Distinct Adipogenic and Fibrogenic Differentiation Capacities of Mesenchymal Stromal Cells from Pancreas and White Adipose Tissue
Abstract
:1. Introduction
2. Results
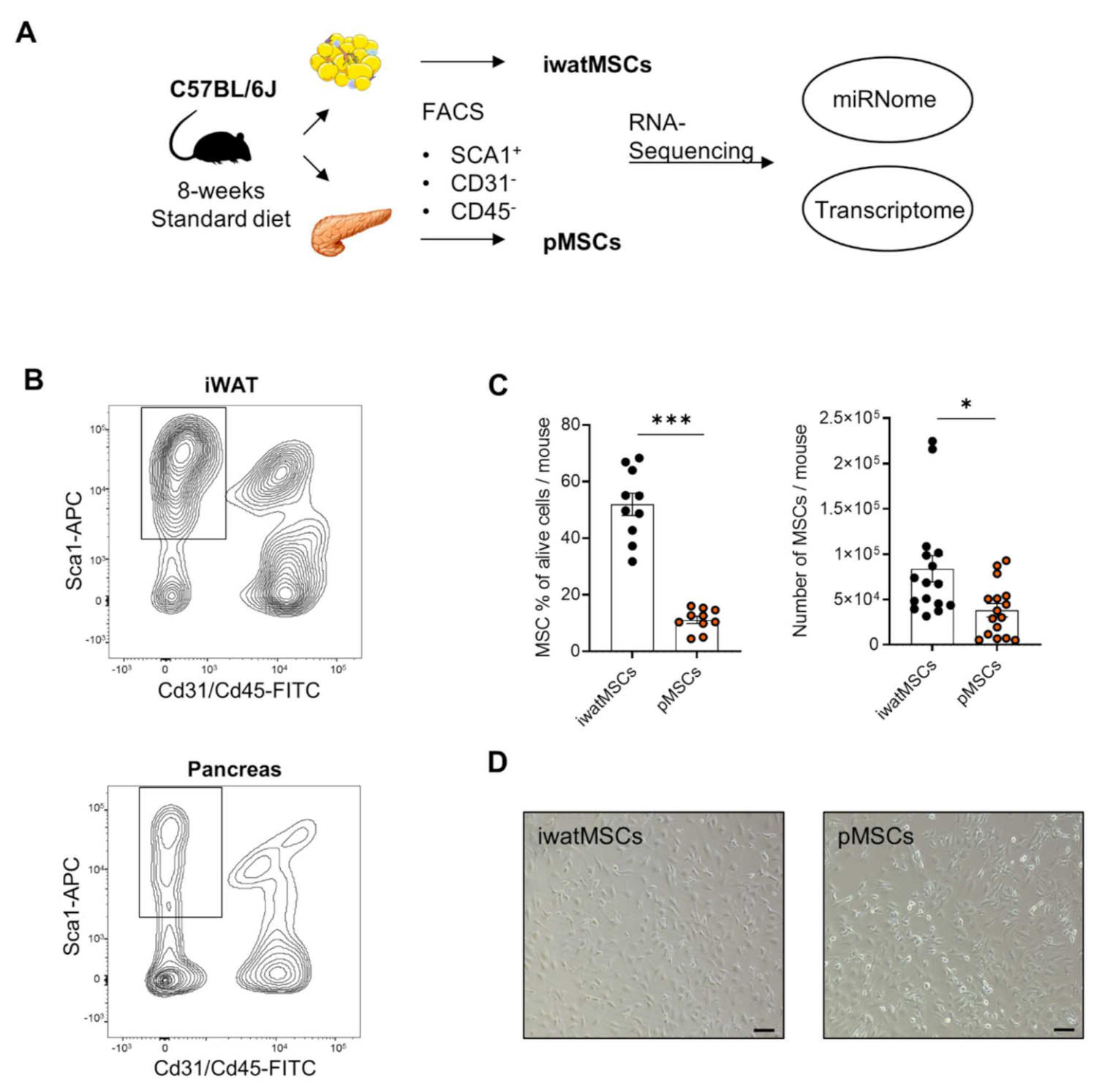
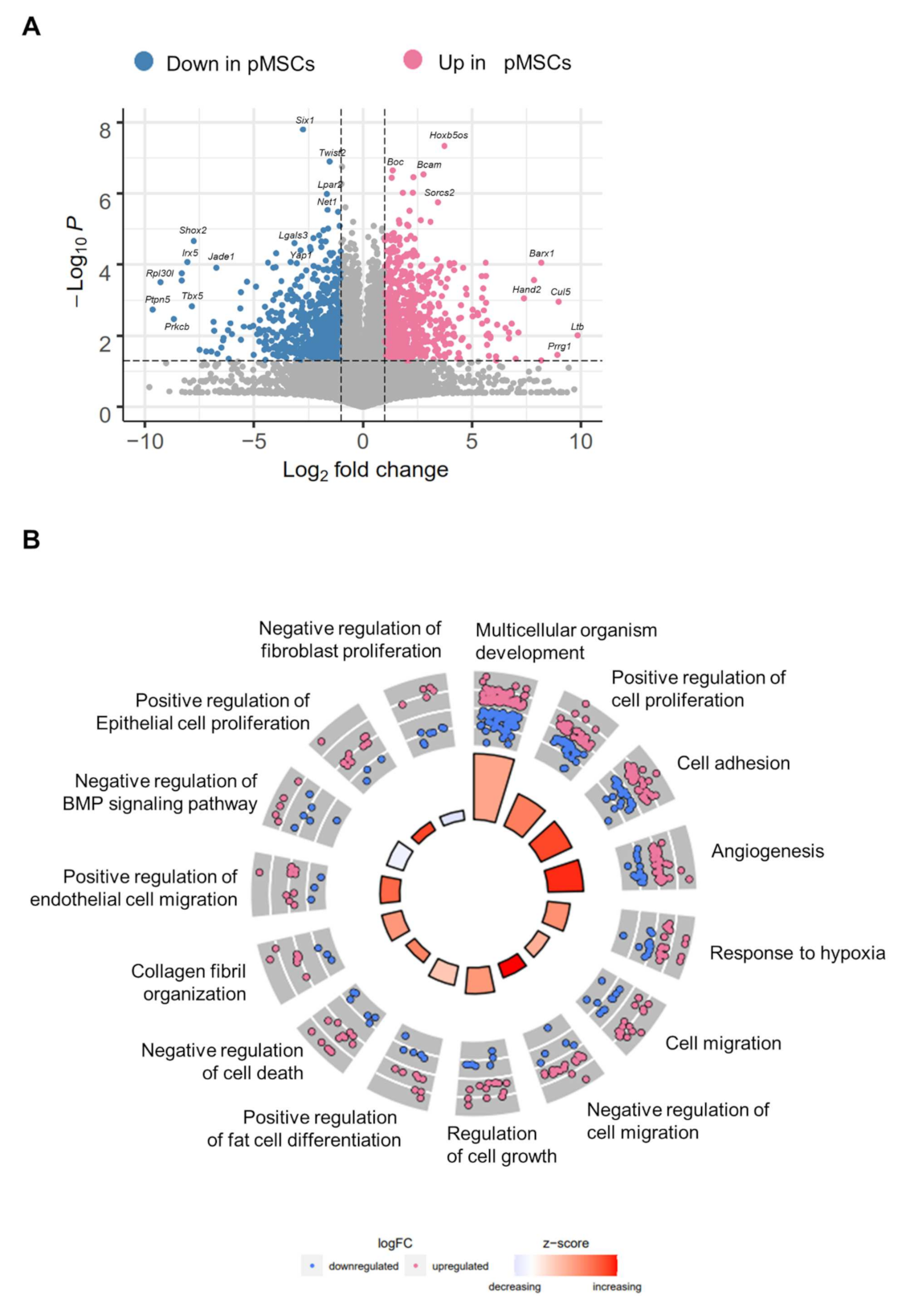
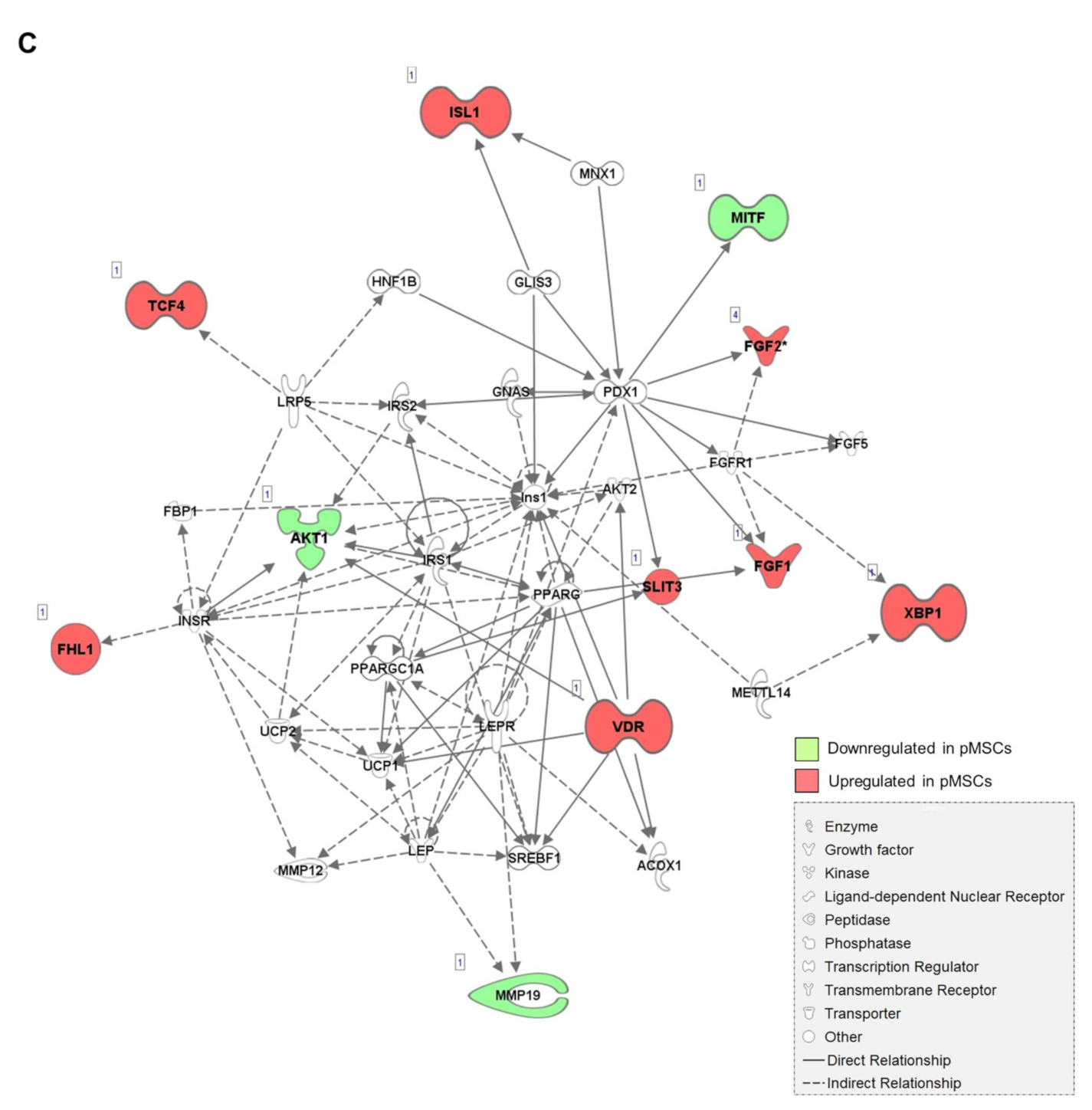
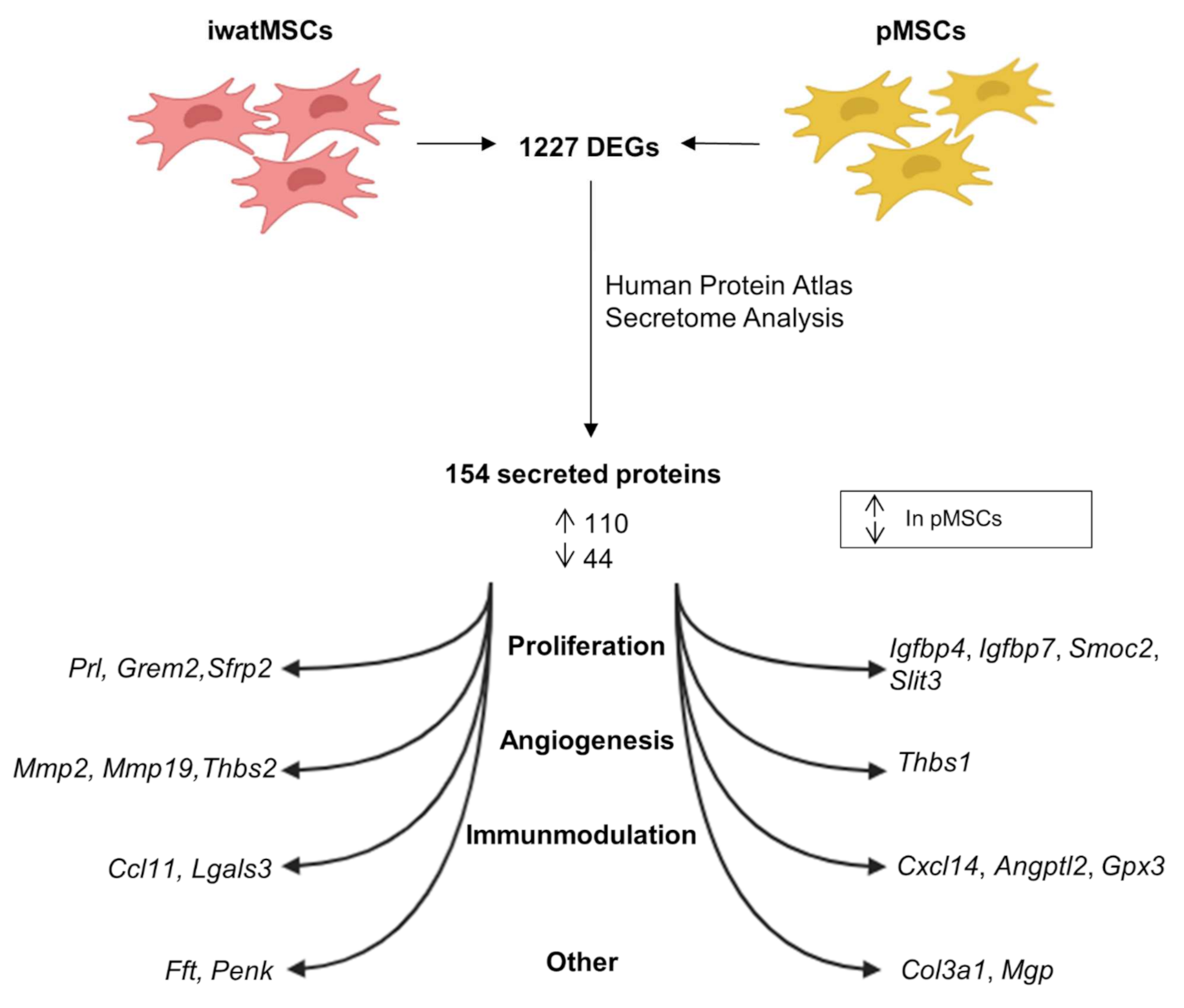
2.1. Distinct Gene Expression Profiles of pMSCs and iwatMSCs
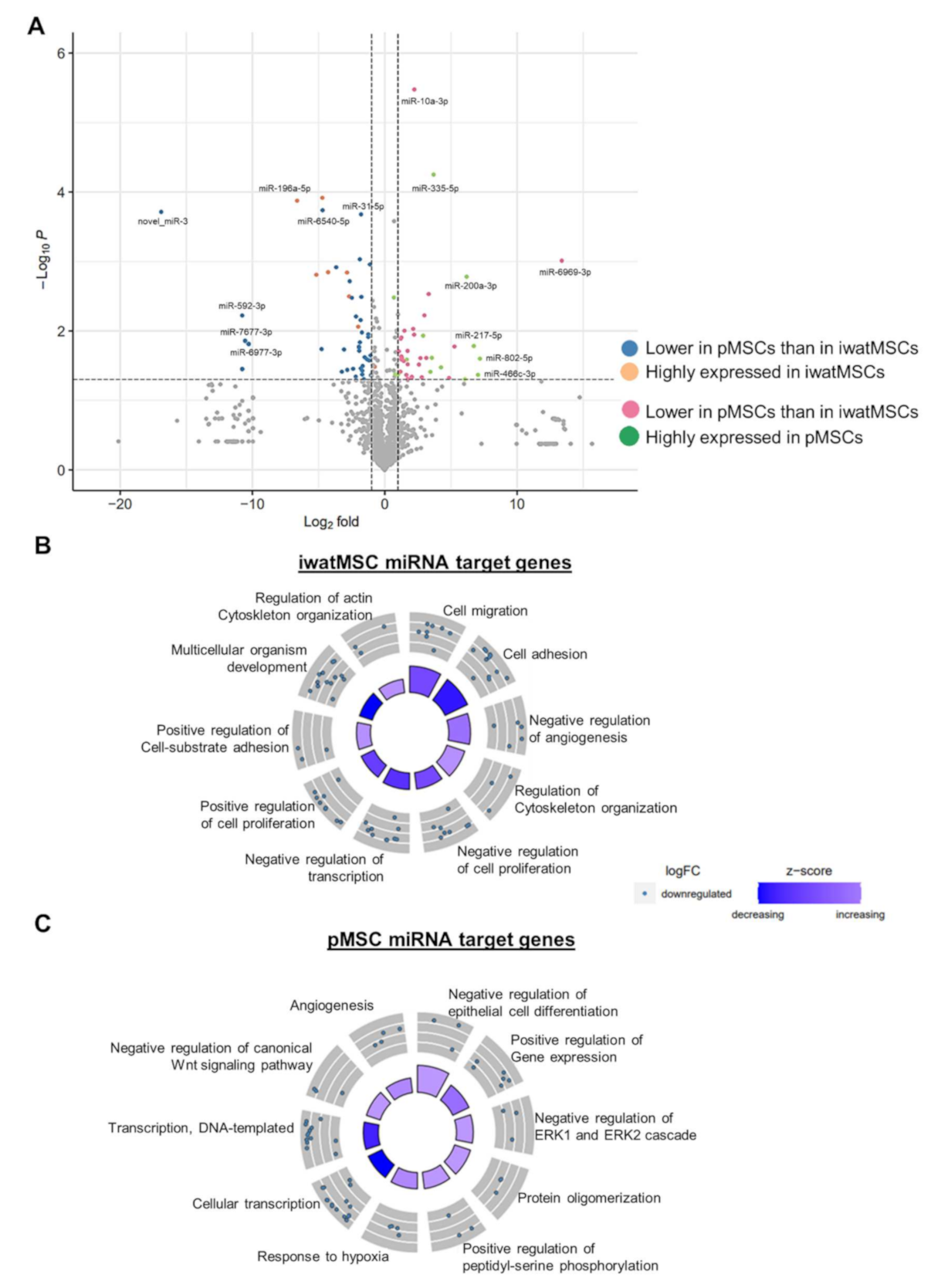
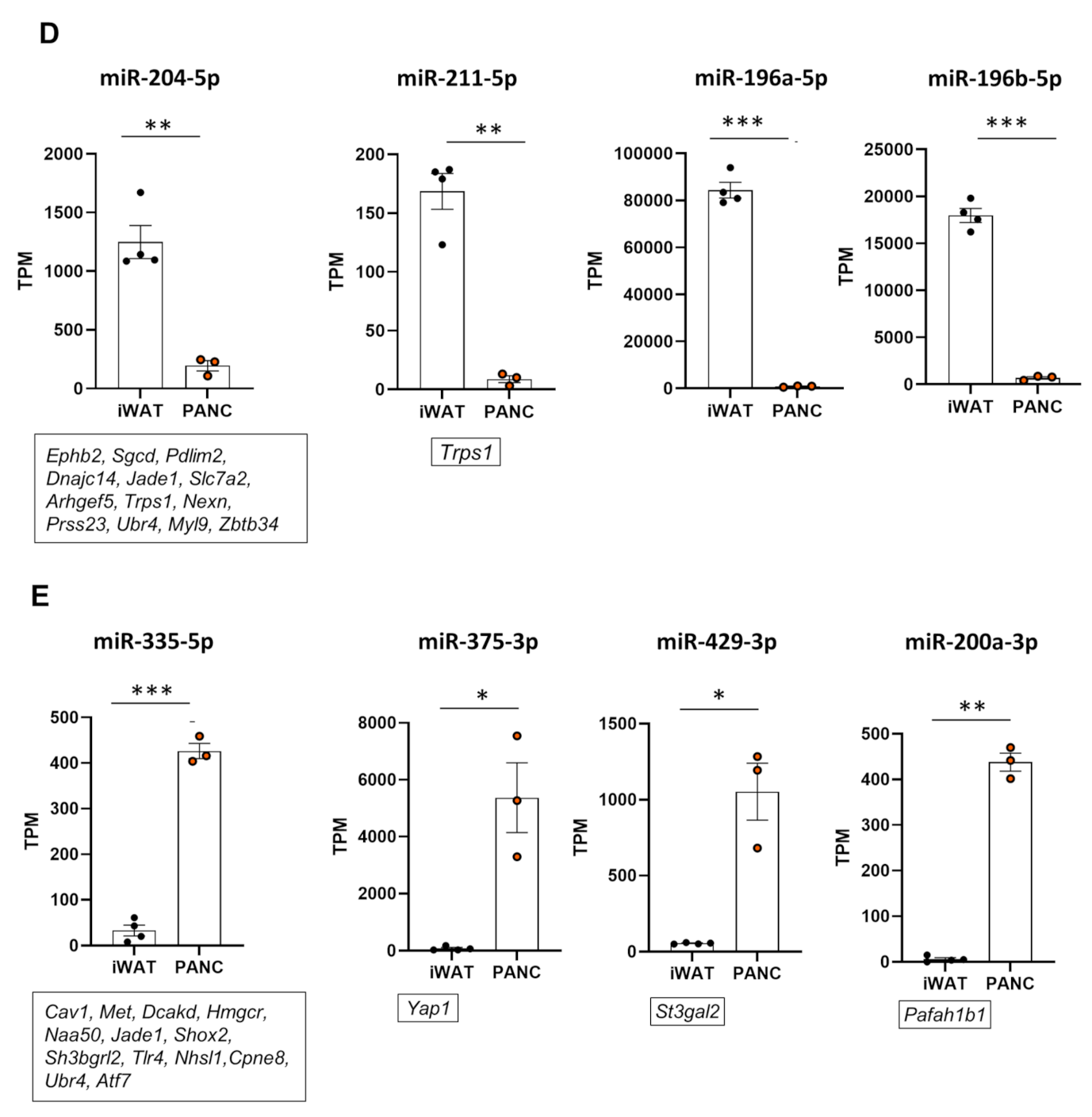
2.2. miRNA Expression Patterns of pMSCs and iwatMSCs
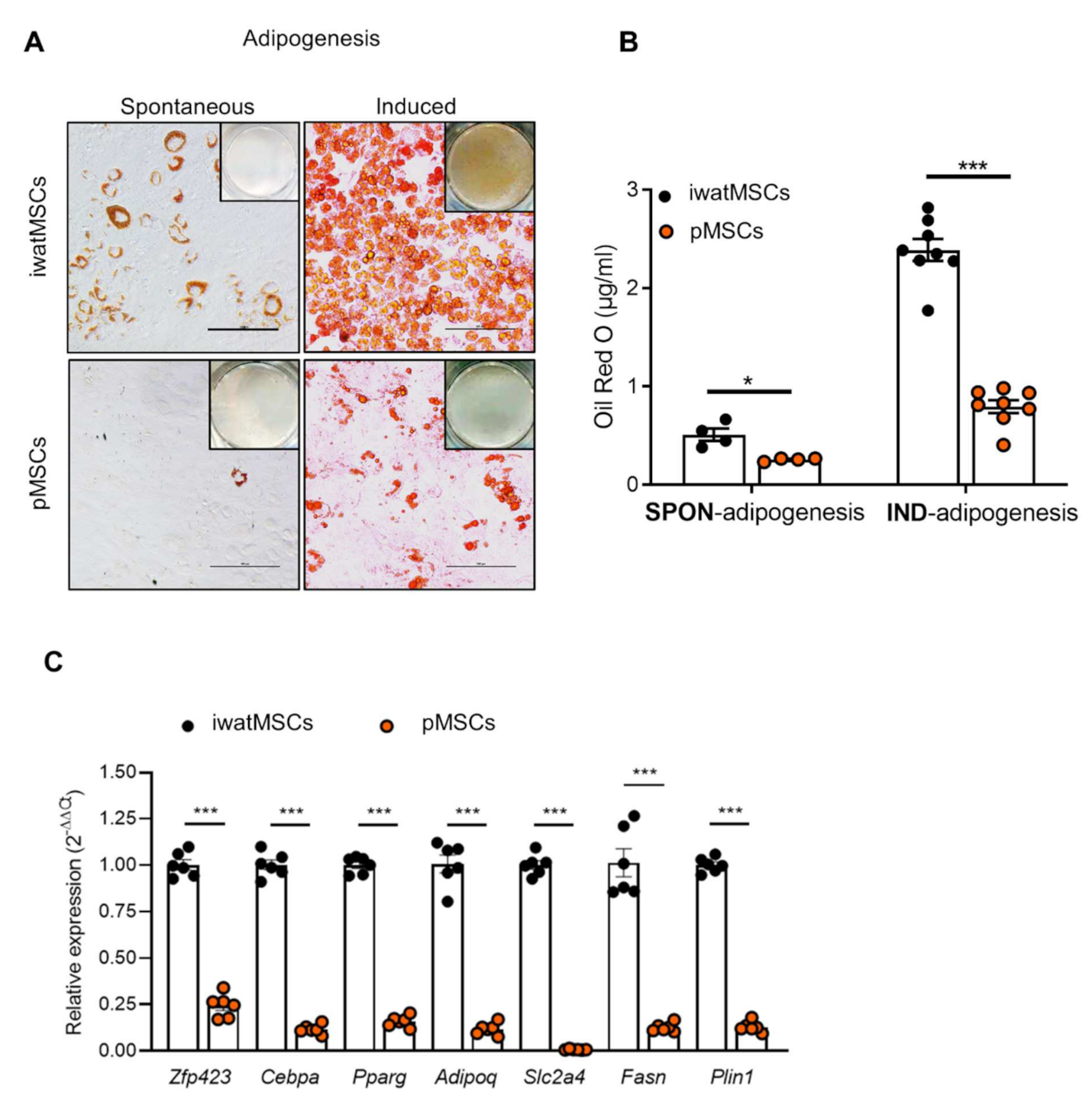
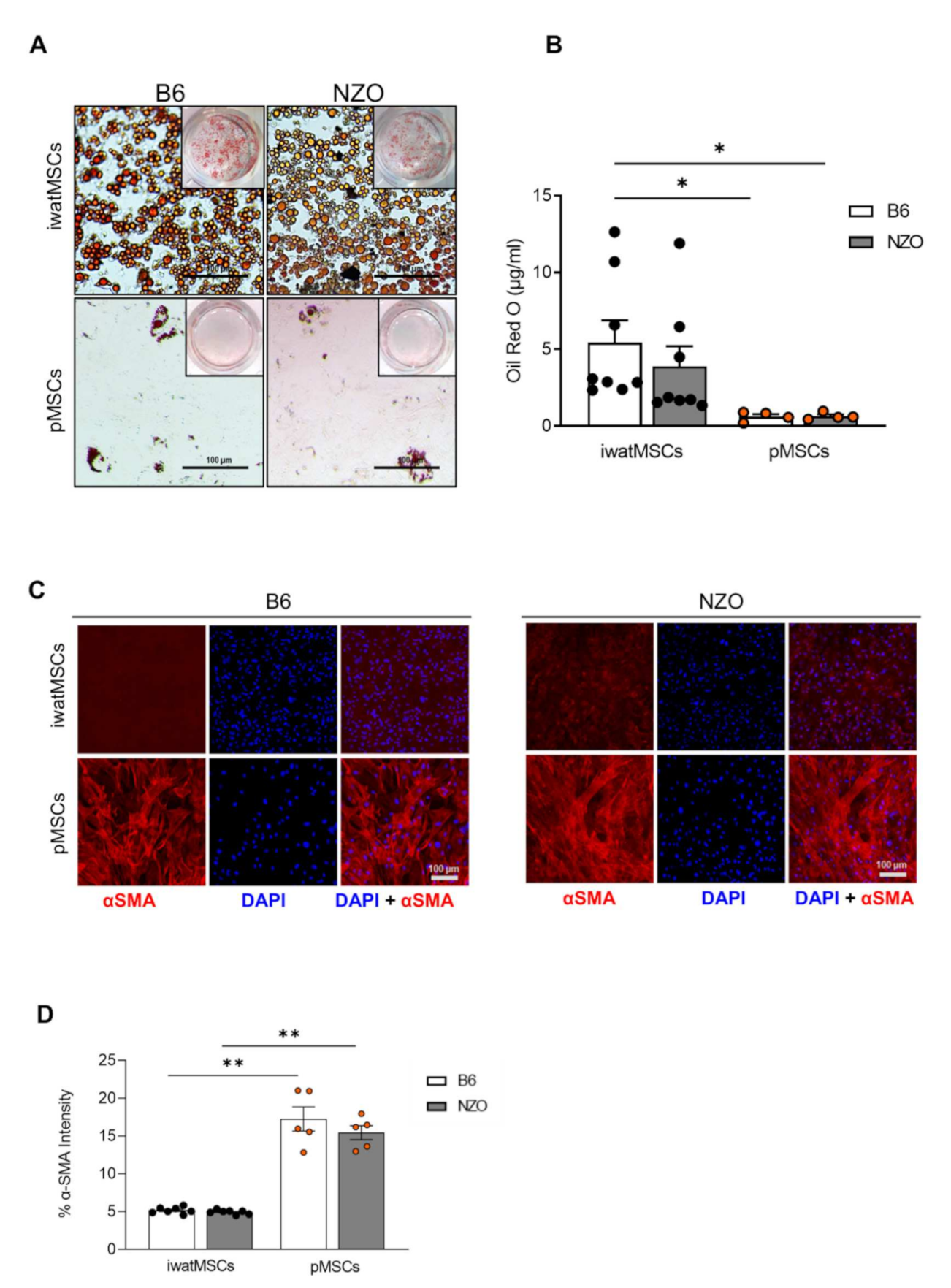
2.3. Decreased Adipogenic Potential of pMSCs
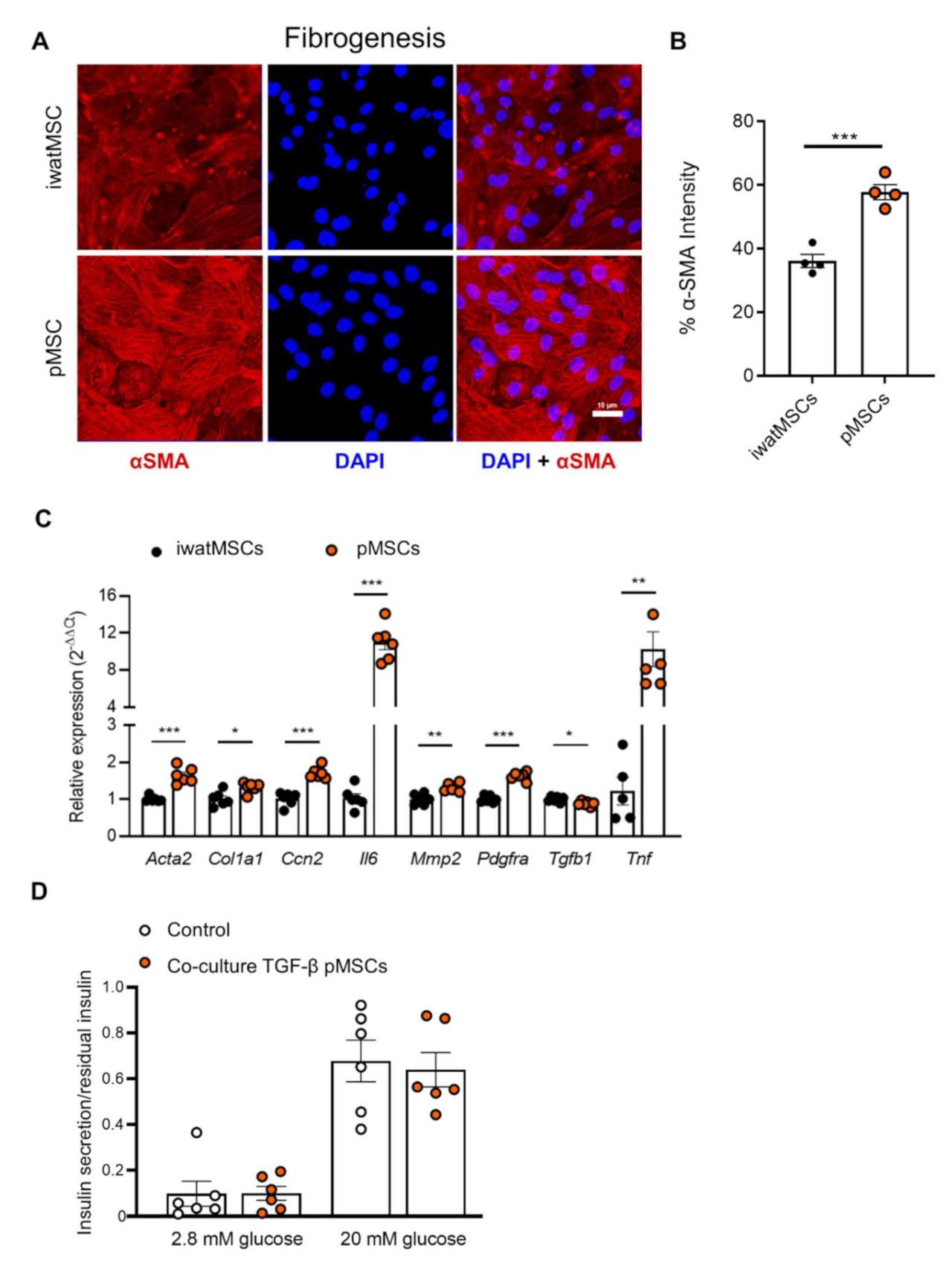
2.4. Increased Fibrogenic Potential in pMSCs
2.5. Similar Cell-Fate Decisions in pMSCs Derived from Diabetes-Susceptible and Diabetes-Resistant Mice
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals
5.2. Isolation of Mesenchymal Stromal Cells with Adipogenic Potential
5.3. Adipogenic Differentiation and Oil Red O Staining
5.4. Fibrogenic Differentiation and Immunocytochemistry
5.5. RNA Isolation and Real-Time qPCR
5.6. RNA and Small RNA Sequencing
5.7. Bioinformatic Analyses
5.8. miRNA Target Prediction
5.9. Statistics and Plotting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stadion, M.; Schwerbel, K.; Graja, A.; Baumeier, C.; Rodiger, M.; Jonas, W.; Wolfrum, C.; Staiger, H.; Fritsche, A.; Haring, H.U. Increased Ifi202b/IFI16 expression stimulates adipogenesis in mice and humans. Diabetologia 2018, 61, 1167–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, R.; Jeffery, E.; Rodeheffer, M.S. Weighing in on adipocyte precursors. Cell Metab. 2014, 19, 8–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Li, D.; Cao, H.; Li, H.; Wang, Y. Expansion and inflammation of white adipose tissue—Focusing on adipocyte progenitors. Biol. Chem. 2021, 402, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Heilbronn, L.; Smith, S.R.; Ravussin, E. Failure of fat cell proliferation, mitochondrial function and fat oxidation results in ectopic fat storage, insulin resistance and type II diabetes mellitus. Int. J. Obes. Relat. Metab. Disord. 