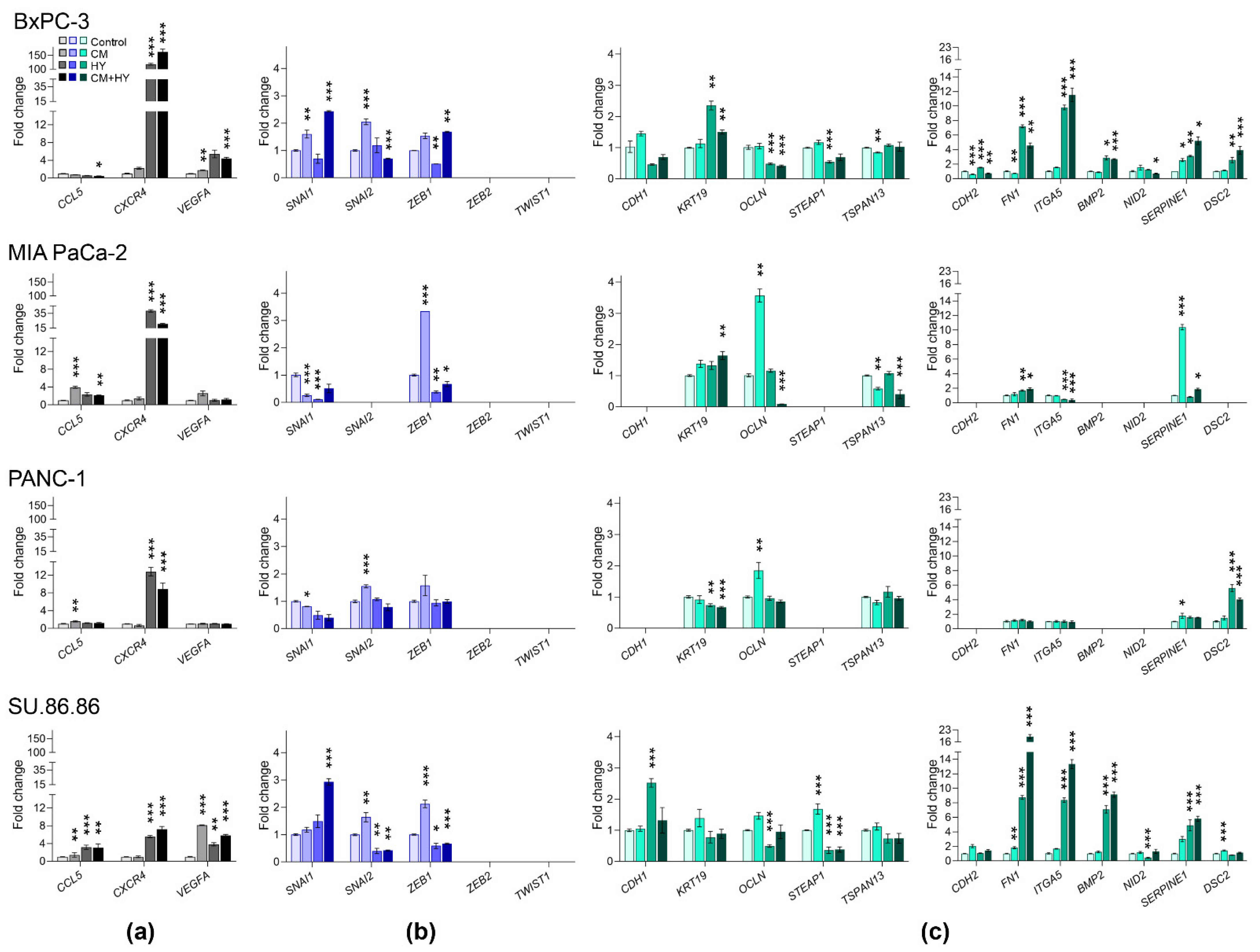
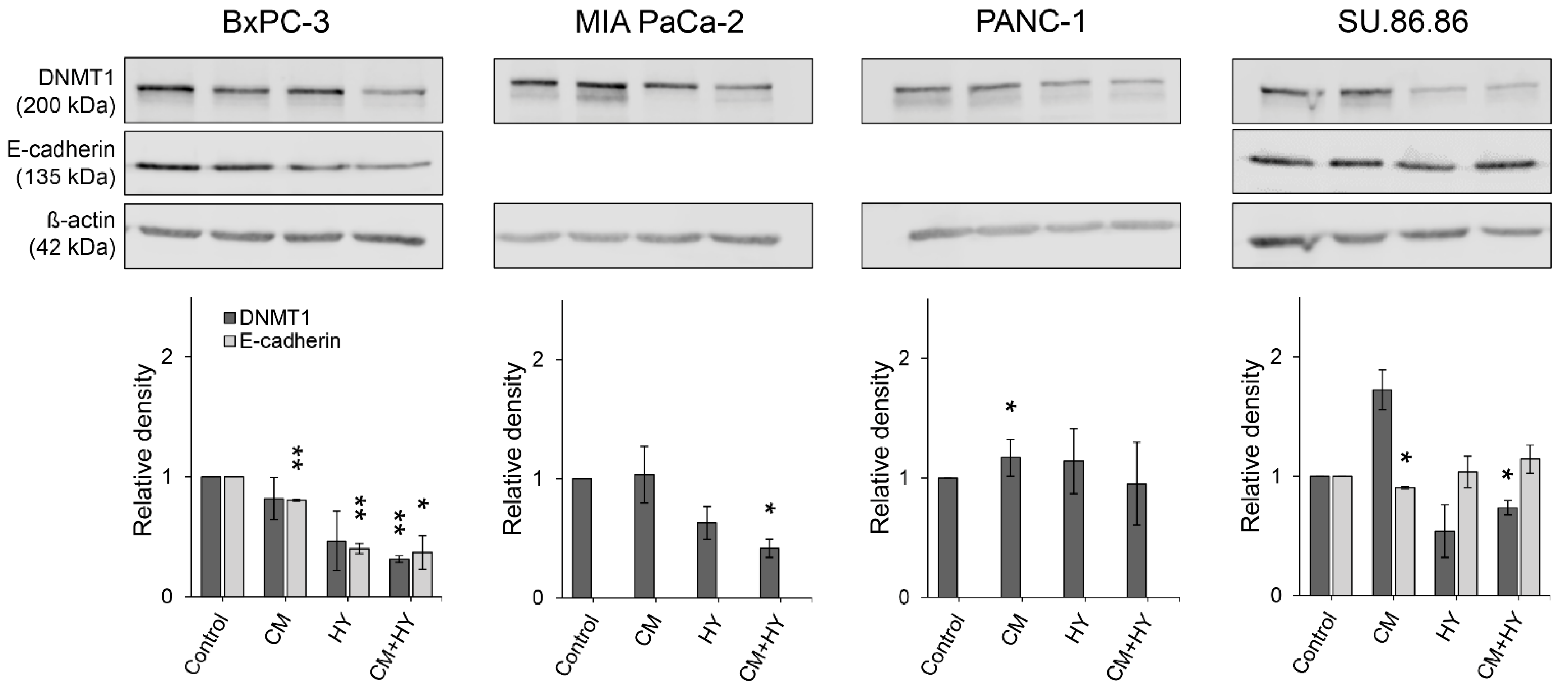
2.3. Prolonged Treatment-Induced EMT-Related Gene Expression and DNA Methylation Changes
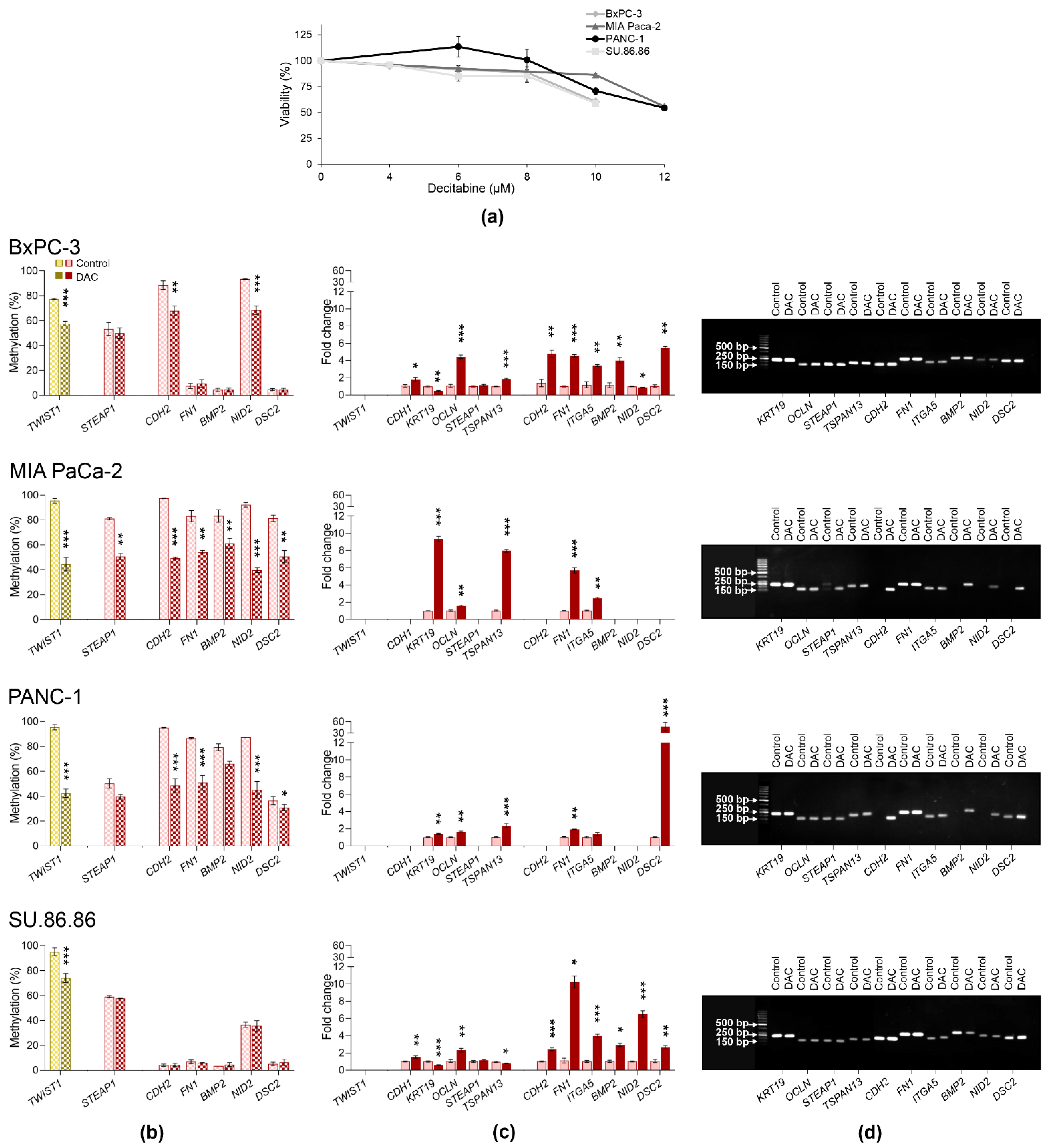
2.4. Gene Expression and DNA Methylation Changes Induced by Decitabine
3. Discussion
4. Materials and Methods
4.1. Pancreatic Cancer Cell Lines
4.2. Cell Viability
4.3. Cell Exposure
4.4. Expression Arrays
4.5. qRT-PCR Analysis
4.6. Western Blot
4.7. DNA Methylation Analysis
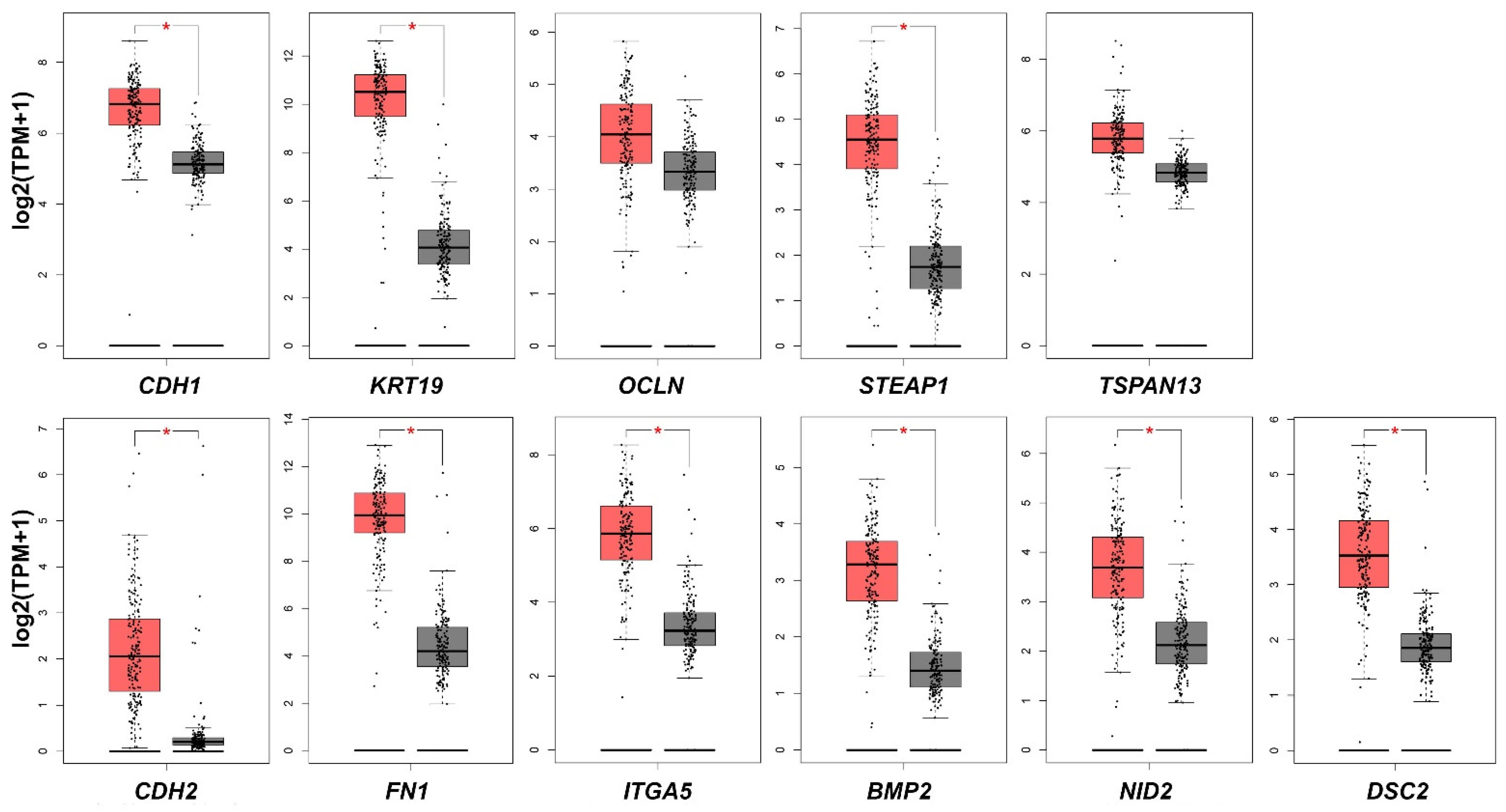
4.8. Validation of mRNA Expression of Studied Genes between PDAC and Normal Tissue
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Wehner, M.R.; Matrisian, L.M.; Nead, K.T. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw. Open 2021, 4, e214708. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cancer.net/cancer-types/pancreatic-cancer/statistics (accessed on 16 December 2021).
- Maitra, A.; Adsay, N.V.; Argani, P.; Iacobuzio-Donahue, C.; De Marzo, A.; Cameron, J.L.; Yeo, C.J.; Hruban, R.H. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod. Pathol. 2003, 16, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, L.; Perna, F.; Nimer, S.D. Beyond transcription factors: How oncogenic signalling reshapes the epigenetic landscape. Nat. Rev. Cancer 2016, 16, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Lomberk, G.; Blum, Y.; Nicolle, R.; Nair, A.; Gaonkar, K.S.; Marisa, L.; Mathison, A.; Sun, Z.; Yan, H.; Elarouci, N.; et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018, 9, 1978. [Google Scholar] [CrossRef]
- Lomberk, G.; Dusetti, N.; Iovanna, J.; Urrutia, R. Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine. Nat. Commun. 2019, 10, 3875. [Google Scholar] [CrossRef] [PubMed]
- Strapcova, S.; Takacova, M.; Csaderova, L.; Martinelli, P.; Lukacikova, L.; Gal, V.; Kopacek, J.; Svastova, E. Clinical and Pre-Clinical Evidence of Carbonic Anhydrase IX in Pancreatic Cancer and Its High Expression in Pre-Cancerous Lesions. Cancers 2020, 12, 2005. [Google Scholar] [CrossRef] [PubMed]
- Benej, M.; Svastova, E.; Banova, R.; Kopacek, J.; Gibadulinova, A.; Kery, M.; Arena, S.; Scaloni, A.; Vitale, M.; Zambrano, N.; et al. CA IX Stabilizes Intracellular pH to Maintain Metabolic Reprogramming and Proliferation in Hypoxia. Front. Oncol. 2020, 10, 1462. [Google Scholar] [CrossRef]
