Heterogenicity within the LPS Structure in Relation to the Chosen Genomic and Physiological Features of the Plant Pathogen Pectobacterium parmentieri
Abstract
1. Introduction
2. Results and Discussion
2.1. The Chemical Structure of OPS of P. parmentieri SCC3193 and IFB5432
| →3)-β-d-Galf-(1→3)-α-d-Galp-(1→8)-β-Pse4Ac5Ac7Ac-(2→6)-α-d-Glcp-(1→6)-β-d-Glcp-(1→ | ||||
| A | B | E | C | D |
2.2. Genomic Structure of P. parmentieri Strains Containing Diverse OPS
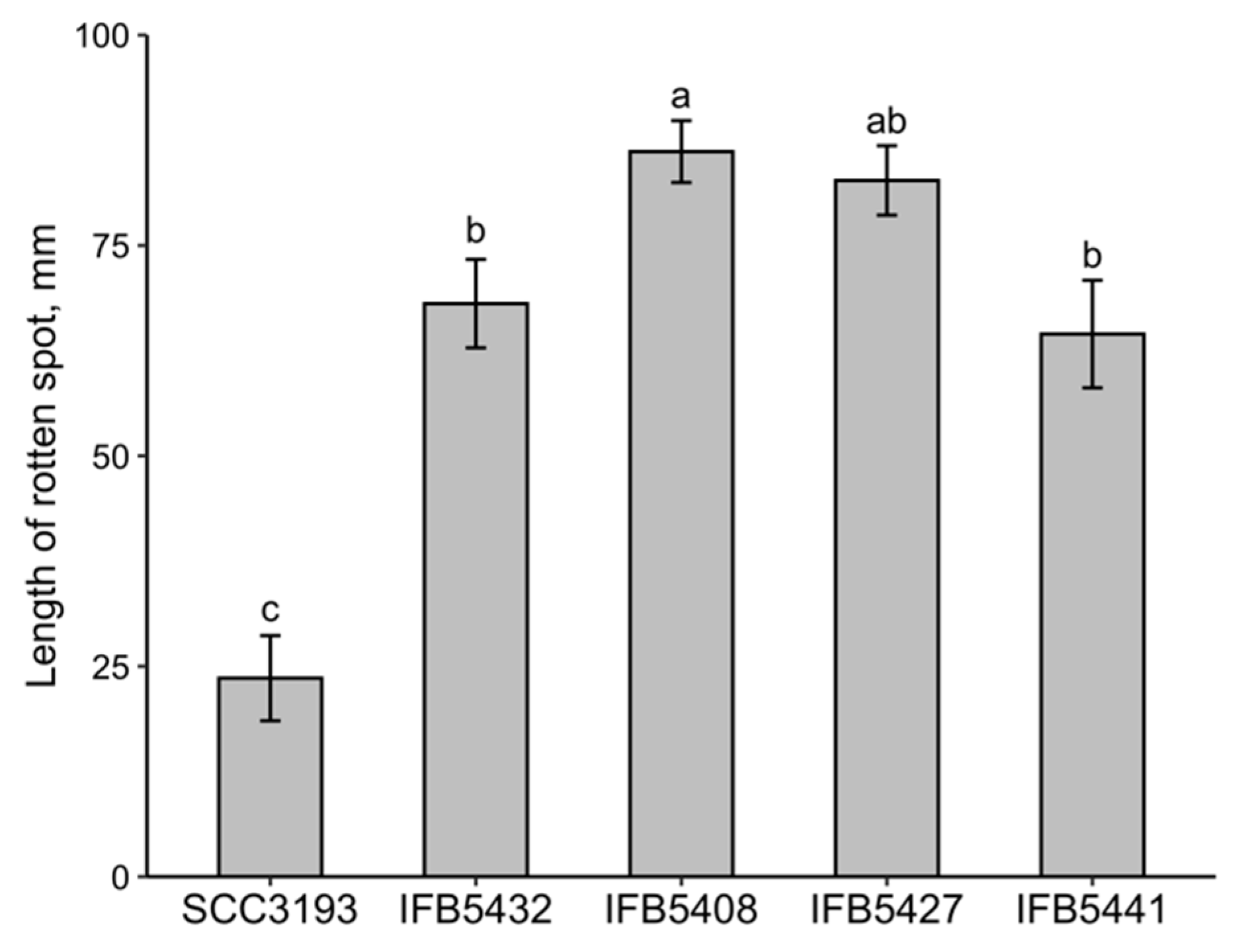
2.3. Analysis of the Phenotypic Features and Ability to Cause Disease Symptoms of P. parmentieri Strains Exhibiting Different OPS Structures
3. Materials and Methods
3.1. Origin and Biomass Production of P. parmentieri SCC3193 and IFB5432 for the OPS Analysis
3.2. Determination of the OPS Structure of P. parmentieri SCC3193 and IFB5432 Strains
3.2.1. LPS Extraction and Purification
3.2.2. Isolation of OPS
3.2.3. Chemical Analysis
3.2.4. GLC and GLC-MS Analyses
3.2.5. NMR Spectroscopy
3.3. Characterization of P. parmentieri Strains of the Already Deciphered OPS Structure
3.3.1. Whole-Genome Comparisons
3.3.2. Phenotypic Assays
3.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [PubMed]
- Nykyri, J.; Niemi, O.; Koskinen, P.; Nokso-Koivisto, J.; Pasanen, M.; Broberg, M. Revised phylogeny and novel horizontally acquired virulence determinants of the model soft rot phytopathogen Pectobacterium wasabiae SCC3193. PLoS Pathog. 2012, 8, e1003013. [Google Scholar] [CrossRef]
- Waleron, M.; Waleron, K.; Lojkowska, E. Occurrence of Pectobacterium wasabiae in potato field samples. Eur. J. Plant Pathol. 2013, 137, 149–158. [Google Scholar] [CrossRef][Green Version]
- Khayi, S.; Cigna, J.; Chong, T.M.; Quêtu-Laurent, A.; Chan, K.-G.; Hélias, V.; Faure, D. Transfer of the potato plant isolates of Pectobacterium wasabiae to Pectobacterium parmentieri sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5379–5383. [Google Scholar] [CrossRef] [PubMed]