2004, 28 (Suppl. S4), S12–S21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrosi, T.H.; Scialdone, A.; Graja, A.; Gohlke, S.; Jank, A.-M.; Bocian, C.; Woelk, L.; Fan, H.; Logan, D.W.; Schurmann, A.; et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 2017, 20, 771–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trouwborst, I.; Bowser, S.M.; Goossens, G.; Blaak, E.E. Ectopic Fat Accumulation in Distinct Insulin Resistant Phenotypes; Targets for Personalized Nutritional Interventions. Front. Nutr. 2018, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef]
- Wang, M.-Y.; Grayburn, P.; Chen, S.; Ravazzola, M.; Orci, L.; Unger, R.H. Adipogenic capacity and the susceptibility to type 2 diabetes and metabolic syndrome. Proc. Natl. Acad. Sci. USA 2008, 105, 6139–6144. [Google Scholar] [CrossRef] [Green Version]
- Bedogni, G.; Gastaldelli, A.; Manco, M.; De Col, A.; Agosti, F.; Tiribelli, C.; Sartorio, A. Relationship between fatty liver and glucose metabolism: A cross-sectional study in 571 obese children. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 120–126. [Google Scholar] [CrossRef] [Green Version]
- Utzschneider, K.M.; Kahn, S.E. Review: The role of insulin resistance in nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2006, 91, 4753–4761. [Google Scholar] [CrossRef] [Green Version]
- Wagner, R.; Jaghutriz, B.A.; Gerst, F.; Oquendo, M.B.; Machann, J.; Schick, F.; Löffler, M.W.; Nadalin, S.; Fend, F.; Königsrainer, A.; et al. Pancreatic Steatosis Associates with Impaired Insulin Secretion in Genetically Predisposed Individuals. J. Clin. Endocrinol. Metab. 2020, 105, 3518–3525. [Google Scholar] [CrossRef] [PubMed]
- Quiclet, C.; Dittberner, N.; Gässler, A.; Stadion, M.; Gerst, F.; Helms, A.; Baumeier, C.; Schulz, T.J.; Schürmann, A. Pancreatic adipocytes mediate hypersecretion of insulin in diabetes-susceptible mice. Metabolism 2019, 97, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Karlsson, M.J.; Hober, A.; Svensson, A.S.; Scheffel, J.; Kotol, D.; Zhong, W.; Tebani, A.; Strandberg, L.; Edfors, F.; et al. The human secretome. Sci. Signal. 2019, 12, eaaz0274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, L.; Li, R.; Wu, A.; Lu, J.; Wu, Q.; Jia, J.; Zhao, M.; Song, H. Hepatic loss of Lissencephaly 1 (Lis1) induces fatty liver and accelerates liver tumorigenesis in mice. J. Biol. Chem. 2018, 293, 5160–5171. [Google Scholar] [CrossRef] [Green Version]