- López-Novoa, J.M.; Nieto, M.A. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 2009, 1, 303–314. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.-Q.; Wang, Z.-F.; Wu, X.-L.; Zhou, C.-H.; Yan, J.-Y.; Hu, B.-Y.; et al. The molecular biology of pancreatic adenocarcinoma: Translational challenges and clinical perspectives. Signal Transduct. Target. Ther. 2021, 6, 249. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Tao, J.; Yang, G.; Zhou, W.; Qiu, J.; Chen, G.; Luo, W.; Zhao, F.; You, L.; Zheng, L.; Zhang, T.; et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J. Hematol. Oncol. 2021, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, A.; Yanai, K.; Onishi, H. Hypoxia and pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 484, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Panisova, E.; Kery, M.; Sedlakova, O.; Brisson, L.; Debreova, M.; Sboarina, M.; Sonveaux, P.; Pastorekova, S.; Svastova, E. Lactate stimulates CA IX expression in normoxic cancer cells. Oncotarget 2017, 8, 77819–77835. [Google Scholar] [CrossRef] [PubMed]
- Takacova, M.; Holotnakova, T.; Barathova, M.; Pastorekova, S.; Kopacek, J.; Pastorek, J. Src induces expression of carbonic anhydrase IX via hypoxia-inducible factor 1. Oncol. Rep. 2010, 23, 869–874. [Google Scholar]
- Zhuang, H.; Wang, S.; Chen, B.; Zhang, Z.; Ma, Z.; Li, Z.; Liu, C.; Zhou, Z.; Gong, Y.; Huang, S.; et al. Prognostic Stratification Based on HIF-1 Signaling for Evaluating Hypoxic Status and Immune Infiltration in Pancreatic Ductal Adenocarcinomas. Front. Immunol. 2021, 12, 790661. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Li, W.; Shan, T.; Lin, W.R.; Ma, J.; Cui, X.; Yang, W.; Cao, G.; Li, Y.; et al. Conversion of epithelial-to-mesenchymal transition to mesenchymal-to-epithelial transition is mediated by oxygen concentration in pancreatic cancer cells. Oncol. Lett. 2018, 15, 7144–7152. [Google Scholar] [CrossRef]
- Miyazono, K.; Ehata, S.; Koinuma, D. Tumor-promoting functions of transforming growth factor-β in progression of cancer. Ups J. Med. Sci. 2012, 117, 143–152. [Google Scholar] [CrossRef]
