IL-6 Cytokine Family: A Putative Target for Breast Cancer Prevention and Treatment
Abstract
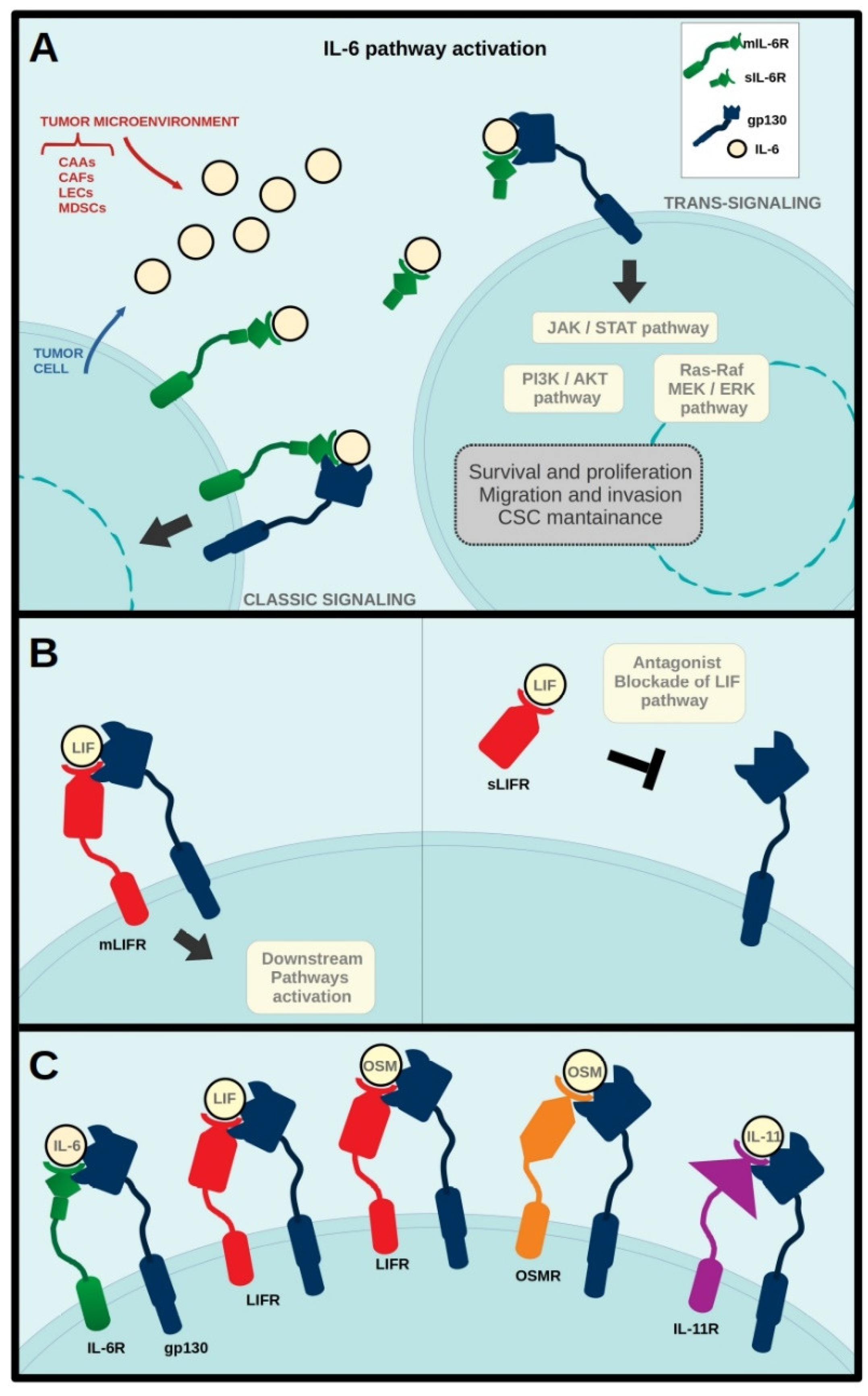
:1. Introduction
2. IL-6 in Breast Cancer
3. LIF in Breast Cancer
4. OSM in Breast Cancer
5. IL-11 in Breast Cancer
6. IL-6 Cytokine Family in Post-Partum Breast Cancer
7. IL-6 Cytokine Family in Mastitis
8. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BC | Breast Cancer |
| CAAs | Cancer Associated Adipocytes, |
| CAFs | Cancer Associated Fibroblasts |
| CSC | Cancer Stem Cells |
| CTCs | Circulating Tumor Cells |
| EMT | Epithelial-to-Mesenchymal Transition |
| ER | Estrogen Receptor |
| ET | Endocrine Therapy |
| gp130 | Glycoprotein130 |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| IL-11 | Interleukin-11 |
| IL-11R | Interleukin-11 Receptor |
| IL-6 | Interleukin-6 |
| IL-6R | Interleukin-6 Receptor |
| LECs | Lymphatic Endothelial Cells |
| LIF | Leukemia Inhibitory Factor |
| LIFR | LIF Receptor |
| MDSCs | Myeloid Derived Stem Cells |
| miR | Micro-RNA |
| OSM | Oncostatin M |
| OSMR | Oncostatin M Receptor |
| PR | Progesterone Receptor |
| pSTAT· | Phosphorylated STAT3 |
| TNBC | Triple Negative Breast Cancer |
References
- Bravo, J.K.; Heath, J.K. Receptor recognition by gp130 cytokines. EMBO J. 2000, 19, 2399–2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulanger, M.J.; Garcia, K.C. Shared cytokine signaling receptors: Structural insights from the gp130 system. Adv. Protein Chem. 2004, 68, 107–146. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Tanaka, T.; Narazaki, M.; Kishimoto, T. Targeting Interleukin-6 Signaling in Clinic. Immunity 2019, 50, 1007–1023. [Google Scholar] [CrossRef]
- Taga, T.; Hibi, M.; Hirata, Y.; Yamasaki, K.; Yasukawa, K.; Matsuda, T.; Hirano, T.; Kishimoto, T. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell 1989, 58, 573–581. [Google Scholar] [CrossRef]
- Hirano, T.; T Yasukawa, K.; Harada, H.; Taga, T.; Watanabe, Y.; Matsuda, T.; Kashiwamura, S.; Nakajima, K.; Koyama, K.; Iwamatsu, A.; et al. Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature 1986, 324, 6092. [Google Scholar] [CrossRef] [PubMed]
- Potter, M.; Boyce, C.R. Induction of plasma-cell neoplasms in strain BALB/c mice with mineral oil and mineral oil adjuvants. Nature 1962, 193, 1086–1087. [Google Scholar] [CrossRef]
- Gauldie, J.; Richards, C.; Harnish, D.; Lansdorp, P.; Baumann, H. Interferon beta 2/B-cell stimulatory factor type 2 shares identity with monocyte-derived hepatocyte-stimulating factor and regulates the major acute phase protein response in liver cells. Proc. Natl. Acad. Sci. USA 1987, 84, 7251–7255. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Boulanger, M.J.; Chow, D.C.; Brevnova, E.E.; Garcia, K.C. Hexameric structure and assembly of the interieukin-6/IL-6 α-recepor/gp130 complex. Science 2003, 300, 2101–2104. [Google Scholar] [CrossRef]