- Kullander, K.; Klein, R. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell. Biol. 2002, 3, 475–486. [Google Scholar] [CrossRef]
- Huna, A.; Griveau, A.; Vindrieux, D.; Jaber, S.; Flaman, J.-M.; Goehrig, D.; Azzi, L.; Médard, J.-J.; Djebali, S.; Hernandez-Vargas, H.; et al. PLA2R1 promotes DNA damage and inhibits spontaneous tumor formation during aging. Cell Death Dis. 2021, 12, 190. [Google Scholar] [CrossRef]
- Parizadeh, S.M.; Jafarzadeh-Esfehani, R.; Fazilatpanah, D.; Hassanian, S.M.; Shahidsales, S.; Khazaei, M.; Parizadeh, S.M.R.; Ghayour-Mobarhan, M.; Ferns, G.A.; Avan, A. The potential therapeutic and prognostic impacts of the c-MET/HGF signaling pathway in colorectal cancer. IUBMB Life 2019, 71, 802–811. [Google Scholar] [CrossRef]
- Wang, W.; Xiao, Z.-D.; Li, X.; Aziz, K.E.; Gan, B.; Johnson, R.L.; Chen, J. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 2015, 17, 490–499. [Google Scholar] [CrossRef] [Green Version]
- Lopez, P.H.; Aja, S.; Aoki, K.; Seldin, M.M.; Lei, X.; Ronnett, G.V.; Wong, G.W.; Schnaar, R.L. Mice lacking sialyltransferase ST3Gal-II develop late-onset obesity and insulin resistance. Glycobiology 2017, 27, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Ekins, T.G.; Mahadevan, V.; Zhang, Y.; D’Amour, J.A.; Akgul, G.; Petros, T.J.; McBain, C.J. Emergence of non-canonical parvalbumin-containing interneurons in hippocampus of a murine model of type I lissencephaly. eLife 2020, 9, e62373. [Google Scholar] [CrossRef]
- Tchoukalova, Y.D.; Votruba, S.B.; Tchkonia, T.; Giorgadze, N.; Kirkland, J.L.; Jensen, M.D. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proc. Natl. Acad. Sci. USA 2010, 107, 18226–18231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamam, D.; Ali, D.; Kassem, M.; Aldahmash, A.; Alajez, N.M. microRNAs as regulators of adipogenic differentiation of mesenchymal stem cells. Stem. Cells Dev. 2015, 24, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badimon, L.; Cubedo, J. Adipose tissue depots and inflammation: Effects on plasticity and resident mesenchymal stem cell function. Cardiovasc. Res. 2017, 113, 1064–1073. [Google Scholar] [CrossRef] [Green Version]
