1. Introduction
Extracellular proteolysis is a major process in tumor biology, thereby regulating the proliferation and invasion of tumor cells into the surrounding tissue, and recruitment of immune cells to the tumor site, eventually shaping a tumor-promoting tumor microenvironment [
1]. In recent studies, A disintegrin and metalloprotease 8 (ADAM8) was identified as an extracellular metalloprotease-disintegrin important for tumor progression, invasion, and metastasis in pancreatic ductal adenocarcinoma (PDAC) [
2]. PDAC is one of the most lethal solid tumors with a 5-year survival rate of less than 8%. In addition, the incidence of PDAC is on the rise and could become a leading cause of cancer deaths by 2030 [
3]. A strong desmoplastic stroma response to tumor growth is a hallmark of PDAC and, at least partially, a cause for the devastating patient prognosis [
4]. In particular, the tumor microenvironment (TME) with its inflammatory nature activates several immune cell types and suppresses the immune competence of the TME, suggesting an intense communication between tumor and immune cells through the extracellular matrix (ECM). In this regard, ADAM proteases, as membrane-located shedding enzymes, are capable of creating these intercellular signals by the controlled release of membrane proteins involved in immune modulation. One such ADAM protease is ADAM8, a metalloprotease-disintegrin with a proven record of tumor-supporting effects when expressed in tumor cells, thereby facilitating tumor progression, invasion, and immune cell recruitment [
2,
5]. Recently, a systematic analysis of tumor-associated immune cells in tumor tissues of PDAC patients revealed the additional expression of ADAM8 in macrophages, neutrophils, and NK cells [
6]. ADAM8
+ immune cells can transmigrate through endothelia and invade the ECM, as demonstrated in several inflammation models (e.g., [
7,
8]). These findings suggest that ADAM8 could also exert its function in immune cells of the TME. Given the strong endogenous expression of ADAM8 in macrophages, we hypothesized that ADAM8 serves important functions in tumor cell–macrophage interactions. These interactions could be mediated by extracellular vesicles (EVs), a defined type of lipid-enclosed particle ranging in size from 30 to 100 nm. ADAM8 was identified as an EV cargo and is of diagnostic value for the early detection of PDAC lesions [
9]. In addition, cargo analyses of ADAM8-positive EVs isolated from PDAC patients revealed diagnostic miRNAs correlated with ADAM8 expression levels, such as miR-451 and miR-720. These miRNAs could play a role in regulating cellular functions in the TME as ADAM8-dependent miRNAs. In the current study, we analyzed EV release from PDAC cells and the cellular interactions between macrophages and PDAC tumor cells in an ADAM8-dependent manner. PDAC cell lines with endogenous expression of ADAM8 or a CRISPR/Cas9 knockout of the
ADAM8 gene were co-cultivated with macrophages, and the resulting changes in gene expression were analyzed.
3. Discussion
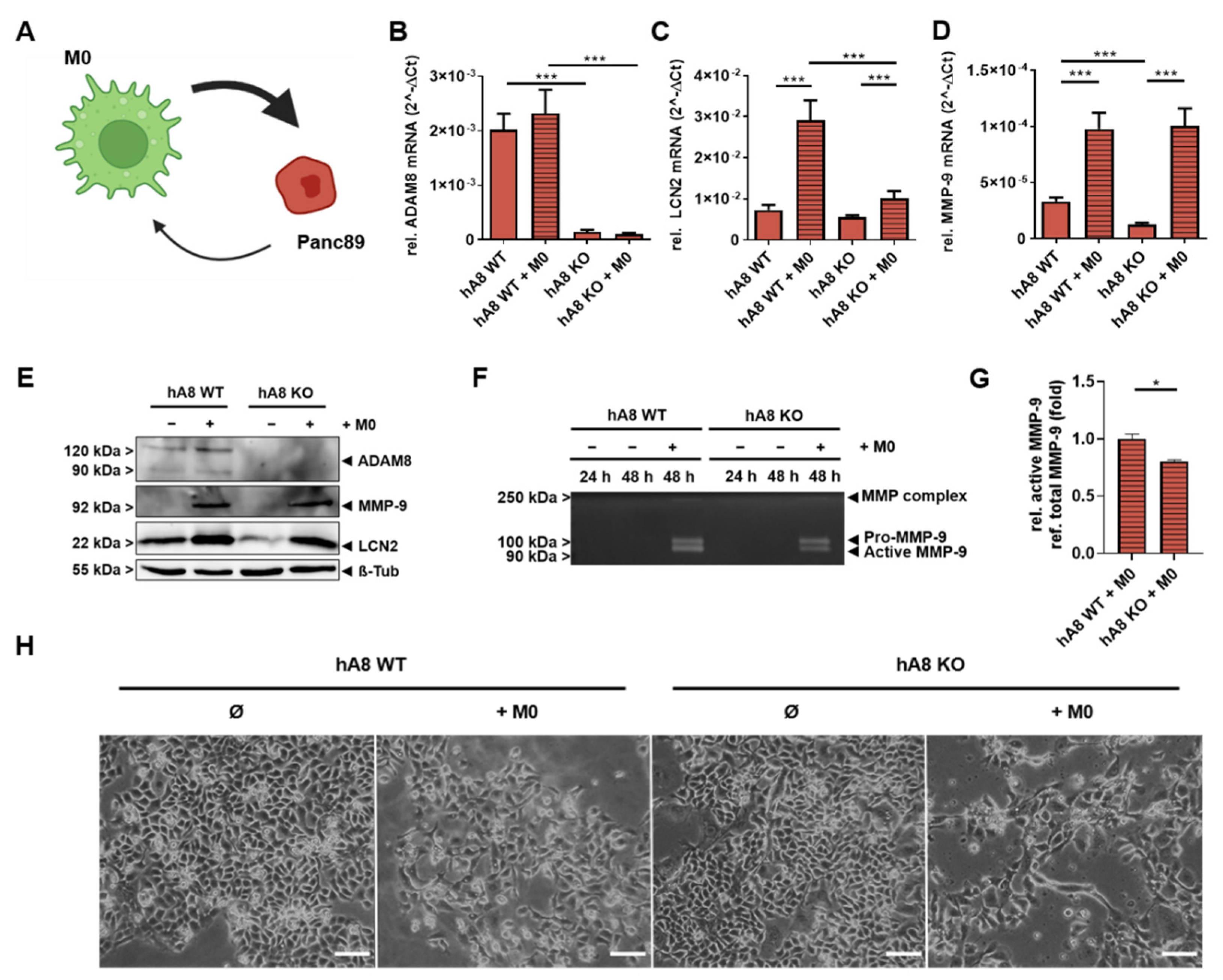
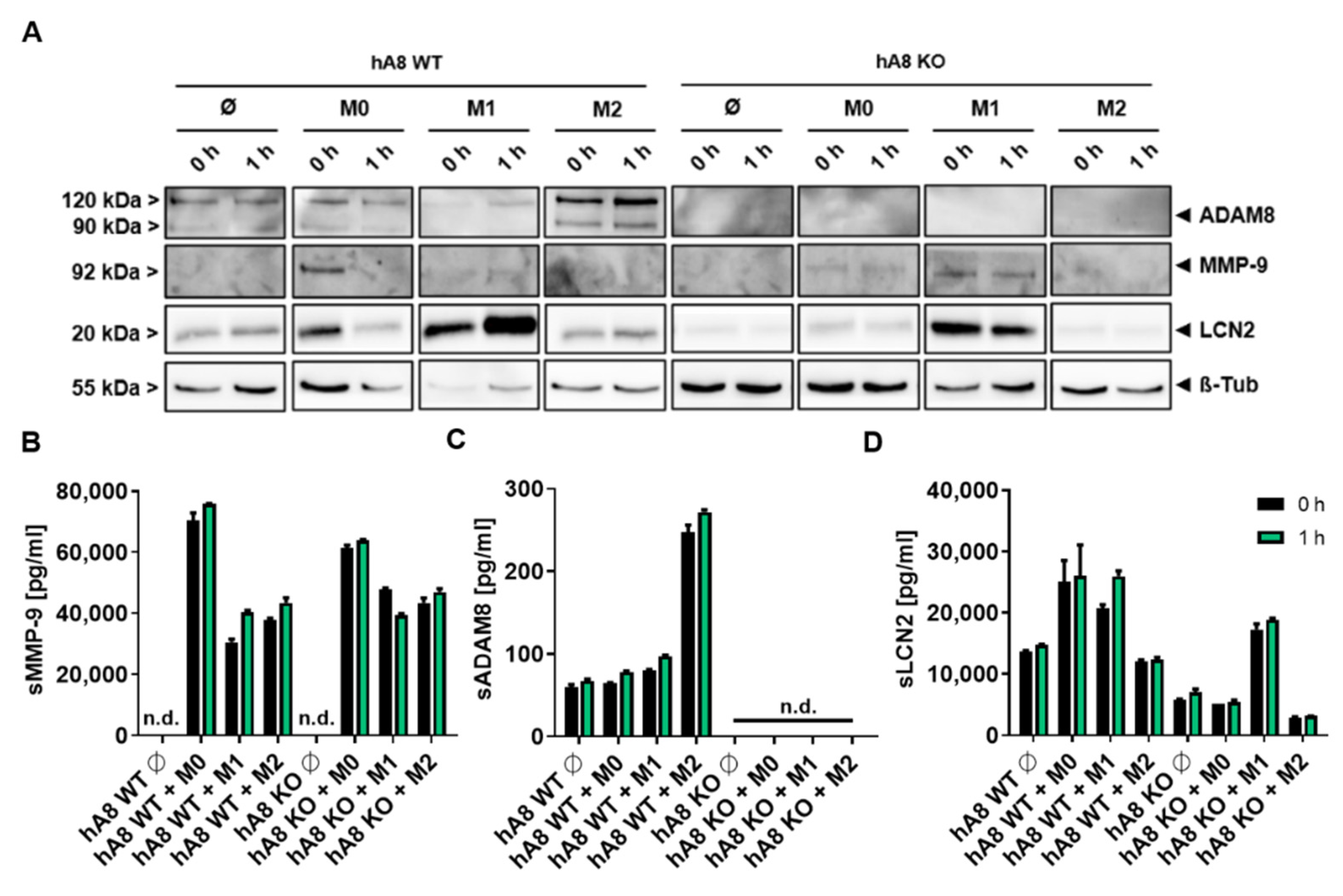
