The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Results
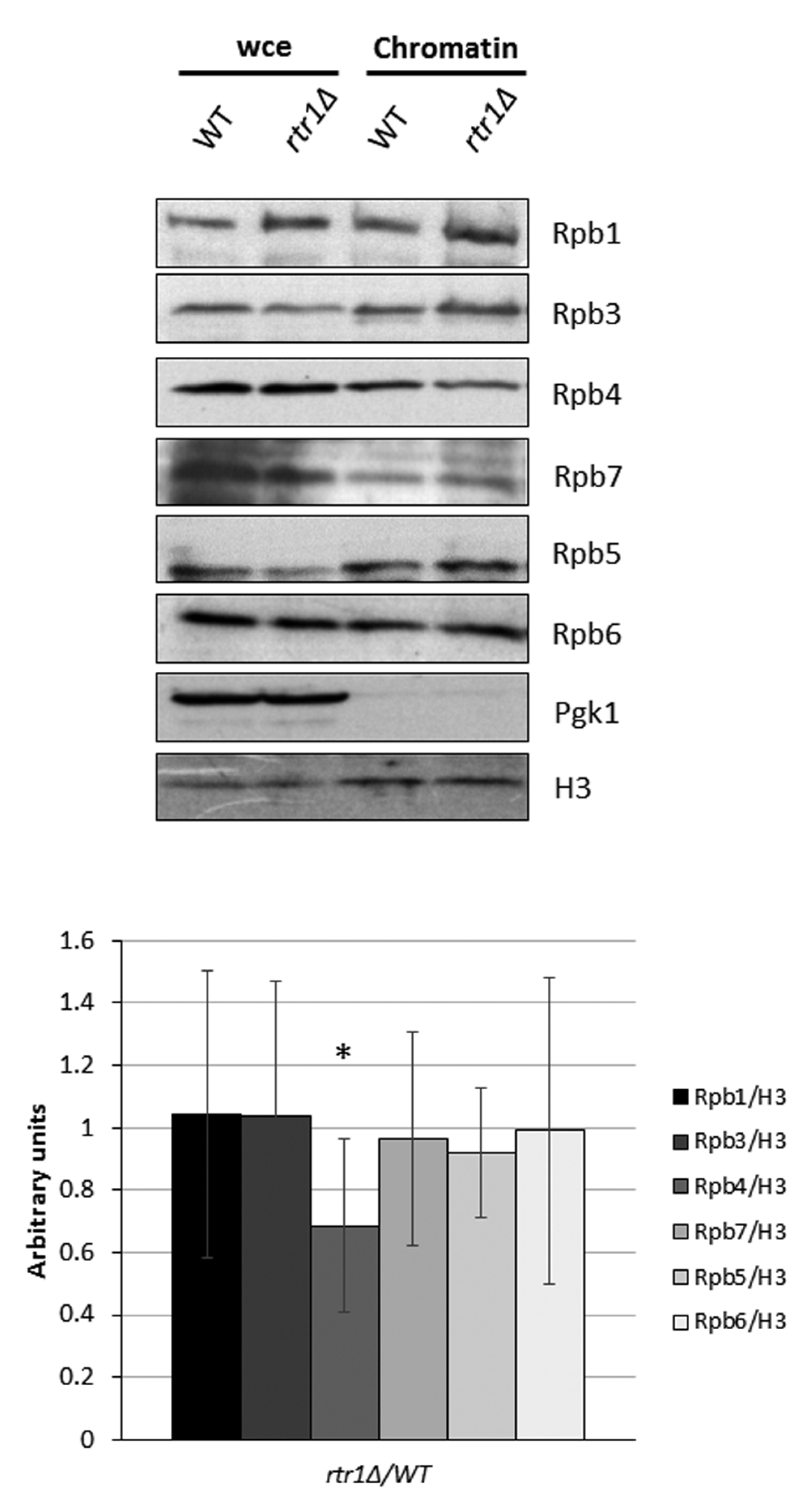
2.1. The Presence of Rpb4 in the Chromatin-Associated RNA Polymerase II Depends on Rtr1
2.2. Lack of Rtr1 Does Not Account for a Global Effect on Rpb4 Dissociation from RNA Pol II during Transcription Elongation
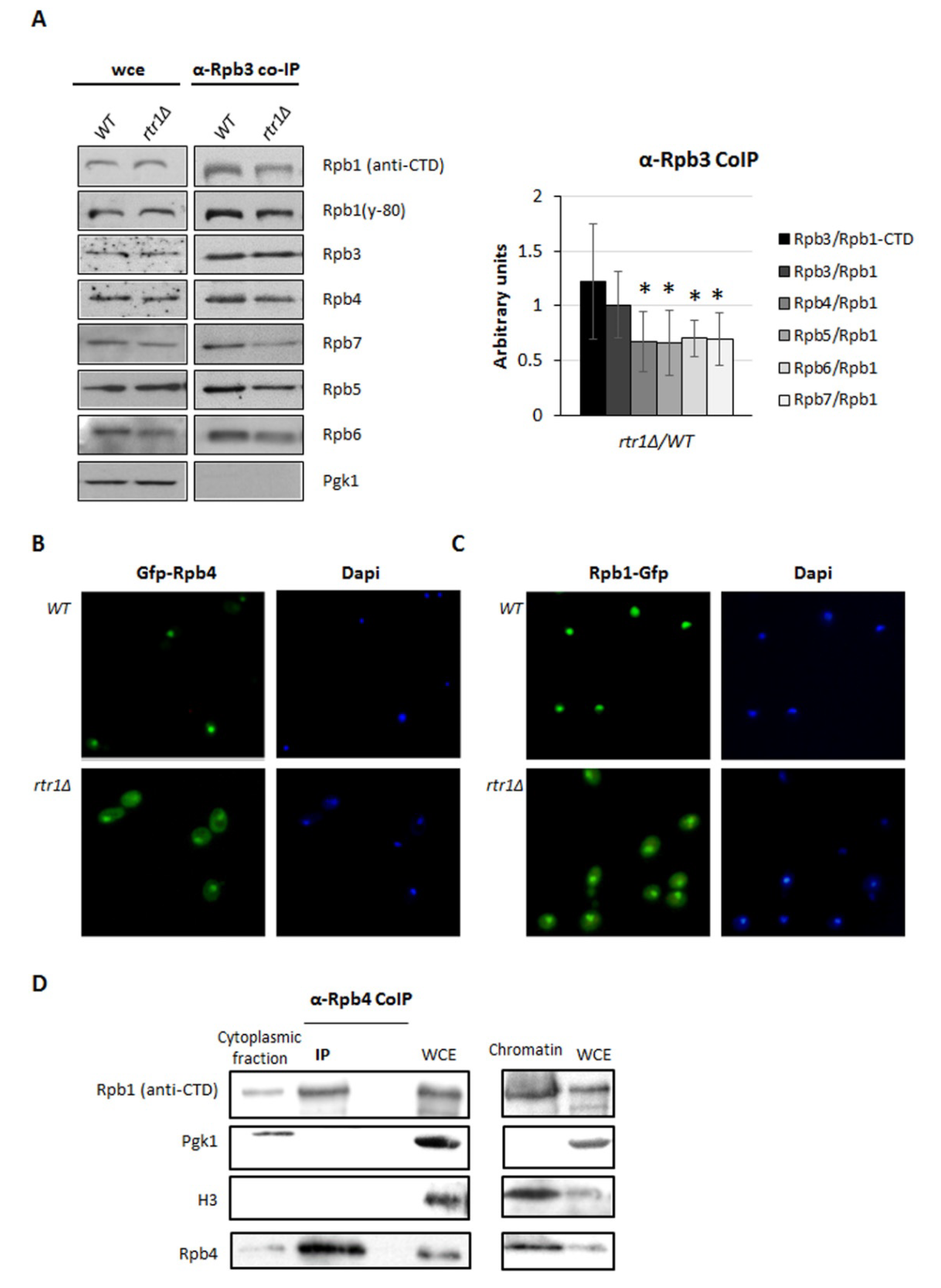
2.3. Rtr1 Mediates the Correct Assembly of the RNA Polymerase II
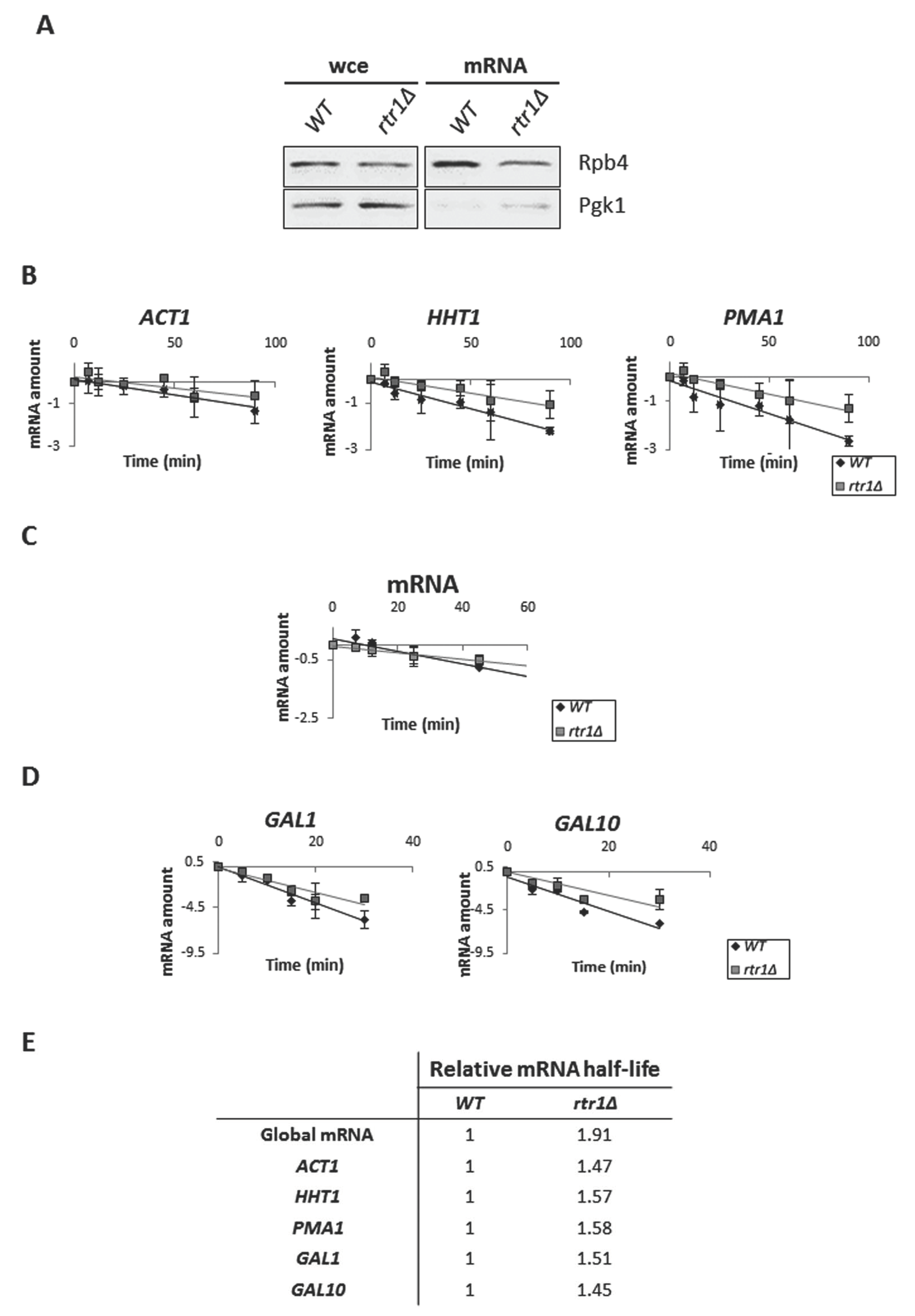
2.4. Defective Association of Rpb4 with the RNA Pol II in the rtr1Δ Mutant Affects mRNA Stability
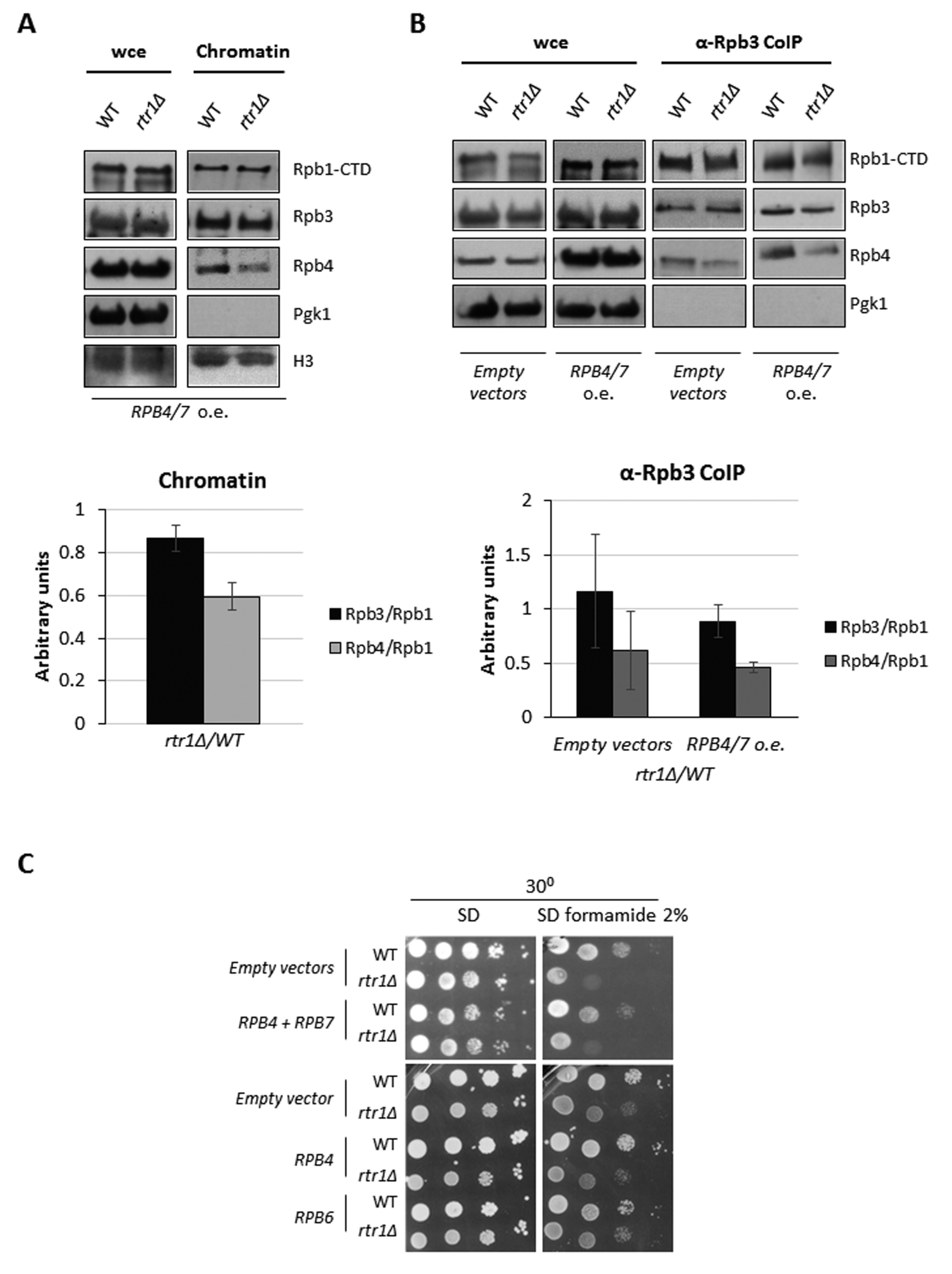
2.5. Rtr1 Cooperates with Rpb4 for mRNA Imprinting
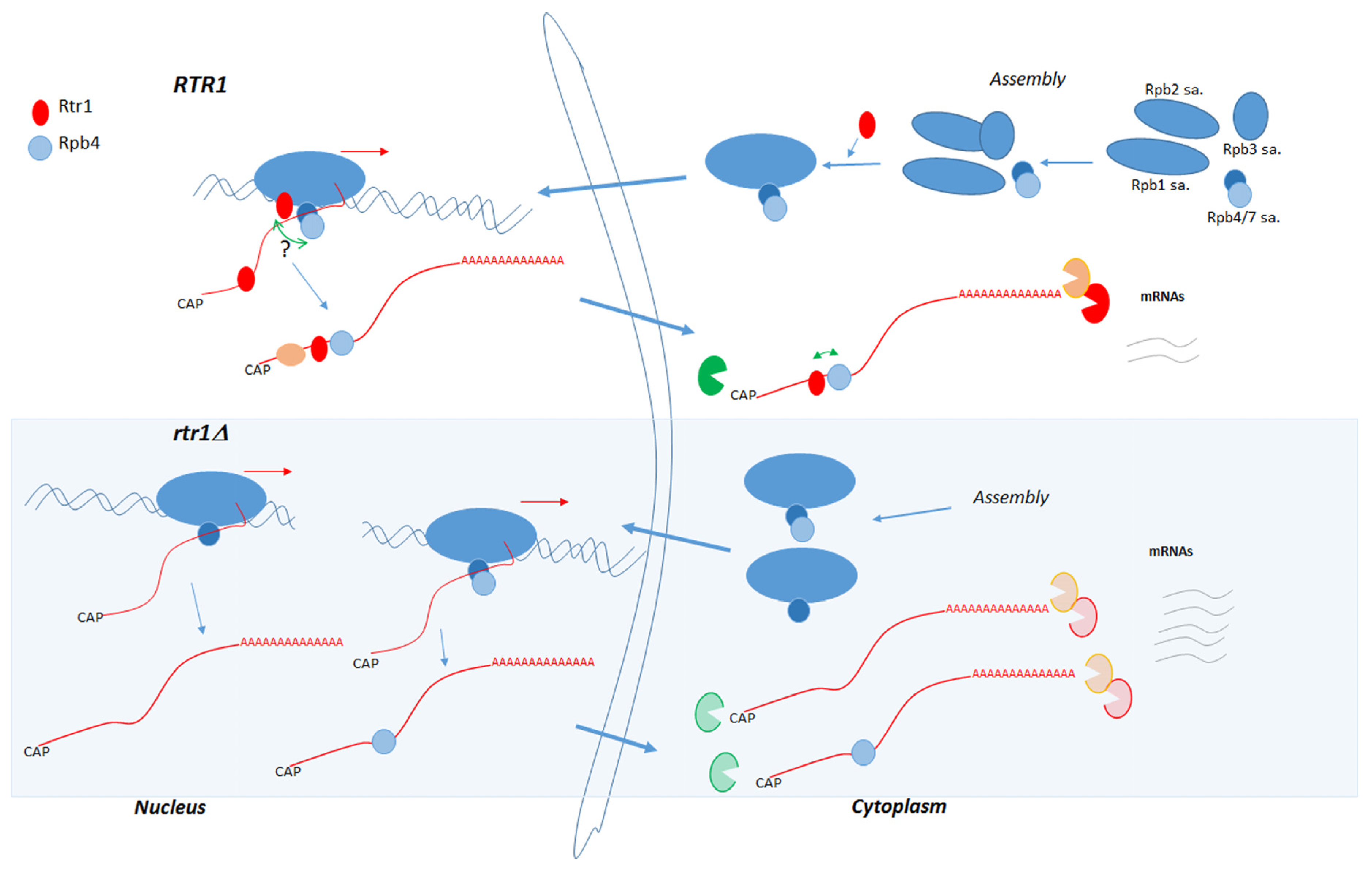
3. Discussion
4. Materials and Methods
4.1. Yeast Strains, Genetic Manipulations, Media, and Genetic Analysis
4.2. Protein Extract Preparation and Immunoprecipitation
4.3. SDS-PAGE, Western Blot Analysis and Immunoreactive Bands Quantification
4.4. Fluorescence Microscopy
4.5. Preparation of Yeast Cytoplasm and Chromatin-Enriched Fractions
4.6. Isolation of the mRNA-Associated Proteins
4.7. mRNA Stability Analysis
4.8. Quantitative Real-Time PCR (RT-qPCR)
4.9. Chromatin Immunoprecipitation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choder, M.; Young, R.A. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell Biol. 1993, 13, 6984–6991. [Google Scholar] [PubMed]
- Bushnell, D.A.; Kornberg, R.D. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: Implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA 2003, 100, 6969–6973. [Google Scholar] [CrossRef] [PubMed]
- Armache, K.J.; Mitterweger, S.; Meinhart, A.; Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005, 280, 7131–7134. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.M.; Kane, C.M.; Young, R.A.; Kornberg, R.D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 1991, 266, 71–75. [Google Scholar] [CrossRef]
- Sampath, V.; Balakrishnan, B.; Verma-Gaur, J.; Onesti, S.; Sadhale, P.P. Unstructured N terminus of the RNA polymerase II subunit Rpb4 contributes to the interaction of Rpb4.Rpb7 subcomplex with the core RNA polymerase II of Saccharomyces cerevisiae. J. Biol. Chem. 2008, 283, 3923–3931. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Pirkl, N.; Lehmann, E.; Cramer, P. Rpb4 subunit functions mainly in mRNA synthesis by RNA polymerase II. J. Biol. Chem. 2014, 289, 17446–17452. [Google Scholar] [CrossRef]
- Allepuz-Fuster, P.; O’Brien, M.J.; González-Polo, N.; Pereira, B.; Dhoondia, Z.; Ansari, A.; Calvo, O. RNA polymerase II plays an active role in the formation of gene loops through the Rpb4 subunit. Nucleic Acids Res. 2019, 47, 8975–8987. [Google Scholar] [CrossRef]