- Roxburgh, J.; Metcalfe, A.D.; Martin, Y.H. The effect of medium selection on adipose-derived stem cell expansion and differentiation: Implications for application in regenerative medicine. Cytotechnology 2016, 68, 957–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, M.; Hori, M.; Ishigamori, R.; Mutoh, M.; Imai, T.; Nakagama, H. Fatty pancreas: A possible risk factor for pancreatic cancer in animals and humans. Cancer Sci. 2018, 109, 3013–3023. [Google Scholar] [CrossRef]
- Ma, X.; Yang, P.; Kaplan, W.H.; Lee, B.H.; Wu, L.E.; Yang, J.Y.; Yasunaga, M.; Sato, K.; Chisholm, D.J.; James, D.E. ISL1 regulates peroxisome proliferator-activated receptor gamma activation and early adipogenesis via bone morphogenetic protein 4-dependent and -independent mechanisms. Mol. Cell Biol. 2014, 34, 3607–3617. [Google Scholar] [CrossRef] [Green Version]
- Garcia de Herreros, A.; Dunach, M. Intracellular Signals Activated by Canonical Wnt Ligands Independent of GSK3 Inhibition and beta-Catenin Stabilization. Cells 2019, 8, 1148. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Meng, K.; Pu, Y.; Wang, C.; Chen, Y.; Wang, L. Hyperglycemia Altered the Fate of Cardiac Stem Cells to Adipogenesis through Inhibiting the beta-Catenin/TCF-4 Pathway. Cell Physiol. Biochem. 2018, 49, 2254–2263. [Google Scholar] [CrossRef]
- Newell, F.S.; Su, H.; Tornqvist, H.; Whitehead, J.P.; Prins, J.B.; Hutley, L.J. Characterization of the transcriptional and functional effects of fibroblast growth factor-1 on human preadipocyte differentiation. FASEB J. 2006, 20, 2615–2617. [Google Scholar] [CrossRef] [Green Version]
- Hutley, L.J.; Newell, F.S.; Kim, Y.-H.; Luo, X.; Widberg, C.H.; Shurety, W.; Prins, J.B.; Whitehead, J.P. A putative role for endogenous FGF-2 in FGF-1 mediated differentiation of human preadipocytes. Mol. Cell Endocrinol. 2011, 339, 165–171. [Google Scholar] [CrossRef]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottmann, P.; Ouni, M.; Saussenthaler, S.; Roos, J.; Stirm, L.; Jähnert, M.; Kamitz, A.; Hallahan, N.; Jonas, W.; Fritsche, A.; et al. A computational biology approach of a genome-wide screen connected miRNAs to obesity and type 2 diabetes. Mol. Metab. 2018, 11, 145–159. [Google Scholar] [CrossRef]