- Terashima, M.; Ishimura, A.; Wanna-Udom, S.; Suzuki, T. MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells. J. Biol. Chem. 2018, 293, 18016–18030. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, J.; Wang, H.; Xue, X.; An, Y.; Tang, D.; Yuan, Z.; Wang, F.; Wu, J.; Zhang, J.; et al. Epithelial mesenchymal transition correlates with CD24+CD44+ and CD133+ cells in pancreatic cancer. Oncol. Rep. 2012, 27, 1599–1605. [Google Scholar] [CrossRef]
- Schito, L.; Semenza, G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2016, 2, 758–770. [Google Scholar] [CrossRef]
- Galván, J.A.; Zlobec, I.; Wartenberg, M.; Lugli, A.; Gloor, B.; Perren, A.; Karamitopoulou, E. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br. J. Cancer 2015, 112, 1944–1950. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.H.; Wu, K.J.; Lee, W.H. Hypoxia, Epithelial-Mesenchymal Transition, and TET-Mediated Epigenetic Changes. J. Clin. Med. 2016, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Camuzi, D.; de Amorim, Í.S.S.; Ribeiro Pinto, L.F.; Oliveira Trivilin, L.; Mencalha, A.L.; Soares Lima, S.C. Regulation Is in the Air: The Relationship between Hypoxia and Epigenetics in Cancer. Cells 2019, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Geismann, C.; Arlt, A. Coming in the Air: Hypoxia Meets Epigenetics in Pancreatic Cancer. Cells 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Dong, C.; Zhou, B.P. Epigenetic regulation of EMT: The Snail story. Curr. Pharm. Des. 2014, 20, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Aghdassi, A.; Sendler, M.; Guenther, A.; Mayerle, J.; Behn, C.O.; Heidecke, C.D.; Friess, H.; Büchler, M.; Evert, M.; Lerch, M.M.; et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 2012, 61, 439–448. [Google Scholar] [CrossRef]
- Von Burstin, J.; Eser, S.; Paul, M.C.; Seidler, B.; Brandl, M.; Messer, M.; von Werder, A.; Schmidt, A.; Mages, J.; Pagel, P.; et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 2009, 137, 361–371. [Google Scholar] [CrossRef]
- Watson, J.A.; Watson, C.J.; McCann, A.; Baugh, J. Epigenetics, the epicenter of the hypoxic response. Epigenetics 2010, 5, 293–296. [Google Scholar] [CrossRef]
- Romero-Garcia, S.; Prado-Garcia, H.; Carlos-Reyes, A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front. Oncol. 2020, 10, 1152. [Google Scholar] [CrossRef]