- Allepuz-Fuster, P.; Martinez-Fernandez, V.; Garrido-Godino, A.I.; Alonso-Aguado, S.; Hanes, S.D.; Navarro, F.; Calvo, O. Rpb4/7 facilitates RNA polymerase II CTD dephosphorylation. Nucleic Acids Res. 2014, 42, 13674–13688. [Google Scholar] [CrossRef]
- Runner, V.M.; Podolny, V.; Buratowski, S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3’ processing factors. Mol. Cell Biol. 2008, 28, 1883–1891. [Google Scholar] [CrossRef]
- Verma-Gaur, J.; Rao, S.N.; Taya, T.; Sadhale, P. Genomewide recruitment analysis of Rpb4, a subunit of polymerase II in Saccharomyces cerevisiae, reveals its involvement in transcription elongation. Eukaryot. Cell 2008, 7, 1009–1018. [Google Scholar] [CrossRef]
- Harel-Sharvit, L.; Eldad, N.; Haimovich, G.; Barkai, O.; Duek, L.; Choder, M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 2010, 143, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Farago, M.; Nahari, T.; Hammel, C.; Cole, C.N.; Choder, M. Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol. Biol. Cell 2003, 14, 2744–2755. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Godino, A.I.; García-López, M.C.; García-Martínez, J.; Pelechano, V.; Medina, D.A.; Pérez-Ortín, J.E.; Navarro, F. Rpb1 foot mutations demonstrate a major role of Rpb4 in mRNA stability during stress situations in yeast. Biochim. Biophys. Acta 2016, 1859, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Godino, A.I.; Gupta, I.; Gutiérrez-Santiago, F.; Martínez-Padilla, A.B.; Alekseenko, A.; Steinmetz, L.M.; Pérez-Ortín, J.E.; Pelechano, V.; Navarro, F. Rpb4 and Puf3 imprint and post-transcriptionally control the stability of a common set of mRNAs in yeast. RNA Biol. 2020, 18, 1206–1220. [Google Scholar] [CrossRef]
- Lotan, R.; Bar-On, V.G.; Harel-Sharvit, L.; Duek, L.; Melamed, D.; Choder, M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 2005, 19, 3004–3016. [Google Scholar] [CrossRef]
- Lotan, R.; Goler-Baron, V.; Duek, L.; Haimovich, G.; Choder, M. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J. Cell Biol. 2007, 178, 1133–1143. [Google Scholar] [CrossRef]
- Goler-Baron, V.; Selitrennik, M.; Barkai, O.; Haimovich, G.; Lotan, R.; Choder, M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008, 22, 2022–2027. [Google Scholar] [CrossRef]
- Haimovich, G.; Medina, D.A.; Causse, S.Z.; Garber, M.; Millán-Zambrano, G.; Barkai, O.; Chávez, S.; Pérez-Ortín, J.E.; Darzacq, X.; Choder, M. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell 2013, 153, 1000–1011. [Google Scholar] [CrossRef]
- Forget, A.; Chartrand, P. Cotranscriptional assembly of mRNP complexes that determine the cytoplasmic fate of mRNA. Transcription 2011, 2, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Villanyi, Z.; Ribaud, V.; Kassem, S.; Panasenko, O.O.; Pahi, Z.; Gupta, I.; Steinmetz, L.; Boros, I.; Collart, M.A. The Not5 subunit of the Ccr4-Not complex connects transcription and translation. PLoS Genet. 2014, 10, e1004569. [Google Scholar] [CrossRef]