In this study, we provide insights into the protease-dependent mechanisms underlying the progression of PDAC as a highly invasive tumor entity. First, we demonstrate the essential function of ADAM8 in the extracellular release of MMP-9 and LCN2, two important mediators of cancer progression in PDAC. For instance, a recent report demonstrated that MMP-9, when derived from macrophages, is essential for EMT in tumor cells via activation of protease-activated receptor (PAR-1, [
18]). Here, we report on the activation of MMP-9 in tumor cells by macrophages and demonstrate that the communication between macrophages and tumor cells can likely cause MMP-9-dependent effects in an autocrine manner, i.e., by self-induction of MMP-9 in tumor cells when associated with macrophages, as our co-culture experiments suggest. Notably, this effect is not dependent on the polarization state of macrophages, as M1- and M2-like macrophages have similar effects on MMP-9 expression in Panc89 cells. Another strong correlation with ADAM8 expression is reported here for LCN2, a siderophore protein involved in pathogen defense and covalent stabilization of MMP-9. From our data, there are some indications that LCN2 can lead to enhanced activation of pro-MMP-9, thereby causing a higher degree of extracellular proteolysis, leading to the tumor-promoting effect of positive co-regulation of MMP-9 and LCN2 by ADAM8. More directly, LCN2 was recently investigated in a mouse model of PDAC. LCN2-deficient mice were crossed with transgenic mice expressing Kras
G12D in acinar cells [
19]. In these mice, the lack of LCN2 caused a significant reduction in immune cell infiltration, PanIN, and tumor growth, suggesting that LCN2 in the TME is an important determinant for PDAC progression and patient prognosis, similar to MMP-9. Thus, ADAM8 could amplify its detrimental function in PDAC by simultaneously affecting MMP-9 and LCN-2 expression. Given the finding that LCN2 can determine MMP-9 activity in vivo, it can be hypothesized that MMP-9 activity in the TME is directly affected. However, in contrast to the regulation of MMP-9, LCN2 is differentially regulated by co-culturing PDAC cells with macrophages. Undifferentiated as well as M1-polarized macrophages cause increased LCN2 release, whereas M2-polarized macrophages suppress LCN2 secretion. Thus, a distinct regulation of MMP-9 and LCN2 expression by macrophages could imply that both proteins serve functions distinct from the regulation of net protease activity in the TME. A limitation of this study is the sole use of THP-1 cells rather than primary macrophages derived from PBMCs. To address this issue, we tried to include these cell sources in our analysis; however, we obtained heterogeneous results that prevent a conclusive interpretation. A further limitation is the analysis of 2D co-cultures that might not reflect the interaction of tumor cells with macrophages in vivo. In this regard, a 3D model using organoid co-cultures will be required to get closer to a more authentic tumor microenvironment. Since we could demonstrate that only MMP-9 and not LCN2 expression is affected in Panc89 cells after treatment with M0-derived supernatants, another limitation of this study is the treatment of Panc89 cells with THP-1 macrophage-derived EVs. As EVs can function as a carrier of signaling molecules, we hypothesize that EV treatment could induce LCN2 expression and therefore MMP-9 activity, which is a matter of further studies.