- Sun, F.; Wang, J.; Pan, Q.; Yu, Y.; Zhang, Y.; Wan, Y.; Wang, J.; Li, X.; Hong, A. Characterization of function and regulation of miR-24-1 and miR-31. Biochem. Biophys. Res. Commun. 2009, 380, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Li, Z.; Fritch, M.R.; Li, L.; Velankar, S.; Liu, Y.; Sohn, J.; Baker, N.; Lin, H.; Tuan, R.S. Caveolin-1 mediates soft scaffold-enhanced adipogenesis of human mesenchymal stem cells. Stem. Cell Res. Ther. 2021, 12, 347. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ng, D.Y.E.; Richards, A.M.; Wang, P. microRNA-221 Inhibits Latent TGF-beta1 Activation through Targeting Thrombospondin-1 to Attenuate Kidney Failure-Induced Cardiac Fibrosis. Mol. Ther. Nucleic. Acids 2020, 22, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Guo, Z.; Xie, W.; Jin, W.; Zhu, D.; Chen, S.; Ren, T. The lncRNA H19 Mediates Pulmonary Fibrosis by Regulating the miR-196a/COL1A1 Axis. Inflammation 2018, 41, 896–903. [Google Scholar] [CrossRef]
- Liu, X.; Guo, B.; Zhang, W.; Ma, B.; Li, Y. MiR-20a-5p overexpression prevented diabetic cardiomyopathy via inhibition of cardiomyocyte apoptosis, hypertrophy, fibrosis, and JNK/NF-kappaB signaling pathway. J. Biochem. 2021, 170, 349–362. [Google Scholar] [CrossRef]
- Li, P.F.; He, R.H.; Shi, S.B.; Li, R.; Wang, Q.T.; Rao, G.T.; Yang, B. Modulation of miR-10a-mediated TGF-beta1/Smads signaling affects atrial fibrillation-induced cardiac fibrosis and cardiac fibroblast proliferation. Biosci. Rep. 2019, 39, BSR20181931. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Xiao, Z.; Guo, H.; Fang, X.; Liang, J.; Zhu, J.; Yang, J.; Li, H.; Pan, R.; Yuan, S.; et al. Novel role of the clustered miR-23b-3p and miR-27b-3p in enhanced expression of fibrosis-associated genes by targeting TGFBR3 in atrial fibroblasts. J. Cell Mol. Med. 2019, 23, 3246–3256. [Google Scholar] [CrossRef]
- Lin, X.; Dong, R.; Diao, S.; Yu, G.; Wang, L.; Li, J.; Fan, Z. SFRP2 enhanced the adipogenic and neuronal differentiation potentials of stem cells from apical papilla. Cell Biol. Int. 2017, 41, 534–543. [Google Scholar] [CrossRef]