- Saito, M.; Yoshida, K.; Hibi, M.; Taga, T.; Kishimoto, T. Molecular cloning of a murine IL-6 receptor-associated signal transducer, gp130, and its regulated expression in vivo. J. Immunol. 1992, 148, 4066–4071. [Google Scholar]
- Chiu, J.J.; Sgagias, M.K.; Cowan, K.H. Interleukin 6 acts as a paracrine growth factor in human mammary carcinoma cell lines. Clin. Cancer Res. 1996, 2, 215–221. [Google Scholar] [PubMed]
- Jostock, T.; Müllberg, J.; Ozbek, S.; Atreya, R.; Blinn, G.; Voltz, N.; Fischer, M.; Neurath, M.F.; Rose-John, S. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur. J. Biochem. 2001, 268, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S.; Heinrich, P.C. Soluble receptors for cytokines and growth factors: Generation and biological function. Biochem. J. 1994, 300, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S. Il-6 trans-signaling via the soluble IL-6 receptor: Importance for the proinflammatory activities of IL-6. Int. J. Biol. Sci. 2012, 8, 1237–1247. [Google Scholar] [CrossRef]
- Gearing, D.P.; Gough, N.M.; King, J.A.; Hilton, D.J.; Nicola, N.A.; Simpson, R.J.; Nice, E.C.; Kelso, A.; Metcalf, D. Molecular cloning and expression of cDNA encoding a murine myeloid leukaemia inhibitory factor (LIF). EMBO J. 1987, 6, 3995–4002. [Google Scholar] [CrossRef]
- Metcalfe, S.M. LIF in the regulation of T-cell fate and as a potential therapeutic. Genes Immun. 2011, 12, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Hirai, H.; Karian, P.; Kikyo, N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J. 2011, 438, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Gearing, D.P.; Thut, C.J.; VandeBos, T.; Gimpel, S.D.; Delaney, P.B.; King, J.; Price, V.; Cosman, D.; Beckmann, M.P. Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J. 1991, 10, 2839–2848. [Google Scholar] [CrossRef]
- Chambers, I.; Cozens, A.; Broadbent, J.; Robertson, M.; Lee, M.; Li, M.; Smith, A. Structure of the mouse leukaemia inhibitory factor receptor gene: Regulated expression of mRNA encoding a soluble receptor isoform from an alternative 5′ untranslated region. Biochem. J. 1997, 328, 879–888. [Google Scholar] [CrossRef] [Green Version]
- Tomida, M. Structural and functional studies on the leukemia inhibitory factor receptor (LIF-R): Gene and soluble form of LIF-R, and cytoplasmic domain of LIF-R required for differentiation and growth arrest of myeloid leukemic cells. Leuk. Lymphoma 2000, 37, 517–525. [Google Scholar] [CrossRef]
- Metz, S.; Naeth, G.; Heinrich, P.C.; Müller-Newen, G. Novel inhibitors for murine and human leukemia inhibitory factor based on fused soluble receptors. J. Biol. Chem. 2008, 283, 5985–5995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicola, N.A.; Babon, J.J. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 2015, 26, 533–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarling, J.M.; Shoyab, M.; Marquardt, H.; Hanson, M.B.; Lioubin, M.N.; Todaro, G.J. Oncostatin M: A growth regulator produced by differentiated histiocytic lymphoma cells. Proc. Natl. Acad. Sci. USA 1986, 83, 9739–9743. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Miyahima, A. Oncostatin M, a multifunctional cytokine. Rev. Physiol. Biochem. Pharmacol. 2003, 149, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Deller, M.C.; Hudson, K.R.; Ikemizu, S.; Bravo, J.; Jones, E.Y.; Heath, J.K. Crystal structure and functional dissection of the cytostatic cytokine oncostatin M. Structure 2000, 8, 863–874. [Google Scholar] [CrossRef]