- Szabova, K.; Bizikova, I.; Mistrik, M.; Bizik, J. Inflammatory environment created by fibroblast aggregates induces growth arrest and phenotypic shift of human myeloma cells. Neoplasma 2015, 62, 938–948. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, C.; Tang, Z.; Zhang, W.; Ye, Z.; Liu, F. GEPIA2021: Integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021, 49, W242–W246. [Google Scholar] [CrossRef] [PubMed]
- Gradiz, R.; Silva, H.C.; Carvalho, L.; Botelho, M.F.; Mota-Pinto, A. MIA PaCa-2 and PANC-1 pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci. Rep. 2016, 6, 21648. [Google Scholar] [CrossRef] [PubMed]
- Deer, E.L.; González-Hernández, J.; Coursen, J.D.; Shea, J.E.; Ngatia, J.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 2010, 39, 425–435. [Google Scholar] [CrossRef]
- Opdenakker, G.; Van Damme, J. The countercurrent principle in invasion and metastasis of cancer cells. Recent insights on the roles of chemokines. Int. J. Dev. Biol. 2004, 48, 519–527. [Google Scholar] [CrossRef]
- Saur, D.; Seidler, B.; Schneider, G.; Algül, H.; Beck, R.; Senekowitsch-Schmidtke, R.; Schwaiger, M.; Schmid, R.M. CXCR4 expression increases liver and lung metastasis in a mouse model of pancreatic cancer. Gastroenterology 2005, 129, 1237–1250. [Google Scholar] [CrossRef]
- Wehler, T.; Wolfert, F.; Schimanski, C.C.; Gockel, I.; Herr, W.; Biesterfeld, S.; Seifert, J.K.; Adwan, H.; Berger, M.R.; Junginger, T.; et al. Strong expression of chemokine receptor CXCR4 by pancreatic cancer correlates with advanced disease. Oncol. Rep. 2006, 16, 1159–1164. [Google Scholar] [CrossRef][Green Version]
- Ding, Y.; Du, Y. Clinicopathological significance and prognostic role of chemokine receptor CXCR4 expression in pancreatic ductal adenocarcinoma, a meta-analysis and literature review. Int. J. Surg. 2019, 65, 32–38. [Google Scholar] [CrossRef]
- Seeber, A.; Kocher, F.; Pircher, A.; Puccini, A.; Baca, Y.; Xiu, J.; Zimmer, K.; Haybaeck, J.; Spizzo, G.; Goldberg, R.M. High CXCR4 expression in pancreatic ductal adenocarcinoma as characterized by an inflammatory tumor phenotype with potential implications for an immunotherapeutic approach. J. Clin. Oncol. 2021, 39, 4021. [Google Scholar] [CrossRef]
- Aldinucci, D.; Borghese, C.; Casagrande, N. The CCL5/CCR5 Axis in Cancer Progression. Cancers 2020, 12, 1765. [Google Scholar] [CrossRef]