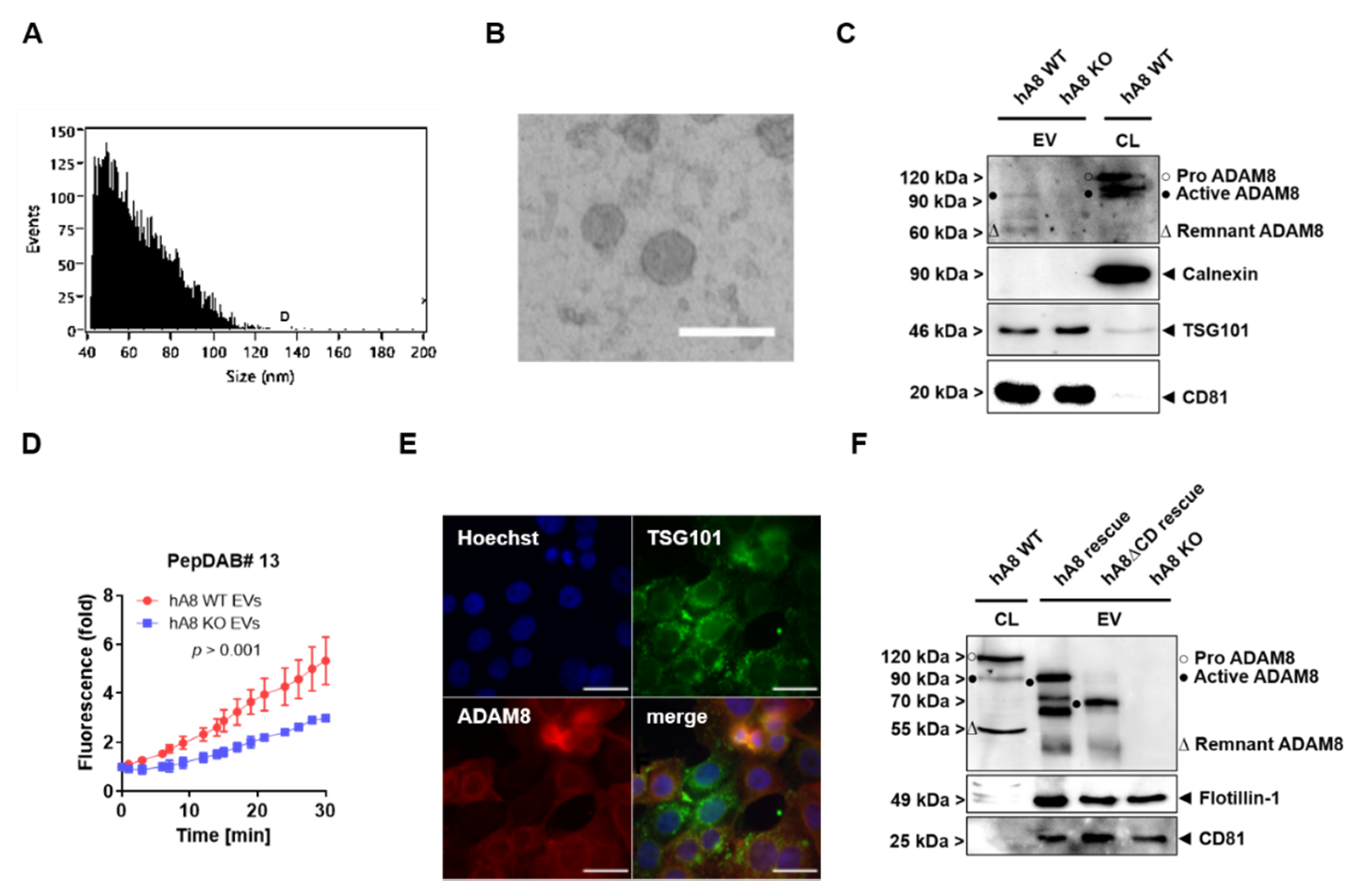
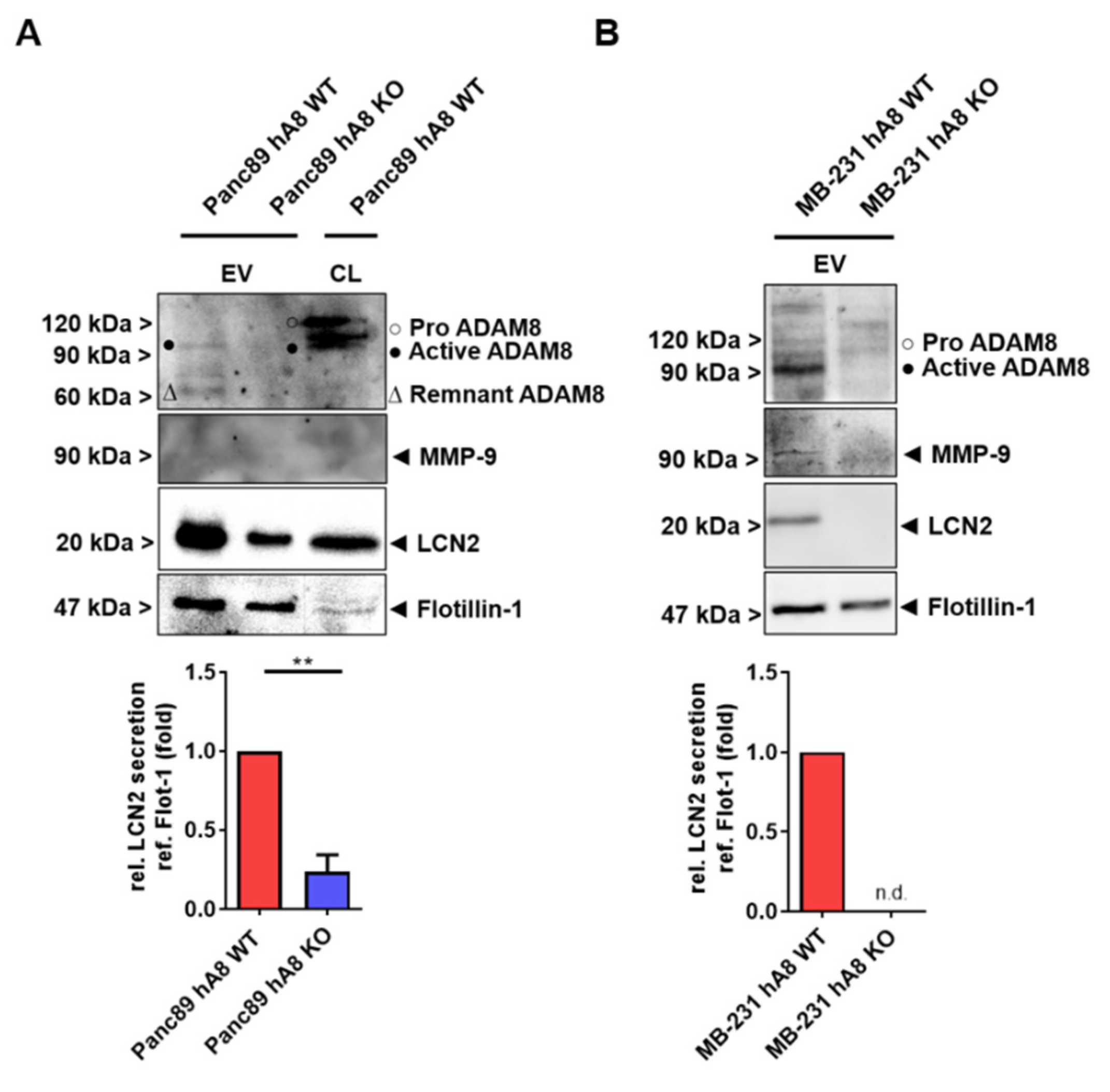
Other important findings in this study are that ADAM8 is sorted into EVs, and the ADAM8 dependence of MMP-9 and LCN2 is found, in addition to lysates, in the EV cargo; thus, we can conclude that ADAM8 is a key regulator of MMP-9 and LCN2 release in the TME. However, the mechanism by which ADAM8 is sorted into EVs remains unclear. Besides the most described ESCRT machinery, many ESCRT-independent cargo and biomolecule sorting pathways in EVs have been reported [
20]. TSG101, as part of the ESCRT complex, can recognize the tetrapeptide protein motif PS/TAP [
14]. The amino acid motif PTAP is found in ADAM8 at the very C-terminus (aa residues 821–824). To demonstrate the function of this motif, we established a Panc89 cell line lacking the C-terminal domain of ADAM8, thereby deleting the PTAP motif of ADAM8, and expected this form of ADAM8 not to be present in EVs. Unexpectedly, the C-terminal mutant form of ADAM8 is still capable of being secreted in EVs. Furthermore, the lack of co-localization of TSG101 with ADAM8 supports the hypothesis that the internalization of ADAM8 in EVs occurs independently of the ESCRT-I protein TSG101. ESCRT-independent pathways such as lipid raft-, tetraspanin-, and ceramide-mediated mechanisms could exhibit potential ADAM8 EV cargo loading alternatives that must be investigated [
21]. Tan et al. [
22] demonstrated that lipid rafts could conduce to a platform for exosomal biogenesis in mesenchymal stem cells. ADAM17 has been described as an active protease associated with lipid rafts [