- Richard, S.; Gross, L.; Fischer, J.; Bendalak, K.; Ziv, T.; Urim, S.; Choder, M. Numerous Post-translational Modifications of RNA Polymerase II Subunit Rpb4/7 Link Transcription to Post-transcriptional Mechanisms. Cell Rep. 2021, 34, 108578. [Google Scholar] [CrossRef]
- Gonzalez-Jimenez, A.; Campos, A.; Navarro, F.; Clemente-Blanco, A.; Calvo, O. Regulation of Eukaryotic RNAPs Activities by Phosphorylation. Front. Mol. Biosci. 2021, 8, 681865. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ortín, J.E.; Alepuz, P.; Chávez, S.; Choder, M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013, 425, 3750–3775. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.A.; Young, E.T. Coupling mRNA synthesis and decay. Mol. Cell Biol. 2014, 34, 4078–4087. [Google Scholar] [CrossRef] [PubMed]
- Begley, V.; Corzo, D.; Jordán-Pla, A.; Cuevas-Bermúdez, A.; Miguel-Jiménez, L.; Pérez-Aguado, D.; Machuca-Ostos, M.; Navarro, F.; Chávez, M.J.; Pérez-Ortín, J.E.; et al. The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4. Nucleic Acids Res. 2019, 47, 9524–9541. [Google Scholar] [CrossRef] [PubMed]
- Choder, M. mRNA imprinting: Additional level in the regulation of gene expression. Cell Logist. 2011, 1, 37–40. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.E.; Tordera, V.; Chávez, S. Homeostasis in the Central Dogma of Molecular Biology: The importance of mRNA instability. RNA Biol. 2019, 16, 1659–1666. [Google Scholar] [CrossRef]
- Medina, D.A.; Jordán-Pla, A.; Millán-Zambrano, G.; Chávez, S.; Choder, M.; Pérez-Ortín, J.E. Cytoplasmic 5’-3’ exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front. Genet. 2014, 5, 1. [Google Scholar] [CrossRef]
- García-Martínez, J.; Troule, K.; Chávez, S.; Pérez-Ortín, J.E. Growth rate controls mRNA turnover in steady and non-steady states. RNA Biol. 2016, 13, 1175–1181. [Google Scholar] [CrossRef]
- Blasco-Moreno, B.; de Campos-Mata, L.; Böttcher, R.; García-Martínez, J.; Jungfleisch, J.; Nedialkova, D.D.; Chattopadhyay, S.; Gas, M.-E.; Oliva, B.; Pérez-Ortín, J.E. The exonuclease Xrn1 activates transcription and translation of mRNAs encoding membrane proteins. Nat. Commun. 2019, 10, 1298. [Google Scholar] [CrossRef]
- Rosaleny, L.E.; Ruiz-Garcia, A.B.; Garcia-Martinez, J.; Perez-Ortin, J.E.; Tordera, V. The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 2007, 8, R119. [Google Scholar] [CrossRef] [PubMed]
- Wild, T.; Cramer, P. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 2012, 37, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Hua, Y.; Liu, X.; Liu, S.; Lao, K.; Zhang, Z.; Kong, D. Gpn2 and Rba50 Directly Participate in the Assembly of the Rpb3 Subcomplex in the Biogenesis of RNA Polymerase II. Mol. Cell Biol. 2018, 38, e00091-18. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Navarro, N.; Peiro-Chova, L.; Rodríguez-Navarro, S.; Polaina, J.; Estruch, F. Rtp1p Is a Karyopherin-Like Protein Required for RNA Polymerase II Biogenesis. Mol. Cell Biol. 2013, 33, 1756–1767. [Google Scholar] [CrossRef] [PubMed]