- Chavey, C.; Mari, B.; Monthouel, M.-N.; Bonnafous, S.; Anglard, P.; Van Obberghen, E.; Tartare-Deckert, S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J. Biol. Chem. 2003, 278, 11888–11896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrance, I.C.; Rogler, G.; Bamias, G.; Breynaert, C.; Florholmen, J.; Pellino, G.; Reif, S.; Speca, S.; Latella, G. Cellular and Molecular Mediators of Intestinal Fibrosis. J. Crohn’s Colitis 2017, 11, 1491–1503. [Google Scholar] [CrossRef] [Green Version]
- Baek, J.H.; Kim, S.J.; Kang, H.G.; Lee, H.W.; Kim, J.H.; Hwang, K.A.; Song, J.; Chun, K.H. Galectin-3 activates PPARgamma and supports white adipose tissue formation and high-fat diet-induced obesity. Endocrinology 2015, 156, 147–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Attar, A.; Antaramian, A.; Noureddin, M. Review of galectin-3 inhibitors in the treatment of nonalcoholic steatohepatitis. Expert Rev. Clin. Pharmacol. 2021, 14, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wu, X.; Yan, X.; Xu, A.; Yang, A.; You, H. Multitranscriptome analyses reveal prioritized genes specifically associated with liver fibrosis progression independent of etiology. Am. J. Physiol. Gastrointest. Liver. Physiol. 2019, 316, G744–G754. [Google Scholar] [CrossRef]
- Jara, P.; Calyeca, J.; Romero, Y.; Plácido, L.; Yu, G.; Kaminski, N.; Maldonado, V.; Cisneros, J.; Selman, M.; Pardo, A. Matrix metalloproteinase (MMP)-19-deficient fibroblasts display a profibrotic phenotype. Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 308, L511–L522. [Google Scholar] [CrossRef] [Green Version]
- Mehta, N.; Zhang, D.; Li, R.; Wang, T.; Gava, A.; Parthasarathy, P.; Gao, B.; Krepinsky, J.C. Caveolin-1 regulation of Sp1 controls production of the antifibrotic protein follistatin in kidney mesangial cells. Cell Commun. Signal. 2019, 17, 37. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Li, J.; He, F.; Li, K.; Li, X.; Zhang, Y. Matrix Gla protein regulates adipogenesis and is serum marker of visceral adiposity. Adipocyte 2020, 9, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Maridas, D.E.; DeMambro, V.E.; Le, P.T.; Mohan, S.; Rosen, C.J. IGFBP4 Is Required for Adipogenesis and Influences the Distribution of Adipose Depots. Endocrinology 2017, 158, 3488–3500. [Google Scholar] [CrossRef]