- Motyka, A.; Zoledowska, S.; Sledz, W.; Lojkowska, E. Molecular methods as tools to control plant diseases caused by Dickeya and Pectobacterium spp: A minireview. New Biotechnol. 2017, 39, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Erbs, G.; Newman, M.A. The role of lipopolysaccharides in induction of plant defence responses. Mol. Plant Pathol. 2003, 4, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Van Gijsegem, F.; Hugouvieux-Cotte-Pattat, N.; Kraepiel, Y.; Lojkowska, E.; Moleleki, L.N.; Gorshkov, V.; Yedidia, I. Molecular Interactions of Pectobacterium and Dickeya with Plants. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., van der Wolf, J.M., Toth, I.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 85–147. [Google Scholar]
- Evans, T.J.; Ind, A.; Komitopoulou, E.; Salmond, G.P.C. Phage-selected lipopolysaccharide mutants of Pectobacterium atrosepticum exhibit different impacts on virulence. J. Appl. Microbiol. 2010, 109, 505–514. [Google Scholar] [CrossRef]
- Zoledowska, S.; Motyka, A.; Zukowska, D.; Sledz, W.; Lojkowska, E. Population structure and biodiversity of Pectobacterium parmentieri isolated from potato fields in temperate climate. Plant Dis. 2018, 102, 154–164. [Google Scholar] [CrossRef]
- Zoledowska, S.; Motyka-Pomagruk, A.; Sledz, W.; Mengoni, A.; Lojkowska, E. High genomic variability in the plant pathogenic bacterium Pectobacterium parmentieri deciphered from de novo assembled complete genomes. BMC Genom. 2018, 19, 751. [Google Scholar] [CrossRef]
- Pitman, A.R.; Harrow, S.A.; Visnovsky, S.B. Genetic characterisation of Pectobacterium wasabiae causing soft rot disease of potato in New Zealand. Eur. J. Plant Pathol. 2010, 126, 423–435. [Google Scholar] [CrossRef]
- De Boer, S.H.; Li, X.; Ward, L.J. Pectobacterium spp. associated with bacterial stem rot syndrome of potato in Canada. Phytopathology 2012, 102, 937–947. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nabhan, S.; Wydra, K.; Linde, M.; Debener, T. The use of two complementary DNA assays, AFLP and MLSA, for epidemic and phylogenetic studies of pectolytic enterobacterial strains with focus on the heterogeneous species Pectobacterium carotovorum. Plant Pathol. 2012, 61, 498–508. [Google Scholar] [CrossRef]