- Minaker, S.W.; Filiatrault, M.C.; Ben-Aroya, S.; Hieter, P.; Stirling, P.C. Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics 2013, 193, 853–864. [Google Scholar] [CrossRef]
- Staresincic, L.; Walker, J.; Dirac-Svejstrup, A.B.; Mitter, R.; Svejstrup, J.Q. GTP-dependent binding and nuclear transport of RNA polymerase II by NPA3. J. Bio. Chem. 2011, 296, 35553–35561. [Google Scholar] [CrossRef]
- Garrido-Godino, A.I.; Gutierrez-Santiago, F.; Navarro, F. Biogenesis of RNA Polymerases in Yeast. Front. Mol. Biosci. 2021, 8, 669300. [Google Scholar] [CrossRef]
- Boulon, S.; Pradet-Balade, B.; Verheggen, C.; Molle, D.; Boireau, S.; Georgieva, M.; Azzag, K.; Robert, M.C.; Ahmad, Y.; Neel, H.; et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 2010, 39, 912–924. [Google Scholar] [CrossRef]
- Forget, D.; Lacombe, A.A.; Cloutier, P.; Al-Khoury, R.; Bouchard, A.; Lavallee-Adam, M.; Faubert, D.; Jeronimo, C.; Blanchette, M.; Coulombe, B. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteom. 2010, 9, 2827–2839. [Google Scholar] [CrossRef]
- Mosley, A.L.; Hunter, G.O.; Sardiu, M.; Smolle, M.E.; Workman, J.L.; Florens, L.; Washburn, M.P. Quantitative Proteomics Demonstrates that the RNA Polymerase II Subunits Rpb4 and Rpb7 Dissociate During Transcription Elongation. Mol. Cell Proteom. 2013, 12, 1530–1538. [Google Scholar] [CrossRef]
- Turowski, T.W.; Boguta, M. Specific Features of RNA Polymerases I and III: Structure and Assembly. Front. Mol. Biosci. 2021, 8, 680090. [Google Scholar] [CrossRef] [PubMed]
- Mirón-García, M.C.; Garrido-Godino, A.I.; García-Molinero, V.; Hernández-Torres, F.; Rodríguez-Navarro, S.; Navarro, F. The prefoldin Bud27 mediates the assembly of the eukaryotic RNA polymerases in an Rpb5-dependent manner. PLoS Genet. 2013, 9, e1003297. [Google Scholar] [CrossRef] [PubMed]
- Vernekar, D.V.; Bhargava, P. Yeast Bud27 modulates the biogenesis of Rpc128 and Rpc160 subunits and the assembly of RNA polymerase III. Biochi. Biophy. Acta 2015, 1849, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Czeko, E.; Seizl, M.; Augsberger, C.; Mielke, T.; Cramer, P. Iwr1 Directs RNA Polymerase II Nuclear Import. Mol. Cell 2011, 42, 261–266. [Google Scholar] [CrossRef]
- Gómez-Navarro, N.; Estruch, F. Different pathways for the nuclear import of yeast RNA polymerase II. Biochim. Biophys. Acta 2015, 1849, 1354–1362. [Google Scholar] [CrossRef]
- Gibney, P.A.; Fries, T.; Bailer, S.M.; Morano, K.A. Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryot. Cell 2008, 7, 938–948. [Google Scholar] [CrossRef]