- Hu, Z.; Wu, J.; Qin, L.; Jin, H.; Cao, Y.; Zhao, Y. IGFBP7 downregulation or overexpression effect on bovine preadipocyte differentiation. Anim. Biotechnol. 2021, 32, 21–30. [Google Scholar] [CrossRef]
- Wang, S.; Shuai, C.; Gao, S.; Jiang, J.; Luan, J.; Lv, X. Chemokine CXCL14 acts as a potential genetic target for liver fibrosis. Int. Immunopharmacol. 2020, 89, 107067. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wei, H.; Li, Z.M.; Dong, X.B.; Wang, P.Y. TGF-beta1 aggravates degenerative nucleus pulposus cells inflammation and fibrosis through the upregulation of angiopoietin-like protein 2 expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12025–12033. [Google Scholar] [PubMed]
- Gong, L.; Wang, S.; Shen, L.; Liu, C.; Shenouda, M.; Li, B.; Liu, X.; Shaw, J.A.; Wineman, A.L.; Yang, Y.; et al. SLIT3 deficiency attenuates pressure overload-induced cardiac fibrosis and remodeling. JCI Insight 2020, 5, e136852. [Google Scholar] [CrossRef] [PubMed]
- Gerarduzzi, C.; Kumar, R.K.; Trivedi, P.; Ajay, A.K.; Iyer, A.; Boswell, S.; Hutchinson, J.N.; Waikar, S.S.; Vaidya, V.S. Silencing SMOC2 ameliorates kidney fibrosis by inhibiting fibroblast to myofibroblast transformation. JCI Insight 2017, 2, e90299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Wang, C.C.; Song, X.P.; Wang, H.M.; Zhou, H.; Sun, Y.; Wang, X.K.; Hou, S.; Pei, F.Y. Suppression of SMOC2 reduces bleomycin (BLM)-induced pulmonary fibrosis by inhibition of TGF-beta1/SMADs pathway. Biomed. Pharm. 2018, 105, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.; Feng, L.; Hao, Q. Secreted modular calcium-binding protein 2 promotes high fat diet (HFD)-induced hepatic steatosis through enhancing lipid deposition, fibrosis and inflammation via targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2019, 509, 48–55. [Google Scholar]