23] and could indicate a possible mechanism for ADAM17 EV encapsulation, and thus for ADAM8. In addition, the release of ADAM10 in EVs by melanoma cells was shown to be linked to a paxillin/integrin interaction and a subsequent shift to lipid rafts [
24], supporting the suggested mechanism for ADAM8 exosomal release. In EVs, ADAM8 can be detected as an active protease, as peptide cleavage assays with WT and KO cells demonstrate. Thus, the topological orientation of ADAM8 in EVs is likely to be directed to the extracellular compartment where ADAM8-dependent cleavage can occur. In some reports, ADAM8 was also discussed as a membrane protein mediating cell–cell fusion, where ADAM8 on the surface of EVs could potentially contribute to the fusion of EVs with adjacent cells to release their cargo [
25]. In addition, as ADAM8 can regulate miRNA expression in tumor cells (Schäfer et al., manuscript submitted), it is likely that ADAM8–ADAM8 interactions between EVs and ADAM8-expressing cells could lead to fusion events and facilitate the transfer of miRNAs into cells to regulate gene expression. In the case of ADAM8-positive EVs in PDAC, such miRNAs were reported to be miRNAs 720 and 451 [
9]. Concomitant with the ADAM8 content in EVs, MMP-9 and LCN2 are affected, suggesting that ADAM8 regulates the amount of these proteins in the TME. A cell-autonomous effect of ADAM8 in tumor cells was extensively described in numerous studies. However, the function of ADAM8 in the TME, given the localization in EVs, is worth further studies involving other immune cell types such as neutrophils and natural killer cells in which ADAM8 is also highly expressed.