- Singh, S.K.; Mishra, M.K.; Eltoum, I.A.; Bae, S.; Lillard, J.W., Jr.; Singh, R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci. Rep. 2018, 8, 1323. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Yashiro, M.; Yamada, N.; Amano, R.; Noda, S.; Hirakawa, K. VEGF-A/VEGFR-2 signaling plays an important role for the motility of pancreas cancer cells. Ann. Surg. Oncol. 2012, 19, 2733–2743. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Qi, Q.; Peng, X.; Hochwald, S.N.; Yan, L.; Takabe, K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci. Rep. 2019, 9, 1310. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.; Wilson, M.B.; Crawford, Y.G.; Reynolds, P.A.; Sigaroudinia, M.; Tlsty, T.D. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 14867–14872. [Google Scholar] [CrossRef]
- Bohl, S.R.; Bullinger, L.; Rücker, F.G. Epigenetic therapy: Azacytidine and decitabine in acute myeloid leukemia. Expert Rev. Hematol. 2018, 11, 361–371. [Google Scholar] [CrossRef]
- Seelan, R.S.; Mukhopadhyay, P.; Pisano, M.M.; Greene, R.M. Effects of 5-Aza-2′-deoxycytidine (decitabine) on gene expression. Drug Metab. Rev. 2018, 50, 193–207. [Google Scholar] [CrossRef]
- Topalovski, M.; Brekken, R.A. Matrix control of pancreatic cancer: New insights into fibronectin signaling. Cancer Lett. 2016, 381, 252–258. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, G.; Zhu, H.; Xu, A. ITGA5 is a prognostic biomarker and correlated with immune infiltration in gastrointestinal tumors. BMC Cancer 2021, 21, 269. [Google Scholar] [CrossRef]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. [Google Scholar] [CrossRef]
- Atay, S. Integrated transcriptome meta-analysis of pancreatic ductal adenocarcinoma and matched adjacent pancreatic tissues. PeerJ 2020, 8, e10141. [Google Scholar] [CrossRef]
- Bendas, G.; Borsig, L. Cancer cell adhesion and metastasis: Selectins, integrins, and the inhibitory potential of heparins. Int. J. Cell Biol. 2012, 2012, 676731. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Ansari, D.; Zhou, Q.; Sasor, A.; Hilmersson, K.S.; Andersson, R. Stromal fibronectin expression in patients with resected pancreatic ductal adenocarcinoma. World J. Surg. Oncol. 2019, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Olmeda, D.; Jordá, M.; Peinado, H.; Fabra, A.; Cano, A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene 2007, 26, 1862–1874. [Google Scholar] [CrossRef]