- Gearing, D.P.; Comeau, M.R.; Friend, D.J.; Gimpel, S.D.; Thut C., J.; McGourty, J.; Brasher K., K.; King J., A.; Gillis, S.; Mosley, B.; et al. The IL-6 signal transducer, gp130: An oncostatin M receptor and affinity converter for the LIF receptor. Science 1992, 255, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.R.; Bennett, F.; Calvetti, J.A.; Kelleher, K.; Wood, C.R.; O’Hara, R.M.; Leary, A.C.; Sibley, B.; Clark, S.C.; Williams, D.A.; et al. Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc. Natl. Acad. Sci. USA 1990, 87, 7512–7516. [Google Scholar] [CrossRef] [Green Version]
- Maroni, P.; Bendinelli, P.; Ferraretto, A.; Lombardi, G. Interleukin 11 (IL-11): Role(s) in Breast Cancer Bone Metastases. Biomedicines 2021, 9, 659. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Uversky, V.N.; Permyakov, S.E. Interleukin-11: A Multifunctional Cytokine with Intrinsically Disordered Regions. Cell Biochem. Biophys. 2016, 74, 285–296. [Google Scholar] [CrossRef]
- Lokau, J.; Nitz, R.; Agthe, M.; Monhasery, N.; Aparicio-Siegmund, S.; Schumacher, N.; Wolf, J.; Möller-Hackbarth, K.; Waetzig, G.H.; Grötzinger, J.; et al. Proteolytic Cleavage Governs Interleukin-11 Trans-signaling. Cell Rep. 2016, 14, 1761–1773. [Google Scholar] [CrossRef] [Green Version]
- Omokehinde, T.; Johnson, R.W. Gp130 cytokines in breast cancer and bone. Cancers 2020, 12, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaiwangyen, W.; Ospina-Prieto, S.; Morales-Prieto, D.M.; Pereira de Sousa, F.L.; Pastuschek, J.; Fitzgerald, J.S.; Schleussner, E.; Markert, U.R. Oncostatin M and leukaemia inhibitory factor trigger signal transducer and activator of transcription 3 and extracellular signal-regulated kinase 1/2 pathways but result in heterogeneous cellular responses in trophoblast cells. Reprod. Fertil. Dev. 2016, 28, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Perugini, J.; Di Mercurio, E.; Tosseta, G.; Severi, I.; Monaco, F.; Reguzzoni, M.; Tomasetti, M.; Dani, C.; Cinti, S.; Giordano, A. Biological Effects of Ciliary Neurotrophic Factor on hMADS Adipocytes. Front. Endocrinol. 2019, 10, 768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24, S26–S35. [Google Scholar] [CrossRef] [Green Version]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, R.; Arima, N. Is triple negative a prognostic factor in breast cancer? Breast Cancer 2008, 15, 303–308. [Google Scholar] [CrossRef]