4. Materials and Methods
4.1. Cell Culture
Panc89 cells were kindly provided by Prof. Anna Trauzold, Kiel University, and were cultivated in RPMI (GibcoTM, Life Technologies, Carlsbad, CA, USA) supplemented with 10% (v/v) Fetal Bovine Serum (FBS; Sigma-Aldrich, Munich, Germany), 0.1 mg/mL Penicillin-Streptomycin (GibcoTM, Life Technologies, Carlsbad, CA, USA), and 1 mM Sodium Pyruvate (GibcoTM, Life Technologies, Carlsbad, CA, USA) in a humidified atmosphere at 37 °C and 5% CO2. MB-231 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and were cultivated in DMEM (GibcoTM, Life Technologies, Carlsbad, CA, USA) with the same additives as described for Panc89 cells.
4.2. Generation of Stable Panc89 and MB-231 ADAM8 Knockout Cells
To generate a genomic ADAM8 knockout in Panc89 and MB-231 cells, the ADAM8 Human Gene Knockout Kit (CRISPR) from OriGene (CAT#: KN213386, Rockville, ML, USA) was used. The transfection of the cells and further necessary steps were performed as previously described [
11]. Single-cell clones were obtained after the selection with 1 μg/mL Puromycin (InvivoGen, San Diego, CA, USA).
4.3. Generation of Stable Panc89 ADAM8 Rescue Cells
For a genomic knock-in of the full-length ADAM8 or an ADAM8 lacking the C-terminal domain in Panc89 hA8 KO cells, the ADAM8 AAVS1 Transgene Kockin kit (BSD) from OriGene (CAT#: GE100036, Rockville, ML, USA) was used. The transfection of the cells and further necessary steps were performed as described in
Section 4.2. Single-cell clones were obtained after the selection with 10 μg/mL Blasticidin (InvivoGen, San Diego, CA, USA).
4.4. THP-1 Cell Differentiation and Polarization
For macrophage differentiation, THP-1 cells were seeded at a density of 500,000 cells per well of a 6-well plate and treated with 10 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Munich, Germany). After 48 h, differentiated THP-1 cells were exposed for another 6 h to either 50 ng/mL LPS (Sigma-Aldrich, Munich, Germany) and 20 ng/mL IFN-γ (PeproTech, Hamburg, Germany) for M1 polarization or 20 ng/mL IL-4 (PeproTech, Hamburg, Germany) for M2 polarization.
4.5. Co-Culture of Panc89 with THP-1
At a density of 500,000 cells per approach, THP-1 cells were seeded in the upper part of ThinCert
TM Cell Culture Insert with a 0.4 µm diameter (Greiner Bio-One GmbH, Frickenhausen, Germany). After 48 h of differentiation, 500,000 Panc89 cells were seeded in the lower compartment (well) and left in co-culture for another 48 h. Cells were either harvested for RNA isolation or protein extraction and subsequent Western blot analysis. For M1 and M2 co-culture, THP-1 cells were treated with 50 ng/mL LPS (Sigma-Aldrich, Munich, Germany) and 20 ng/mL IFN-γ (PeproTech, Hamburg, Germany) for M1 polarization or 20 ng/mL IL-4 (PeproTech, Hamburg, Germany) for M2 polarization 6 h after differentiation and before co-culture. The described co-culture procedure was performed considering different established protocols, as described, for example, in Landmann and Buchholz, Department of Gastroenterology, Endocrinology, Metabolism, and Infectiology, Philipps University Marburg
https://archiv.ub.uni-marburg.de/ubfind/Record/urn:nbn:de:hebis:04-z2019-0310 (accessed on 29 December 2021) and [
26,
27].
4.6. EV Isolation and Characterization by NanoFCM and Electron Microscopy
After the tumor cells reached 80% confluency, the medium was changed to RPMI medium without Phenol Red (Gibco
TM, Life Technologies, Carlsbad, CA, USA) but supplemented with 1% (
v/
v) Insulin-Transferrin-Selenium (Gibco
TM, Life Technologies, Carlsbad, CA, USA). After 72 h, the supernatants were centrifuged at 300×
g for 10 min followed by an additional centrifugation step at 3000×
g for 10 min. The resulting supernatant was filtered (0.2 μm) and subsequently centrifuged at 10,000×
g for 1 h at 4 °C. EVs were pelleted at 100,000×
g for 90 min at 4 °C in an Optima XPN-80 ultracentrifuge (Beckman Coulter, Krefeld, Germany) with an SW32Ti swing-out rotor, resuspended in 700 μm HBSS (Gibco
TM, Life Technologies, Carlsbad, CA, USA), and again centrifuged at 100,000×
g for 90 min at 4 °C in an Optima MAX-XP (Beckman Coulter, Krefeld, Germany) ultracentrifuge with a TLA-55 fixed angle rotor. Finally, the EVs were resuspended in 50 μL HBSS (Gibco™, Life Technologies, Carlsbad, CA, USA). For particle size and quantity determination, EVs were applied to a flow nano analyzer (NanoFCM Co. Ltd., Nottingham, UK). Electron microscopy was conducted as previously described [
9].