- Jeronimo, C.; Forget, D.; Bouchard, A.; Li, Q.; Chua, G.; Poitras, C.; Therien, C.; Bergeron, D.; Bourassa, S.; Greenblatt, J.; et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 2007, 27, 262–274. [Google Scholar] [CrossRef]
- Mosley, A.L.; Pattenden, S.G.; Carey, M.; Venkatesh, S.; Gilmore, J.M.; Florens, L.; Workman, J.L.; Washburn, M.P. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 2009, 34, 168–178. [Google Scholar] [CrossRef]
- Hunter, G.O.; Fox, M.J.; Smith-Kinnaman, W.R.; Gogol, M.; Fleharty, B.; Mosley, A.L. Phosphatase Rtr1 Regulates Global Levels of Serine 5 RNA Polymerase II C-Terminal Domain Phosphorylation and Cotranscriptional Histone Methylation. Mol. Cell Biol. 2016, 36, 2236–2245. [Google Scholar] [CrossRef]
- Hsu, P.L.; Yang, F.; Smith-Kinnaman, W.; Yang, W.; Song, J.E.; Mosley, A.L.; Varani, G. Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J. Mol. Biol. 2014, 426, 2970–2981. [Google Scholar] [CrossRef][Green Version]
- Forget, D.; Lacombe, A.A.; Cloutier, P.; Lavallee-Adam, M.; Blanchette, M.; Coulombe, B. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res. 2013, 41, 6881–6891. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.T.; Koh, J.L.; Friesen, H.; Duffy, S.K.; Cox, M.J.; Moses, A.; Moffat, J.; Boone, C.; Andrews, B.J. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell 2015, 161, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Smith-Kinnaman, W.R.; Berna, M.J.; Hunter, G.O.; True, J.D.; Hsu, P.; Cabello, G.I.; Fox, M.J.; Varani, G.; Mosley, A.L. The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae. Mol. Biosyst. 2014, 10, 1730–1741. [Google Scholar] [CrossRef]
- Egloff, S.; Zaborowska, J.; Laitem, C.; Kiss, T.; Murphy, S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell 2012, 45, 111–122. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Giannoni, F.; Dubois, M.F.; Seo, S.J.; Vigneron, M.; Kedinger, C.; Bensaude, O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996, 24, 2924–2929. [Google Scholar] [CrossRef]
- Wani, S.; Hirose, Y.; Ohkuma, Y. Human RNA polymerase II-associated protein 2 (RPAP2) interacts directly with the RNA polymerase II subunit Rpb6 and participates in pre-mRNA 3’-end formation. Drug Discov. Ther. 2014, 8, 255–261. [Google Scholar] [CrossRef]
- Fianu, I.; Dienemann, C.; Aibara, S.; Schilbach, S.; Cramer, P. Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2. Commun. Biol. 2021, 4, 606. [Google Scholar] [CrossRef]
- Braunwarth, A.; Fromont-Racine, M.; Legrain, P.; Bischoff, F.R.; Gerstberger, T.; Hurt, E.; Künzler, M. Identification and characterization of a novel RanGTP-binding protein in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 15397–15405. [Google Scholar] [CrossRef]