- Wagner, R.; Eckstein, S.S.; Yamazaki, H.; Gerst, F.; Machann, J.; Jaghutriz, B.A.; Schürmann, A.; Solimena, M.; Singer, S.; Königsrainer, A.; et al. Metabolic implications of pancreatic fat accumulation. Nat. Rev. Endocrinol. 2022, 18, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.; Huang, T.L.; Tran, T.T.; Zhang, H.; Townsend, K.L.; Shadrach, J.L.; Cerletti, M.; McDougall, L.E.; Giorgadze, N.; Tchkonia, T.; et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. USA 2011, 108, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kordes, C.; Sawitza, I.; Götze, S.; Häussinger, D. Stellate cells from rat pancreas are stem cells and can contribute to liver regeneration. PLoS ONE 2012, 7, e51878. [Google Scholar] [CrossRef] [PubMed]
- Kordes, C.; Sawitza, I.; Gotze, S.; Haussinger, D. Hepatic stellate cells support hematopoiesis and are liver-resident mesenchymal stem cells. Cell Physiol. Biochem. 2013, 31, 290–304. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aga, H.; Soultoukis, G.; Stadion, M.; Garcia-Carrizo, F.; Jähnert, M.; Gottmann, P.; Vogel, H.; Schulz, T.J.; Schürmann, A. Distinct Adipogenic and Fibrogenic Differentiation Capacities of Mesenchymal Stromal Cells from Pancreas and White Adipose Tissue. Int. J. Mol. Sci. 2022, 23, 2108. https://doi.org/10.3390/ijms23042108
Aga H, Soultoukis G, Stadion M, Garcia-Carrizo F, Jähnert M, Gottmann P, Vogel H, Schulz TJ, Schürmann A. Distinct Adipogenic and Fibrogenic Differentiation Capacities of Mesenchymal Stromal Cells from Pancreas and White Adipose Tissue. International Journal of Molecular Sciences. 2022; 23(4):2108. https://doi.org/10.3390/ijms23042108
Chicago/Turabian StyleAga, Heja, George Soultoukis, Mandy Stadion, Francisco Garcia-Carrizo, Markus Jähnert, Pascal Gottmann, Heike Vogel, Tim J. Schulz, and Annette Schürmann. 2022. "Distinct Adipogenic and Fibrogenic Differentiation Capacities of Mesenchymal Stromal Cells from Pancreas and White Adipose Tissue" International Journal of Molecular Sciences 23, no. 4: 2108. https://doi.org/10.3390/ijms23042108
APA StyleAga, H., Soultoukis, G., Stadion, M., Garcia-Carrizo, F., Jähnert, M., Gottmann, P., Vogel, H., Schulz, T. J., & Schürmann, A. (2022). Distinct Adipogenic and Fibrogenic Differentiation Capacities of Mesenchymal Stromal Cells from Pancreas and White Adipose Tissue. International Journal of Molecular Sciences, 23(4), 2108. https://doi.org/10.3390/ijms23042108