- Studebaker, A.W.; Storci, G.; Werbeck, J.L.; Sansone, P.; Sasser, A.K.; Tavolari, S.; Huang, T.; Chan, M.W.; Marini, F.C.; Rosol, T.J.; et al. Fibroblasts Isolated from Common Sites of Breast Cancer Metastasis Enhance Cancer Cell Growth Rates and Invasiveness in an Interleukin-6–Dependent Manner. Cancer Res. 2008, 68, 9087–9095. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.A.; Jenkins, B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018, 18, 773–789. [Google Scholar] [CrossRef]
- Berishaj, M.; Gao, S.P.; Ahmed, S.; Leslie, K.; Al-Ahmadie, H.; Gerald, W.L.; Bornmann, W.; Bromberg, J.F. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, K.; Watson, C.J. The multifaceted role of STAT3 in mammary gland involution and breast cancer. Int. J. Mol. Sci. 2018, 19, 1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ásgeirsson, K.S.; Ólafsdóttir, K.; Jónasson, J.G.; Ógmundsdóttir, H.M. The effects of IL-6 on cell adhesion and E-cadherin expression in breast cancer. Cytokine 1998, 10, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Kozłowski, L.; Zakrzewska, I.; Tokajuk, P.; Wojtukiewicz, M.Z. Concentration of interleukin-6 (IL-6), interleukin-8 (IL-8) and interleukin-10 (IL-10) in blood serum of breast cancer patients. Rocz. Akad. Med. Białymstoku 2003, 48, 82–84. [Google Scholar] [PubMed]
- Salgado, R.; Junius, S.; Benoy, I.; Van Dam, P.; Vermeulen, P.; Van Marck, E.; Huget, P.; Dirix, L.Y. Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. Int. J. Cancer 2003, 103, 642–646. [Google Scholar] [CrossRef]
- Ahmad, N.; Ammar, A.; Storr, S.J.; Green, A.R.; Rakha, E.; Ellis, I.O.; Martin, S.G. IL-6 and IL-10 are associated with good prognosis in early stage invasive breast cancer patients. Cancer Immunol. Immunother. 2018, 67, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Sasser, A.K.; Sullivan, N.J.; Studebaker, A.W.; Hendey, L.F.; Axel, A.E.; Hall, B.M. Interleukin-6 is a potent growth factor for ER-α-positive human breast cancer. FASEB J. 2007, 21, 3763–3770. [Google Scholar] [CrossRef]
- Liu, H.; Liu, K.; Bodenner, D.L. Estrogen receptor inhibits interleukin-6 gene expression by disruption of nuclear factor kappaB transactivation. Cytokine 2005, 31, 251–257. [Google Scholar] [CrossRef]
- Tamm, I.; Cardinale, I.; Krueger, J.; Murphy, J.S.; May, L.T.; Sehgal, P.B. Interleukin 6 decreases cell-cell association and increases motility of ductal breast carcinoma cells. J. Exp. Med. 1989, 170, 1649–1669. [Google Scholar] [CrossRef]
- Badache, A.; Hynes, N.E. Interleukin 6 Inhibits Proliferation and, in Cooperation with an Epidermal Growth Factor Receptor Autocrine Loop, Increases Migration of T47D Breast Cancer Cells 1. Cancer Res. 2001, 61, 383–391. [Google Scholar]
- Sansone, P.; Storci, G.; Tavolari, S.; Guarnieri, T.; Giovannini, C.; Taffurelli, M.; Ceccarelli, C.; Santini, D.; Paterini, P.; Marcu, K.B.; et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. Am. Soc. Clin. Investig. 2007, 117, 3988–4002. Available online: https://www.jci.org/articles/view/32533 (accessed on 1 January 2022). [CrossRef] [PubMed]