- Guerrero-Serrano, G.; Castanedo, L.; Cristobal-Mondragon, G.R.; Montalvo-Arredondo, J.; Riego-Ruiz, L.; DeLuna, A.; De Las Penas, A.; Castano, I.; Calera, M.R.; Sanchez-Olea, R. Npa3/ScGpn1 carboxy-terminal tail is dispensable for cell viability and RNA polymerase II nuclear targeting but critical for microtubule stability and function. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 451–462. [Google Scholar] [CrossRef]
- Hodko, D.; Ward, T.; Chanfreau, G. The Rtr1p CTD phosphatase autoregulates its mRNA through a degradation pathway involving the REX exonucleases. RNA 2016, 22, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Godino, A.I.; García-López, M.C.; Navarro, F. Correct assembly of RNA polymerase II Depends on the foot domain and Is required for multiple steps of transcription in Saccharomyces cerevisiae. Mol. Cell Biol. 2013, 33, 3611–3626. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Bermúdez, A.; Garrido-Godino, A.; Navarro, F. A novel yeast chromatin-enriched fractions purification approach, yChEFs, for the chromatin-associated protein analysis used for chromatin-associated and RNA-dependent chromatin-associated proteome studies from Saccharomyces cerevisiae. Gene Rep. 2019, 16, 100450. [Google Scholar] [CrossRef]
- Cuevas-Bermúdez, A.; Garrido-Godino, A.I.; Gutiérrez-Santiago, F.; Navarro, F. A Yeast Chromatin-enriched Fractions Purification Approach, yChEFs, from Saccharomyces cerevisiae. Bio-protocol 2020, 10, e3471. [Google Scholar] [CrossRef]
- Tan, Q.; Prysak, M.H.; Woychik, N.A. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I., II, and III. Mol. Cell Biol. 2003, 23, 3329–3338. [Google Scholar] [CrossRef]
- McKune, K.; Richards, K.L.; Edwards, A.M.; Young, R.A.; Woychik, N.A. RPB7, one of two dissociable subunits of yeast RNA polymerase II, is essential for cell viability. Yeast 1993, 9, 295–299. [Google Scholar] [CrossRef]
- Choder, M. Rpb4 and Rpb7: Subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 2004, 29, 674–681. [Google Scholar] [CrossRef]
- Duek, L.; Barkai, O.; Elran, R.; Adawi, I.; Choder, M. Dissociation of Rpb4 from RNA polymerase II is important for yeast functionality. PLoS ONE 2018, 13, e0206161. [Google Scholar] [CrossRef]
- Armache, K.J.; Kettenberger, H.; Cramer, P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 2003, 100, 6964–6968. [Google Scholar] [CrossRef]
- Victorino, J.F.; Fox, M.J.; Smith-Kinnaman, W.R.; Peck Justice, S.A.; Burriss, K.H.; Boyd, A.K.; Zimmerly, M.A.; Chan, R.R.; Hunter, G.O.; Liu, Y.; et al. RNA Polymerase II CTD phosphatase Rtr1 fine-tunes transcription termination. PLoS Genet. 2020, 16, e1008317. [Google Scholar] [CrossRef]
- Shalem, O.; Groisman, B.; Choder, M.; Dahan, O.; Pilpel, Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: A role for RNA Pol II. PLoS Genet. 2011, 7, e1002273. [Google Scholar] [CrossRef]