4.7. RNA Isolation and Quantitative Real-Time PCR
For total RNA isolation, cells were resuspended in 1 mL Qiazol (Qiagen, Hilden, Germany) and 200 μL Chloroform was added. After mixing the samples, the lysed cells were incubated at RT for 5 min and subsequently centrifuged at 12,000×
g for 15 min at 4 °C. The RNA in the upper aqueous phase was then precipitated by adding 500 μL Isopropanol. After repeated centrifugation, the RNA was washed with 75% (
v/
v) Ethanol, dried, and dissolved in 10–30 μL RNase-Free Water. The resulting concentration of the isolated RNA was determined by NanoPhotometer
® NP80 (Implen, Munich, Germany). An amount of 2 µg of isolated RNA was applied to RNA to cDNA EcoDry
TM kit (Takara Bio Inc., Kusatsu, Japan) for reverse transcription. The procedure was conducted according to the manufacturer’s instructions. Quantitative real-time PCR was performed using iTaq
TM Universal SYBR
® Green Supermix (Bio-Rad Laboratories GmbH, Feldkirchen, Germany), cDNA equivalent of 20 ng total RNA, and either forward and reverse primers or QuantiTect Primer Assay (Qiagen, Hilden, Germany) in a 20 µL total PCR reaction volume in a StepOnePlusTM Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). Relative changes in mRNA expression were calculated using the ΔCt method using XS13 as a housekeeping gene (the sequence was described elsewhere) [
2].
4.8. rLCN2 and rTGF-α Stimulation of Panc89 Cells
A total of 500,000 cells were seeded in each well of a 6-well plate. After 48 h, the medium was changed to RPMI (GibcoTM, Life Technologies, Carlsbad, CA, USA) Medium supplemented with 0.5% (v/v) FBS (Sigma-Aldrich, Munich, Germany) and cultivated for another 6 h. Starved cells were then treated with either rTGF-α (PeproTech, Hamburg, Germany), rLCN2 (R&D Systems, Minneapolis, MN, USA), or their combination for 1 h. Stimulated cells were then applied to total protein extraction and Western blot analysis.
4.9. Protein Extraction and Western Blot Analysis
Total protein extraction was performed by detaching the cells with a cell scraper. Cells were washed with PBS, resuspended in 50 μL RIPA (50 mM HEPES pH 7.4; 150 mM NaCl; 1% (v/v) NP-40; 0.5% (w/v) Natriumdeoxycholate; 0.1% (w/v) SDS; 10 mM Phenantrolin; 10 mM EDTA; PierceTM Protease Inhibitor Mini Tablets, EDTA-free, Thermo Scientific, Waltham, MA, USA; PierceTM Phosphatase Inhibitor Mini Tablets, Thermo Scientific, Waltham, MA, USA) and incubated for 30 min on ice. After centrifugation at 12,000× g for 5 min at 4 °C, the protein concentration of the supernatant containing the total protein was determined by PierceTM BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA). Proteins from equal amounts of lysate were then boiled in 1× loading buffer (5× loading buffer: 60 mM Tris-HCl pH 6.8; 2% (w/v) SDS; 10% (w/v) Glycerol; 5% (v/v) ß-Mercaptoethanol; 0.01% (w/v) Bromphenol-Blue) for 5 min. The proteins were separated by 10% SDS-PAGE and subsequently transferred to a nitrocellulose membrane (GE Healthcare, Chicago, IL, USA). The membrane was then blocked with 5% (w/v) milk powder (MP) in TBST (50 mM Tris, pH 7.5; 150 mM NaCl; 0.1% (w/v) Tween-20) for 1 h and probed with the following primary antibodies: anti-ADAM8 (PA5-47047, Thermo Fisher Scientific, Waltham, MA, USA; 1:1000 in 5% MP in TBST), anti-beta Tubulin (NB600-936, Novus Biological, Littleton, CO, USA; 1:2000), anti-LCN2 (AF1757, R&D Systems, Minneapolis, MN, USA; 1:1000), anti-EGFR (4267, Cell Signaling Technology, Danvers, MA, USA; 1:1000), anti-pEGFR (3777, Cell Signaling Technology, Danvers, MA, USA; 1:1000), anti-MAPK (4696, Cell Signaling Technology, Danvers, MA, USA; 1:2000), anti-pMAPK (4370, Cell Signaling Technology, Danvers, MA, USA; 1:2000), Calnexin (2679, Cell Signaling Technology, Danvers, MA, USA; 1:1000), anti-Flotillin-1 (PA5-18053, Thermo Scientific, Waltham, MA, USA; 1:2000), anti-TSG101 (ab83, Abcam, Cambridge, UK; 1:1000), MMP-9 (AF911, R&D Systems, Minneapolis, MN, USA; 1:2000), and anti-CD81 (sc166029, Santa Cruz Biotechnology, Dallas, TX, USA; 1:500), at 4 °C overnight. After washing 3 times with TBST, the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h (Abcam, Cambridge, UK; 1:5000) followed by a repeated washing step. Detection was performed by using Chemiluminescent HRP Substrate, Western Bright Sirius (Advansta Inc., San Jose, CA, USA).