- Murphy, P.A.; Begum, S.; Hynes, R.O. Tumor angiogenesis in the absence of fibronectin or its cognate integrin receptors. PLoS ONE 2015, 10, e0120872. [Google Scholar] [CrossRef]
- Kuninty, P.R.; Bansal, R.; De Geus, S.W.L.; Mardhian, D.F.; Schnittert, J.; van Baarlen, J.; Storm, G.; Bijlsma, M.F.; van Laarhoven, H.W.; Metselaar, J.M.; et al. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci. Adv. 2019, 5, eaax2770. [Google Scholar] [CrossRef]
- Fang, Z.; Yao, W.; Xiong, Y.; Zhang, J.; Liu, L.; Li, J.; Zhang, C.; Wan, J. Functional elucidation and methylation-mediated downregulation of ITGA5 gene in breast cancer cell line MDA-MB-468. J. Cell. Biochem. 2010, 110, 1130–1141. [Google Scholar] [CrossRef] [PubMed]
- Ohgami, R.S.; Campagna, D.R.; McDonald, A.; Fleming, M.D. The Steap proteins are metalloreductases. Blood 2006, 108, 1388–1394. [Google Scholar] [CrossRef]
- Gomes, I.M.; Maia, C.J.; Santos, C.R. STEAP proteins: From structure to applications in cancer therapy. Mol. Cancer Res. 2012, 10, 573–587. [Google Scholar] [CrossRef]
- Huo, S.-F.; Shang, W.-L.; Yu, M.; Ren, X.-P.; Wen, H.-X.; Chai, C.-Y.; Sun, L.; Hui, K.; Liu, L.-H.; Wei, S.-H. Steap1 facilitates metastasis and epithelial–mesenchymal transition of lung adenocarcinoma via the JAK2/STAT3 signaling pathway. Biosci. Rep. 2020, 40, BSR20193169. [Google Scholar] [CrossRef]
- Xie, J.; Yang, Y.; Sun, J.; Jiao, Z.; Zhang, H.; Chen, J. STEAP1 inhibits breast cancer metastasis and is associated with epithelial–mesenchymal transition procession. Clin. Breast Cancer 2019, 19, e195–e207. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, W.B.; Zhang, C.; Tan, Y.E.; Zhang, D.D.; An, W.; Pan, S.W.; Wu, W.D.; Chen, Q.C.; Xu, H.M. A research of STEAP1 regulated gastric cancer cell proliferation, migration and invasion in vitro and in vivos. J. Cell. Mol. Med. 2020, 24, 14217–14230. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Merritt, A.; Rouhi-Parkouhi, M.; Tabernero, L.; Garrod, D. Membrane-impermeable Cross-linking Provides Evidence for Homophilic, Isoform-specific Binding of Desmosomal Cadherins in Epithelial Cells. J. Biol. Chem. 2011, 286, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, L.; Yang, X.-X.; Jiang, Y.-H.; Guo, X.