- Moleleki, L.N.; Onkendi, E.M.; Mongae, A.; Kubheka, G.C. Characterisation of Pectobacterium wasabiae causing blackleg and soft rot diseases in South Africa. Eur. J. Plant Pathol. 2013, 135, 279–288. [Google Scholar] [CrossRef]
- Ozturk, M.; Aksoy, H.M.; Potrykus, M.; Lojkowska, E. Genotypic and phenotypic variability of Pectobacterium strains causing blackleg and soft rot on potato in Turkey. Eur. J. Plant Pathol. 2018, 152, 143–155. [Google Scholar] [CrossRef]
- Li-ping, W.; Min, Z.; Min-hua, R.; Xing-wei, L.; Qiong-guang, L. Pectobacterium carotovorum subsp. brasiliensis and Pectobacterium parmentieri as causal agents of potato blackleg and soft rot in China. J. Plant Pathol. 2020, 102, 871–879. [Google Scholar]
- Pirhonen, M.; Heino, P.; Helander, I.; Harju, P.; Palva, E.T. Bacteriophage T4 resistant mutants of the plant pathogen Erwinia carotovora. Microb. Pathog. 1988, 4, 359–367. [Google Scholar] [CrossRef]
- Ossowska, K.; Czerwicka, M.T.; Sledz, W.; Żołędowska, S.; Motyka-Pomagruk, A.; Szulta, S.; Lojkowska, E.; Kaczyński, Z. The structure of O-polysaccharides isolated from plant pathogenic bacteria Pectobacterium wasabiae IFB5408 and IFB5427. Carbohydr. Res. 2016, 426, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.A.; Shashkov, A.S.; Tsvetkov, Y.E.; Jansson, P.-E.; Zahringer, U. 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: Chemistry and biochemistry. Adv. Carbohydr. Chem. Biochem. 2013, 58, 371–414. [Google Scholar]
- L’vov, V.L.; Shashkov, A.S.; Dmitriev, B.A. Antigenic determinant of bacteria. The structure of the repeating unit of a specific polysaccharide from Shigella boydii type 7. Bioorg. Khimiia 1987, 13, 223–233. [Google Scholar]