- Ishihama, A. Subunit of assembly of Escherichia coli RNA polymerase. Adv. Biophys. 1981, 14, 1–35. [Google Scholar] [PubMed]
- Maillet, I.; Buhler, J.M.; Sentenac, A.; Labarre, J. Rpb4p is necessary for RNA polymerase II activity at high temperature. J. Biol. Chem. 1999, 274, 22586–22590. [Google Scholar] [CrossRef] [PubMed]
- Pillai, B.; Verma, J.; Abraham, A.; Francis, P.; Kumar, Y.; Tatu, U.; Brahmachari, S.K.; Sadhale, P.P. Whole genome expression profiles of yeast RNA polymerase II core subunit, Rpb4, in stress and nonstress conditions. J. Biol. Chem. 2003, 278, 3339–3346. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Kumari, R. Rpb4 and Rpb7: Multifunctional subunits of RNA polymerase II. Crit. Rev. Microbiol. 2013, 39, 362–372. [Google Scholar] [CrossRef]
- Sampath, V.; Sadhale, P. Rpb4 and Rpb7: A sub-complex integral to multi-subunit RNA polymerases performs a multitude of functions. IUBMB Life 2005, 57, 93–102. [Google Scholar] [CrossRef]
- Qiu, Z.; Jiang, R. Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7. Biotechnol. Biofuels 2017, 10, 125. [Google Scholar] [CrossRef]
- Selitrennik, M.; Duek, L.; Lotan, R.; Choder, M. Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot. Cell 2006, 5, 2092–2103. [Google Scholar] [CrossRef]
- Grohmann, D.; Nagy, J.; Chakraborty, A.; Klose, D.; Fielden, D.; Ebright, R.H.; Michaelis, J.; Werner, F. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell 2011, 43, 263–274. [Google Scholar] [CrossRef]
- Grohmann, D.; Werner, F. Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: Structure, function and evolution of archaeal RNA polymerase. Res. Microbiol. 2011, 162, 10–18. [Google Scholar] [CrossRef]
- Li, W.; Giles, C.; Li, S. Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 2014, 42, 7069–7083. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Perez-Ortin, J.E. The transcriptional inhibitor thiolutin blocks mRNA degradation in yeast. Yeast 2008, 25, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Kebaara, B.W.; Nielsen, L.E.; Nickerson, K.W.; Atkin, A.L. Determination of mRNA half-lives in Candida albicans using thiolutin as a transcription inhibitor. Genome 2006, 49, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Rossini, C.; Taylor, W.; Fagan, T.; Hastings, J.W. Lifetimes of mRNAs for clock-regulated proteins in a dinoflagellate. Chronobiol. Int. 2003, 20, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Grigull, J.; Mnaimneh, S.; Pootoolal, J.; Robinson, M.D.; Hughes, T.R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell Biol. 2004, 24, 5534–5547. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Malik, I.; Arora, P.; Laperuta, A.J.; Pavlovic, E.M.; Ugochuckwu, S.; Naik, M.; Kaplan, C. Thiolutin is a direct inhibitor of RNA Polymerase II. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lauinger, L.; Li, J.; Shostak, A.; Cemel, I.A.; Ha, N.; Zhang, Y.; Merkl, P.E.; Obermeyer, S.; Stankovic-Valentin, N.; Schafmeier, T.; et al. Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat. Chem. Biol. 2017, 13, 709–714. [Google Scholar] [CrossRef]