-L. The aberrant expression or disruption of desmocollin2 in human diseases. Int. J. Biol. Macromol. 2019, 131, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Hamidov, Z.; Altendorf-Hofmann, A.; Chen, Y.; Settmacher, U.; Petersen, I.; Knösel, T. Reduced expression of desmocollin 2 is an independent prognostic biomarker for shorter patients survival in pancreatic ductal adenocarcinoma. J. Clin. Pathol. 2011, 64, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.-H.; Wang, Y.-M.; Jiang, Y.-Z.; Ma, S.-J.; Zhong, Q.; Wan, Y.-Y.; Wang, X.-W. NID2 can serve as a potential prognosis prediction biomarker and promotes the invasion and migration of gastric cancer. Pathol. Res. Pract. 2019, 215, 152553. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Wang, W.; Tong, Y.; Zhang, J. Genome-Scale Analysis Identified NID2, SPARC, and MFAP2 as Prognosis Markers of Overall Survival in Gastric Cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021, 27, e929558-1. [Google Scholar] [CrossRef]
- Strelnikov, V.V.; Kuznetsova, E.B.; Tanas, A.S.; Rudenko, V.V.; Kalinkin, A.I.; Poddubskaya, E.V.; Kekeeva, T.V.; Chesnokova, G.G.; Trotsenko, I.D.; Larin, S.S. Abnormal promoter DNA hypermethylation of the integrin, nidogen, and dystroglycan genes in breast cancer. Sci. Rep. 2021, 11, 2264. [Google Scholar] [CrossRef]
- Kleeff, J.; Maruyama, H.; Ishiwata, T.; Sawhney, H.; Friess, H.; Büchler, M.W.; Korc, M. Bone morphogenetic protein 2 exerts diverse effects on cell growth in vitro and is expressed in human pancreatic cancer in vivo. Gastroenterology 1999, 116, 1202–1216. [Google Scholar] [CrossRef]
- Rao, A.D.; Liu, Y.; von Eyben, R.; Hsu, C.C.; Hu, C.; Rosati, L.M.; Parekh, A.; Ng, K.; Hacker-Prietz, A.; Zheng, L. Multiplex proximity ligation assay to identify potential prognostic biomarkers for improved survival in locally advanced pancreatic cancer patients treated with stereotactic body radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 486–489. [Google Scholar] [CrossRef]
- Li, C.-S.; Tian, H.; Zou, M.; Zhao, K.-W.; Li, Y.; Lao, L.; Brochmann, E.J.; Duarte, M.E.L.; Daubs, M.D.; Zhou, Y.-H. Secreted phosphoprotein 24 kD (Spp24) inhibits growth of human pancreatic cancer cells caused by BMP-2. Biochem. Biophys. Res. Commun. 2015, 466, 167–172. [Google Scholar] [CrossRef]