4.10. Human Protease/Protease Inhibitor Array
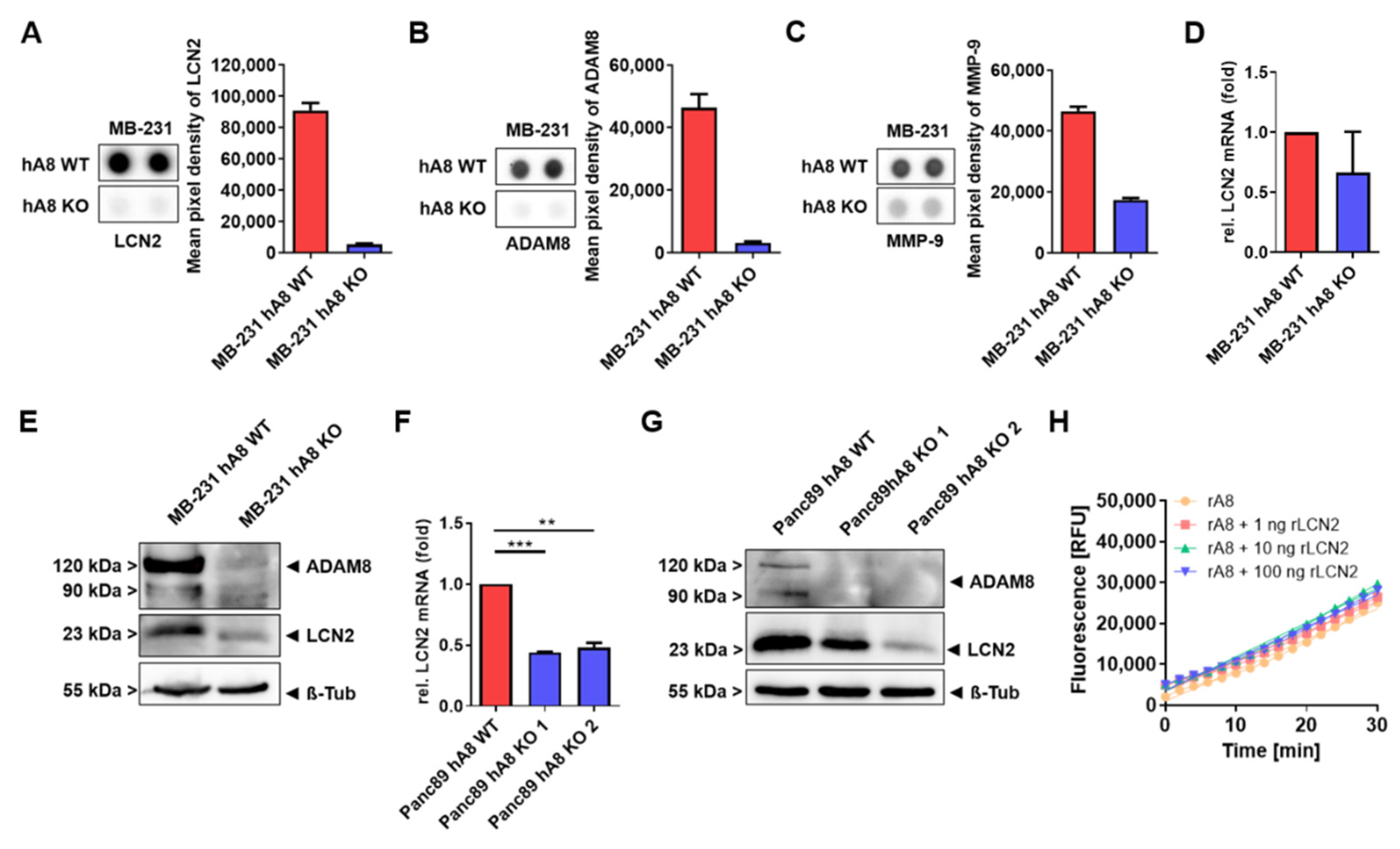
For the Proteome Profiler Human Protease/Protease Inhibitor Array (ARY025, R&D Systems, Minneapolis, MN, USA), 500,000 MB-231 cells were seeded in each well of a 6-well plate overnight. The next day, the medium was changed to GibcoTM, Life Technologies, Carlsbad, CA, USA) supplemented with 2% (v/v) FBS (Sigma-Aldrich, Munich, Germany) for 48 h. The supernatants were collected, and protein concentration was determined by NanoPhotometer® NP80 (Implen, Munich, Germany). Concentrations were adjusted, and the array was carried out according to the manufacturer’s protocol.
4.11. Immunofluorescence Staining
Before the procedure, coverslips were coated with 50 µg/mL Collagen Type I from rat tail (Sigma-Aldrich, Munich, Germany) and incubated for 1 h at 37 °C. The coating was then washed 2 times with PBS, and subsequently, Panc89 cells were seeded at a density of 100,000 cells per 0.5 mL on the coated coverslips. After 24 h, cells were washed 3 times with PBS and then fixed with 4% (w/v) PFA for 15 min followed by 1 h blocking with 5% (w/v) BSA in PBS. Primary ADAM8 (PA5-47047, Thermo Fisher Scientific, Waltham, MA, USA; 1:100) or TSG101 (ab83, Abcam, Cambridge, UK; 1:100) antibody was added for overnight incubation at 4 °C. The next day, cells were washed 3 times with PBS and incubated with secondary antibody Alexa Fluor 488 (A-11055, Thermo Fisher Scientific, Waltham, MA, USA; 1:500), Alexa Fluor 488 (ab150105, Abcam, Cambridge, UK; 1:500), or Texas Red (PA1-28662, Thermo Fisher Scientific, Waltham, MA, USA; 1:1000) for 1 h at RT. After another washing step, cells were incubated with Hoechst 33,342 dye (Sigma-Aldrich, Munich, Germany) for counterstaining for 20 min. Images were obtained using a Leica DM 5500 microscope (Leica Microsystems, Wetzlar, Germany).
4.12. Gelatin Zymography
After 48 h of co-culture, cells were cultivated in monoculture for another 24 h with 1 mL RPMI (GibcoTM, Life Technologies, Carlsbad, CA, USA) without phenol red and FBS, but supplemented with 1% (v/v) Insulin-Transferrin-Selenium (GibcoTM, Life Technologies, Carlsbad, CA, USA). The supernatants were collected, centrifuged at 4000× g for 10 min, and concentrated with Vivaspin (Sartorius AG, Göttingen, Germany). Protein concentration determination was performed as described above. Before the electrophoresis, the samples were either treated with 0.6 mM APMA (Sigma-Aldrich, Munich, Germany), with recombinant LCN2 (R&D Systems, Minneapolis, MN, USA), or with both for 1 h at 37 °C. The samples were then diluted with 2x non-reducing loading dye (1 M Tris-HCl, pH 6.8; 20% (w/v) Glycerol; 10% SDS (w/v); 0.1% (w/v) Bromphenol-Blue) and loaded on a gel containing 0.1% (w/v) Gelatin in the separating gel. After the run, the gel was initially washed 2 times with renaturation buffer (2.5% (v/v) Triton-X-100) for 30 min each, followed by equilibration with developing buffer (1 M Tris-HCl, pH 7.5; 200 mM NaCl; 4 mM CaCl2; 0.02% (v/v) Brij-35) for 30 min and subsequent incubation in a fresh developing buffer for 16 h at 37 °C. The next day, the gel was stained with Coomassie staining solution (50% (v/v) Methanol, 10% (v/v) Acetic acid, 0.5% (w/v) Coomassie Brilliant Blue G250) for 1 h and destained with destaining solution (50% (v/v) Methanol, 10% (v/v) Acetic acid) until the bands of the active enzyme were visible.