- Bergmann, R. Thiolutin inhibits utilization of glucose and other carbon sources in cells of Escherichia coli. Antonie Van Leeuwenhoek 1989, 55, 143–152. [Google Scholar] [CrossRef]
- Monje-Casas, F.; Michan, C.; Pueyo, C. Absolute transcript levels of thioredoxin- and glutathione-dependent redox systems in Saccharomyces cerevisiae: Response to stress and modulation with growth. Biochem. J. 2004, 383 Pt 1, 139–147. [Google Scholar] [CrossRef]
- García-López, M.C.; Mirón-García, M.C.; Garrido-Godino, A.I.; Mingorance, C.; Navarro, F. Overexpression of SNG1 causes 6-azauracil resistance in Saccharomyces cerevisiae. Curr. Genet. 2010, 56, 251–263. [Google Scholar] [CrossRef]
- Thomas, B.J.; Rothstein, R. Elevated recombination rates in transcriptionally active DNA. Cell 1989, 56, 619–630. [Google Scholar] [CrossRef]
- Voth, W.P.; Jiang, Y.W.; Stillman, D.J. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 2003, 20, 985–993. [Google Scholar] [CrossRef]
- Gari, E.; Piedrafita, L.; Aldea, M.; Herrero, E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 1997, 13, 837–848. [Google Scholar] [CrossRef]
- Shpakovski, G.V.; Acker, J.; Wintzerith, M.; Lacroix, J.F.; Thuriaux, P.; Vigneron, M. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 4702–4710. [Google Scholar] [CrossRef]
- Zaros, C.; Thuriaux, P. Rpc25, a conserved RNA polymerase III subunit, is critical for transcription initiation. Mol. Microbiol. 2005, 55, 104–114. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido-Godino, A.I.; Cuevas-Bermúdez, A.; Gutiérrez-Santiago, F.; Mota-Trujillo, M.d.C.; Navarro, F. The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 2002. https://doi.org/10.3390/ijms23042002
Garrido-Godino AI, Cuevas-Bermúdez A, Gutiérrez-Santiago F, Mota-Trujillo MdC, Navarro F. The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2022; 23(4):2002. https://doi.org/10.3390/ijms23042002
Chicago/Turabian StyleGarrido-Godino, Ana I., Abel Cuevas-Bermúdez, Francisco Gutiérrez-Santiago, Maria del Carmen Mota-Trujillo, and Francisco Navarro. 2022. "The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae" International Journal of Molecular Sciences 23, no. 4: 2002. https://doi.org/10.3390/ijms23042002
APA StyleGarrido-Godino, A. I., Cuevas-Bermúdez, A., Gutiérrez-Santiago, F., Mota-Trujillo, M. d. C., & Navarro, F. (2022). The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae. International Journal of Molecular Sciences, 23(4), 2002. https://doi.org/10.3390/ijms23042002