- Czerwicka, M.T.; Marszewska, K.; Bychowska, A.; Dziadziuszko, H.; Brzozowski, K.; Lojkowska, E.; Stepnowski, P.; Kaczyński, Z. Chemical structure of the O-polysaccharide isolated from Pectobacterium atrosepticum SCRI 1039. Carbohydr. Res. 2011, 346, 2978–2981. [Google Scholar] [CrossRef]
- Gorshkov, V.; Islamov, B.; Mikshina, P.; Petrova, O.; Burygin, G.L.; Sigida, E.; Shashkov, A.; Daminova, A.; Ageeva, M.; Idiyatullin, B.; et al. Pectobacterium atrosepticum exopolysaccharides: Identification, molecular structure, formation under stress and in planta conditions. Glycobiology 2017, 27, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Shneider, M.M.; Lukianova, A.A.; Evseev, P.V.; Shpirt, A.M.; Kabilov, M.R.; Tokmakova, A.D.; Miroshnikov, K.K.; Obraztsova, E.A.; Baturina, O.A.; Shashkov, A.S.; et al. Autographivirinae Bacteriophage Arno 160 Infects Pectobacterium carotovorum via Depolymerization of the Bacterial O-Polysaccharide. Int. J. Mol. Sci. 2020, 21, 3170. [Google Scholar] [CrossRef]
- Lukianova, A.A.; Shneider, M.M.; Evseev, P.V.; Shpirt, A.M.; Bugaeva, E.N.; Kabanova, A.P.; Obraztsova, E.A.; Miroshnikov, K.K.; Senchenkova, S.N.; Shashkov, A.S.; et al. Morphologically Different Pectobacterium Brasiliense Bacteriophages PP99 and PP101: Deacetylation of O-Polysaccharide by the Tail Spike Protein of Phage PP99 Accompanies the Infection. Front. Microbiol. 2020, 10, 3147. [Google Scholar] [CrossRef] [PubMed]
- Ossowska, K.; Czerwicka, M.T.; Sledz, W.; Żołędowska, S.; Motyka-Pomagruk, A.; Golanowska, M.; Condemine, G.; Lojkowska, E.; Kaczyński, Z. The uniform structure of O-polysaccharides isolated from Dickeya solani strains of different origin. Carbohydr. Res. 2017, 445, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Kabanova, A.P.; Shneider, M.M.; Korzhenkov, A.A.; Bugaeva, E.N.; Miroshnikov, K.K.; Zdorovenko, E.L.; Kulikov, E.E.; Toschakov, S.V.; Ignatov, A.N.; Knirel, Y.A.; et al. Host Specificity of the Dickeya Bacteriophage PP35 Is Directed by a Tail Spike Interaction with Bacterial O-Antigen, Enabling the Infection of Alternative Non-pathogenic Bacterial Host. Front. Microbiol. 2019, 9, 3288. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.; Szpakowska, N.; Sledz, W.; Motyka-Pomagruk, A.; Ossowska, K.; Lojkowska, E.; Kaczynski, Z. The structure of the O-polysaccharide isolated from pectinolytic gram-negative bacterium Dickeya aquatica IFB0154 is different from the O-polysaccharides of other Dickeya species. Carbohydr. Res. 2020, 497, 108135. [Google Scholar] [CrossRef]
- Duprey, A.; Taib, N.; Leonard, S.; Garin, T.; Flandrois, J.-P.; Nasser, W.; Brochier-Armanet, C.; Reverchon, S. The phytopathogenic nature of Dickeya aquatica 174/2 and the dynamic early evolution of Dickeya pathogenicity. Environ. Microbiol. 2019, 21, 2809–2835. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of procedure. Methods Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N. The RhaS activator controls the Erwinia chrysanthemi 3937 genes rhiN, rhiT and rhiE involved in rhamnogalacturonan catabolism. Mol. Microbiol. 2004, 51, 1361–1374. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.R-project.org/ (accessed on 30 December 2021).
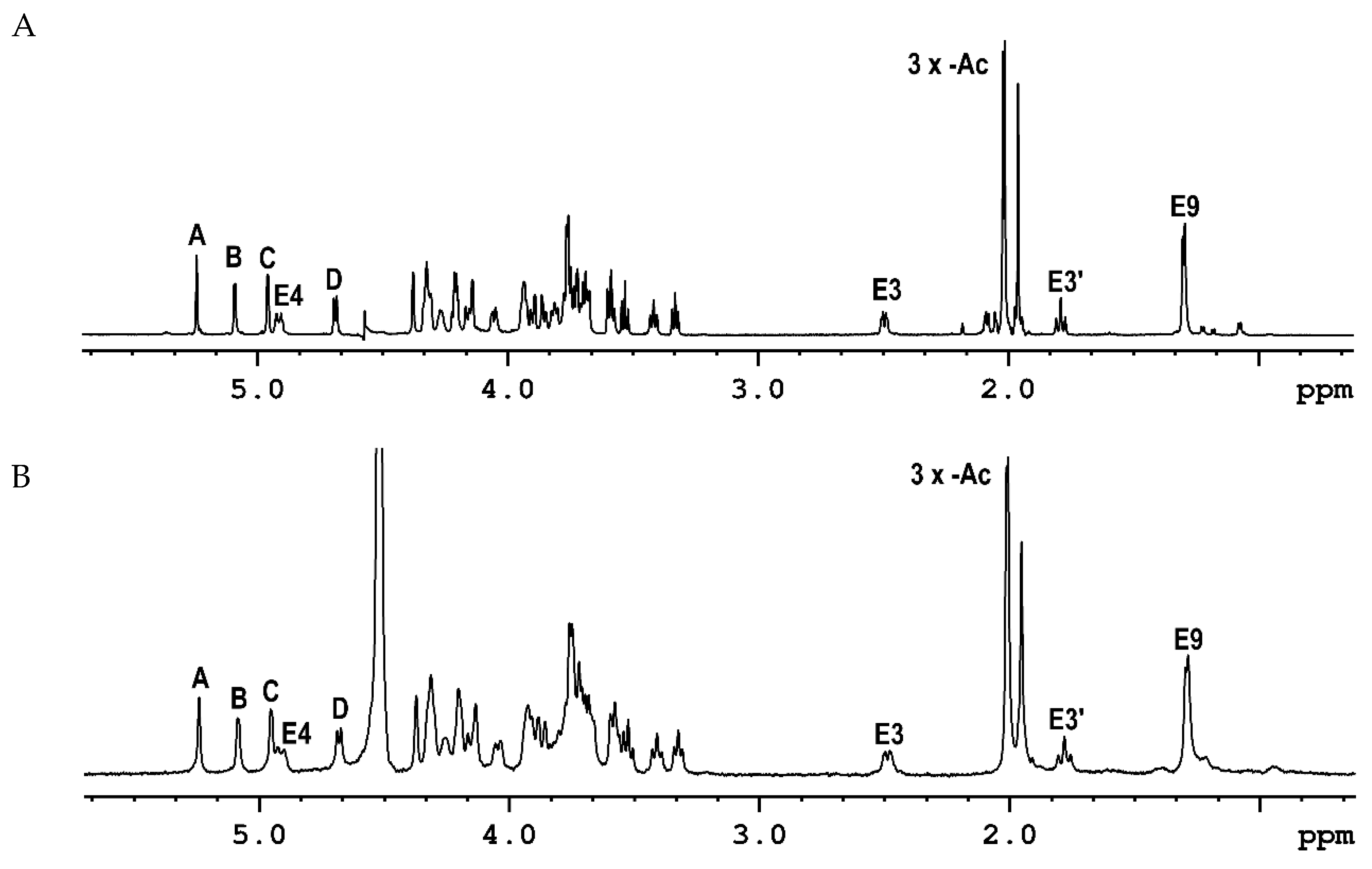
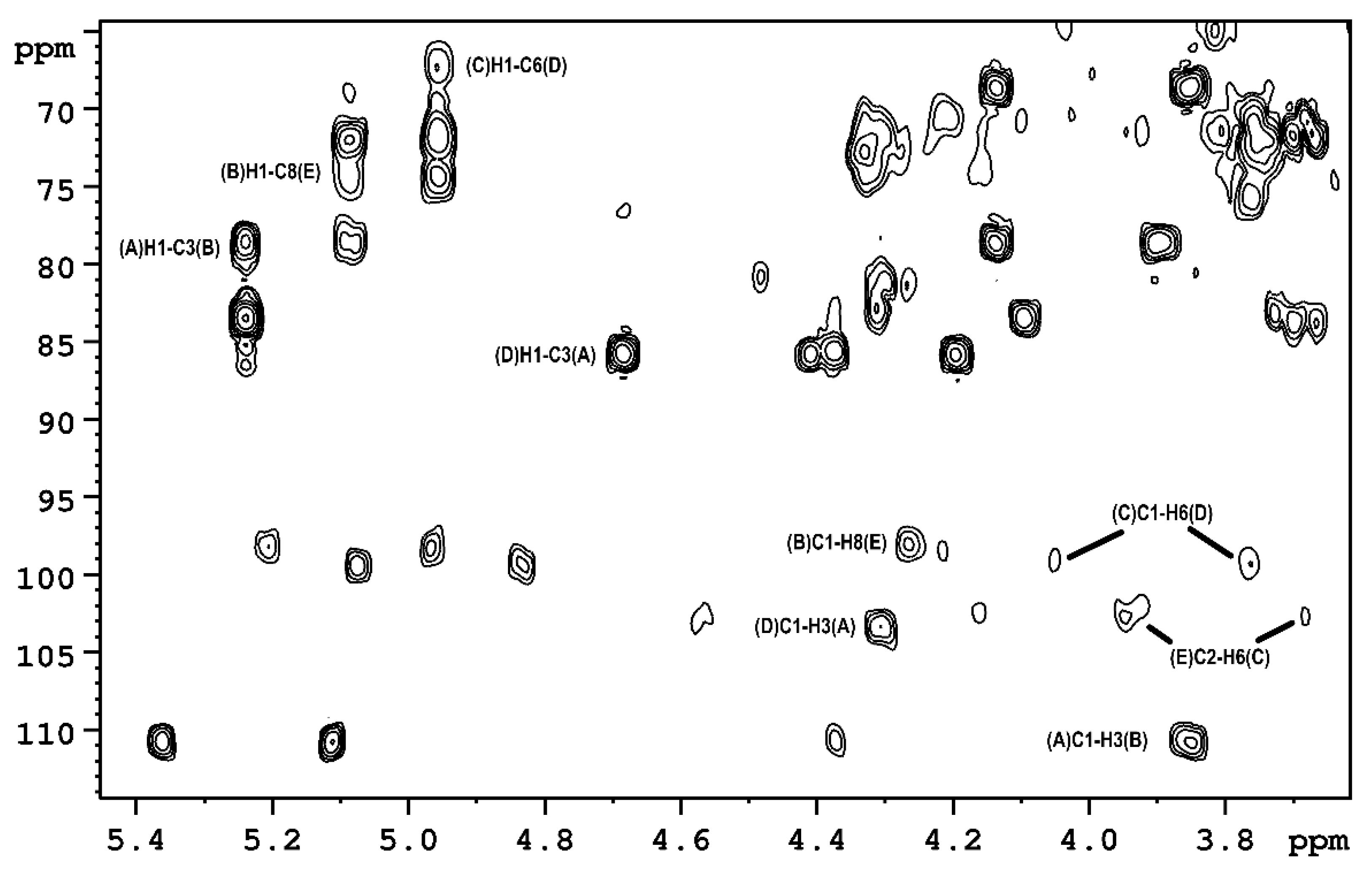

| Residue [1JC1-H1; 3JH1-H2 (Hz)] | Chemical Shifts of 1H and 13C [ppm] | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| H1 C1 | H2 C2 | H3 C3 | H4 C4 | H5 C5 | H6 C6 | H7 C7 | H8 C8 | H9 C9 | |
| →3)-β-D-Galf (A) [172; <1.5] | 5.241 110.71 | 4.373 81.25 | 4.303 85.77 | 4.199 83.39 | 3.932 71.87 | 3.722/3.686 64.14 | - | - | - |
| →3)-α-D-Galp (B) [171; 3.7] | 5.088 98.06 | 3.892 68.56 | 3.854 78.64 | 4.138 70.52 | 4.209 71.96 | 3.756/3.756 62.36 | - | - | - |
| →6)-α-D-Glcp (C) [172; 3.4] | 4.959 99.27 | 3.592 72.84 | 3.737 74.45 | 3.410 71.39 | 3.809 71.87 | 3.928/3.709 64.78 | - | - | - |
| →6)-β-D-Glcp (D) [161; 7.8] | 4.689 103.29 | 3.328 74.29 | 3.528 77.19 | 3.581 70.49 | 3.676 75.74 | 4.051/3.762 66.79 | - | - | - |
| →8)-β-Pse4Ac5Ac7Ac-(2→ (E) -O-Ac -N-Ac -N-Ac | - 174.38 - 174.50 - 175.57 - 174.13 | - 102.33 1.956 23.48 2.008 23.21 2.014 21.76 | 2.489/1.784 34.09 | 4.911 70.59 | 4.325 47.12 | 4.157 72.59 | 4.325 54.15 | 4.267 74.13 | 1.293 14.19 |