- Iliopoulos, D.; Hirsch, H.A.; Wang, G.; Struhl, K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc. Natl. Acad. Sci. USA 2011, 108, 1397–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, K.; Lee, O.Y.; Shon, S.Y.; Nam, O.; Ryu, P.M.; Seo, M.W.; Lee, D.S. A mutual activation loop between breast cancer cells and myeloid-derived suppressor cells facilitates spontaneous metastasis through IL-6 trans-signaling in a murine model. Breast Cancer Res. 2013, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, K.; Pandey, N.B.; Popel, A.S. Simultaneous blockade of IL-6 and CCL5 signaling for synergistic inhibition of triple-negative breast cancer growth and metastasis. Breast Cancer Res. 2018, 20, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dirat, B.; Bochet, L.; Dabek, M.; Daviaud, D.; Dauvillier, S.; Majed, B.; Wang, Y.Y.; Meulle, A.; Salles, B.; Le Gonidec, S.; et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011, 71, 2455–2465. [Google Scholar] [CrossRef] [Green Version]
- Gyamfi, J.; Eom, M.; Koo, J.S.; Choi, J. Multifaceted Roles of Interleukin-6 in Adipocyte–Breast Cancer Cell Interaction. Transl. Oncol. 2018, 11, 275–285. [Google Scholar] [CrossRef]
- Hoene, M.; Weigert, C. The role of interleukin-6 in insulin resistance, body fat distribution and energy balance. Obes. Rev. 2008, 9, 20–29. [Google Scholar] [CrossRef]
- Knüpfer, H.; Preiß, R. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res. Treat. 2007, 102, 129–135. [Google Scholar] [CrossRef]
- Walter, M.; Liang, S.; Ghosh, S.; Hornsby, P.J.; Li, R. Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene 2009, 28, 2745–2755. [Google Scholar] [CrossRef] [Green Version]
- Matutino, A.; Joy, A.A.; Brezden-Masley, C.; Chia, S.; Verma, S. Hormone receptor-positive, HER2-negative metastatic breast cancer: Redrawing the lines. Curr. Oncol. 2018, 25, S131–S141. [Google Scholar] [CrossRef] [Green Version]
- Masjedi, A.; Hashemi, V.; Hojjat-Farsangi, M.; Ghalamfarsa, G.; Azizi, G.; Yousefi, M.; Jadidi-Niaragh, F. The significant role of interleukin-6 and its signaling pathway in the immunopathogenesis and treatment of breast cancer. Biomed. Pharmacother. 2018, 108, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Kim, G.I.; Davis, A.; Malik, F.; Henry, N.L.; Ithimakin, S.; Quraishi, A.A.; Tawakkol, N.; D’Angelo, R.; Paulson, A.K.; et al. Activation of an IL6 Inflammatory Loop Mediates Trastuzumab Resistance in HER2+ Breast Cancer by Expanding the Cancer Stem Cell Population. Mol. Cell 2012, 47, 570–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoni, M.; Romagnoli, E.; Saladino, T.; Foghini, L.; Guarino, S.; Capponi, M.; Giannini, M.; Cognigni, P.D.; Ferrara, G.; Battelli, N. Triple negative breast cancer: Key role of tumor-associated macrophages in regulating the activity of anti-PD-1/PD-L1 agents. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Z.C.; Poage, G.M.; den Hollander, P.; Tsimelzon, A.; Hill, J.; Panupinthu, N.; Zhang, Y.; Mazumdar, A.; Hilsenbeck, S.G.; Mills, G.B.; et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013, 73, 3470–3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.; Chen, Z.; Jiang, G.; Zhou, Y.; Liu, Q.; Su, Q.; Wei, W.; Du, J.; Wang, H. Activation of GPER suppresses migration and angiogenesis of triple negative breast cancer via inhibition of NF-κB/IL-6 signals. Cancer Lett. 2017, 386, 12–23. [Google Scholar] [CrossRef] [PubMed]