- Chen, X.; Liao, J.; Lu, Y.; Duan, X.; Sun, W. Activation of the PI3K/Akt pathway mediates bone morphogenetic protein 2-induced invasion of pancreatic cancer cells Panc-1. Pathol. Oncol. Res. 2011, 17, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Ishiguro, M.; Ishikawa, T.; Okazaki, S.; Baba, H.; Kikuchi, A.; Yamauchi, S.; Matsuyama, T.; Uetake, H.; Kinugasa, Y. Methylation of bone morphogenetic protein 2 is associated with poor prognosis in colorectal cancer. Oncol. Lett. 2020, 19, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Kankuri, E.; Babusikova, O.; Hlubinova, K.; Salmenperä, P.; Boccaccio, C.; Lubitz, W.; Harjula, A.; Bizik, J. Fibroblast nemosis arrests growth and induces differentiation of human leukemia cells. Int. J. Cancer 2008, 122, 1243–1252. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbanova, M.; Buocikova, V.; Trnkova, L.; Strapcova, S.; Kajabova, V.H.; Melian, E.B.; Novisedlakova, M.; Tomas, M.; Dubovan, P.; Earl, J.; et al. DNA Methylation Mediates EMT Gene Expression in Human Pancreatic Ductal Adenocarcinoma Cell Lines. Int. J. Mol. Sci. 2022, 23, 2117. https://doi.org/10.3390/ijms23042117
Urbanova M, Buocikova V, Trnkova L, Strapcova S, Kajabova VH, Melian EB, Novisedlakova M, Tomas M, Dubovan P, Earl J, et al. DNA Methylation Mediates EMT Gene Expression in Human Pancreatic Ductal Adenocarcinoma Cell Lines. International Journal of Molecular Sciences. 2022; 23(4):2117. https://doi.org/10.3390/ijms23042117
Chicago/Turabian StyleUrbanova, Maria, Verona Buocikova, Lenka Trnkova, Sabina Strapcova, Viera Horvathova Kajabova, Emma Barreto Melian, Maria Novisedlakova, Miroslav Tomas, Peter Dubovan, Julie Earl, and et al. 2022. "DNA Methylation Mediates EMT Gene Expression in Human Pancreatic Ductal Adenocarcinoma Cell Lines" International Journal of Molecular Sciences 23, no. 4: 2117. https://doi.org/10.3390/ijms23042117
APA StyleUrbanova, M., Buocikova, V., Trnkova, L., Strapcova, S., Kajabova, V. H., Melian, E. B., Novisedlakova, M., Tomas, M., Dubovan, P., Earl, J., Bizik, J., Svastova, E., Ciernikova, S., & Smolkova, B. (2022). DNA Methylation Mediates EMT Gene Expression in Human Pancreatic Ductal Adenocarcinoma Cell Lines. International Journal of Molecular Sciences, 23(4), 2117. https://doi.org/10.3390/ijms23042117