4.13. Protease Activity Assay in Panc89-Derived Supernatants or EVs
Both supernatants and EVs derived from Panc89 cells were tested for ADAM and MMP activities by determining the cleavage of FRET-based polypeptide substrates with a high Kcat/Km for different proteases (PEPDab5, PEPDab8, PEPDab10, PEPDab13, PEPDab14, BioZyme, Inc., Apex, NC, USA), as previously described [
28]. For the conditioned supernatants, 500,000 cells were seeded in each well of a 6-well plate. After 24 h, the medium was changed to 1 mL RPMI (Gibco
TM, Life Technologies, Carlsbad, CA, USA)without phenol red and FBS, but supplemented with 1% (
v/
v) Insulin-Transferrin-Selenium (Gibco
TM, Life Technologies, Carlsbad, CA, USA) for another 24 h. After the incubation, the supernatants were centrifuged at 4000×
g for 10 min. Briefly, 10 µM of each PEPDab in 50 µL assay buffer (1 mM ZnCl2, 20 mM Tris-HCl pH 8.0, 10 mM CaCl2, 150 mM NaCl, 0.0006% Brij-35) was incubated with either 50 µL conditioned supernatant or 5 × 10
9 EVs in a total volume of 100 µL. The resulting fluorescence was monitored every 2 min for 6 h at 37 °C with a multiwell plate reader (FLUOstar OPTIMA, BMG Labtech, Offenburg, Germany) using a λex of 485 nm and a λem of 530 nm.
4.14. ELISA
Soluble ADAM8 (DY1031, R&D Systems, Minneapolis, MN, USA), LCN2 (DY1737, R&D Systems, Minneapolis, MN, USA), and MMP-9 (DY911, R&D Systems, Minneapolis, MN, USA) levels in Panc89-derived supernatants were determined by using DuoSet ELISA kits. A total of 500,000 cells were incubated in a normal growth medium (as described above) in each well of a 6-well plate for 48 h. Before the procedure, the collected supernatants were centrifuged at 4000× g for 10 min and subjected to ELISA according to the manufacturer’s protocol.
4.15. Proliferation Assay
A total of 5000 cells per well were seeded in triplicates in a 96-well plate. After 24 h, 48 h, and 72 h, viability was determined by adding 50 µL CellTiter-Glo® 3D Cell Viability Assay (Promega, Walldorf, Germany), followed by a shaking step for 15 min and subsequent incubation for 15 min at RT in the dark. The resulting luminescence was measured using a multiwell plate reader (FLUOstar OPTIMA, BMG Labtech, Offenburg, Germany).
4.16. Scratch Assay
The scratch assay was performed by seeding 30,000 cells per well of a Culture-Insert 2 Well (ibidi GmbH, Gräfelfing, Germany) inserted in a 24-well plate. After 6 h and allowing the cells to attach, the normal growth medium was changed to RPMI Medium (Gibco
TM, Life Technologies, Carlsbad, CA, USA) supplemented with 0.5% (
v/
v) FBS (Sigma-Aldrich, Munich, Germany) for overnight starvation. The Culture-Insert 2 Well was removed, and cells were washed with a fresh normal growth medium to remove non-adherent cells. From this moment, images at each edge of the gap were taken at time points 0 h and 10 h. The images were analyzed using the Image J software plugin “Wound_ healing_size_tool_updated” published by [
29].
4.17. Invasion Assay
To examine the cells’ invasive behavior, an invasion assay was performed using ThinCertTM Cell Culture Inserts with an 8 µm pore diameter (Greiner Bio-One GmbH, Frickenhausen, Germany). Before seeding the cells, the upper side of the Thincert was coated with 50 µL Basement Membrane Matrix Growth Factor Reduced Matrigel (Corning®, Corning, NY, USA). After 1 h incubation at 37 °C, 25,000 cells were seeded in 50 µL RPMI Medium (GibcoTM, Life Technologies, Carlsbad, CA, USA) supplemented with 0.5% (v/v) FBS (Sigma-Aldrich, Munich, Germany) at the lower side of the Thincert. Cells were allowed to attach to the membrane for a maximum of 4 h. Subsequently, 250 µL RPMI Medium (GibcoTM, Life Technologies, Carlsbad, CA, USA) supplemented with 20% (v/v) FBS (Sigma-Aldrich, Munich, Germany) was added to the upper part of the Thincert and 750 µL RPMI Medium (GibcoTM, Life Technologies, Carlsbad, CA, USA) supplemented with 0.5% (v/v) FBS (Sigma-Aldrich, Munich, Germany) to the well. Cells were then allowed to invade the Matrigel due to an FBS gradient. After 24 h, the Thincerts were treated with 4% (w/v) PFA for 30 min, and subsequently, cells were permeabilized with 0.3% (v/v) Triton-X for another 30 min. For counting and quantification, cells were stained with Hoechst 33,342 (Thermo Scientific, Waltham, MA, USA) dye overnight. Z-Stacks of five random viewing fields were recorded, and cells on the lower part of the Thincert (non-invasive) and cells in the matrigel (invasive) were counted.
4.18. Statistical Analysis
Student’s t-test or two-way ANOVA was applied for statistical analysis. Data were considered not significant (p > 0.05, no asterisk), significant * (p < 0.05), highly significant ** (p < 0.01), or very highly significant *** (p < 0.001) and are expressed as the mean ± S.D.