| SCC3193 | IFB5432 | IFB5408 | IFB5427 | IFB5441 | |
|---|---|---|---|---|---|
| SCC3193 | - | 98.81 [90.29] | 98.69 [90.50] | 98.70 [89.58] | 98.78 [90.14] |
| IFB5432 | 98.91 [93.22] | - | 98.77 [93.27] | 98.76 [93.14] | 99.89 [97.47] |
| IFB5408 | 98.80 [93.00] | 98.76 [92.98] | - | 99.94 [98.41] | 98.77 [92.94] |
| IFB5427 | 98.74 [90.64] | 98.73 [91.22] | 99.93 [96.57] | - | 98.69 [91.25] |
| IFB5441 | 98.79 [91.61] | 99.77 [96.05] | 98.68 [91.73] | 98.60 [91.70] | - |
| SCC3193 | IFB5432 | IFB5408 | IFB5427 | IFB5441 | |
|---|---|---|---|---|---|
| SCC3193 | - | 99.13 [90.98] | 99.06 [91.15] | 99.06 [90.22] | 99.11 [90.87] |
| IFB5432 | 99.13 [94.02] | - | 99.01 [93.99] | 99.03 [93.83] | 99.90 [97.78] |
| IFB5408 | 99.06 [93.75] | 99.01 [93.63] | - | 99.99 [98.57] | 99.02 [93.62] |
| IFB5427 | 99.06 [91.35] | 99.03 [91.84] | 99.99 [96.69] | - | 99.04 [91.63] |
| IFB5441 | 99.12 [92.38] | 99.91 [96.37] | 99.02 [92.53] | 99.04 [92.30] | - |
| SCC3193 | IFB5432 | IFB5408 | IFB5427 | IFB5441 | |
|---|---|---|---|---|---|
| SCC3193 | - | 0.99975 | 0.99973 | 0.99967 | 0.99975 |
| IFB5432 | 0.99975 | - | 0.99987 | 0.99987 | 0.99991 |
| IFB5408 | 0.99973 | 0.99987 | - | 0.9999 | 0.99984 |
| IFB5427 | 0.99967 | 0.99987 | 0.9999 | - | 0.99979 |
| IFB5441 | 0.99975 | 0.99991 | 0.99984 | 0.99979 | - |
| Biochemical Feature | SCC3193 | IFB5432 | IFB5408 | IFB5427 | IFB5441 |
|---|---|---|---|---|---|
| β-galactosidase activity | + | + | + | + | + |
| Arginine dihydrolase activity | - | - | - | - | - |
| Lysine decarboxylase activity | - | - | - | - | - |
| Ornithine decarboxylase activity | - | - | - | - | - |
| Citrate utilization | - | - | - | - | - |
| H2S production | - | - | - | - | - |
| Urease production | - | - | - | - | - |
| Tryptophane deaminase activity | - | - | - | - | - |
| Indole production | - | - | - | - | - |
| Acetoin production (VP) | - | - | - | - | - |
| Liquefaction of gelatin | - | - | - | - | - |
| Fermentation of glucose | + | + | + | + | + |
| Fermentation of mannitol | + | + | + | + | + |
| Fermentation of inositol | - | - | - | - | - |
| Fermentation of sorbitol | - | - | - | - | - |
| Fermentation of rhamnose | + | + | + | + | + |
| Fermentation of saccharose | + | + | + | + | + |
| Fermentation of melibiose | + | + | + | + | + |
| Fermentation of amygdalin | + | + | + | + | + |
| Fermentation of arabinose | + | + | + | + | + |
| NO3 reduction to NO2 | + | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ossowska, K.; Motyka-Pomagruk, A.; Kaczyńska, N.; Kowalczyk, A.; Sledz, W.; Lojkowska, E.; Kaczyński, Z. Heterogenicity within the LPS Structure in Relation to the Chosen Genomic and Physiological Features of the Plant Pathogen Pectobacterium parmentieri. Int. J. Mol. Sci. 2022, 23, 2077. https://doi.org/10.3390/ijms23042077
Ossowska K, Motyka-Pomagruk A, Kaczyńska N, Kowalczyk A, Sledz W, Lojkowska E, Kaczyński Z. Heterogenicity within the LPS Structure in Relation to the Chosen Genomic and Physiological Features of the Plant Pathogen Pectobacterium parmentieri. International Journal of Molecular Sciences. 2022; 23(4):2077. https://doi.org/10.3390/ijms23042077
Chicago/Turabian StyleOssowska, Karolina, Agata Motyka-Pomagruk, Natalia Kaczyńska, Agnieszka Kowalczyk, Wojciech Sledz, Ewa Lojkowska, and Zbigniew Kaczyński. 2022. "Heterogenicity within the LPS Structure in Relation to the Chosen Genomic and Physiological Features of the Plant Pathogen Pectobacterium parmentieri" International Journal of Molecular Sciences 23, no. 4: 2077. https://doi.org/10.3390/ijms23042077
APA StyleOssowska, K., Motyka-Pomagruk, A., Kaczyńska, N., Kowalczyk, A., Sledz, W., Lojkowska, E., & Kaczyński, Z. (2022). Heterogenicity within the LPS Structure in Relation to the Chosen Genomic and Physiological Features of the Plant Pathogen Pectobacterium parmentieri. International Journal of Molecular Sciences, 23(4), 2077. https://doi.org/10.3390/ijms23042077