- García-Tuñón, I.; Ricote, M.; Ruiz, A.; Fraile, B.; Paniagua, R.; Royuela, M. OSM, LIF, its receptors, and its relationship with the malignance in human breast carcinoma (in situ and in infiltrative). Cancer Investig. 2008, 26, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Zhao, Y.; Zhang, C.; Li, J.; Liu, Z.; Liu, J.; Hu, W. Leukemia inhibitory factor promotes EMT through STAT3-dependent miR-21 induction. Oncotarget 2016, 7, 3777–3790. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, Q.; Yu, H.; Wu, L.; Zhao, Y.; Zhang, C.; Yue, X.; Liu, Z.; Wu, H.; Haffty, B.G.; et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014, 5, 788–801. [Google Scholar] [CrossRef] [Green Version]
- Quaglino, A.; Schere-Levy, C.; Romorini, L.; Meiss, R.P.; Kordon, E.C. Mouse mammary tumors display Stat3 activation dependent on leukemia inhibitory factor signaling. Breast Cancer Res. 2007, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Albrengues, J.; Bourget, I.; Pons, C.; Butet, V.; Hofman, P.; Tartare-Deckert, S.; Feral, C.C.; Meneguzzi, G.; Gaggioli, C. LIF mediates proinvasive activation of stromal fibroblasts in cancer. Cell Rep. 2014, 7, 1664–1678. [Google Scholar] [CrossRef] [Green Version]
- Iorns, E.; Ward, T.M.; Dean, S.; Jegg, A.; Thomas, D.; Murugaesu, N.; Sims, D.; Mitsopoulos, C.; Fenwick, K.; Kozarewa, I.; et al. Whole genome in vivo RNAi screening identifies the leukemia inhibitory factor receptor as a novel breast tumor suppressor. Breast Cancer Res. Treat. 2012, 135, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, Y.; Wei, Y.; Zhang, P.; Rezaeian, A.H.; Teruya-Feldstein, J.; Gupta, S.; Liang, H.; Lin, H.K.; Hung, M.C.; et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 2012, 18, 1511–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, R.W.; Finger, E.C.; Olcina, M.M.; Vilalta, M.; Aguilera, T.; Miao, Y.; Merkel, A.R.; Johnson, J.R.; Sterling, J.A.; Wu, J.Y.; et al. Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nat. Cell Biol. 2016, 18, 1078–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Qu, J.; Jin, N.; Xu, J.; Lin, C.; Chen, Y.; Yang, X.; He, X.; Tang, S.; Lan, X.; et al. Feedback Activation of Leukemia Inhibitory Factor Receptor Limits Response to Histone Deacetylase Inhibitors in Breast Cancer. Cancer Cell 2016, 30, 459–473. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Hunter, S.; Hunter, T. Stem Cell Factor LIFted as a Promising Clinical Target for Cancer Therapy. Mol. Cancer Ther. 2019, 18, 1337–1340. [Google Scholar] [CrossRef] [Green Version]
- Viswanadhapalli, S.; Luo, Y.; Sareddy, G.R.; Santhamma, B.; Zhou, M.; Li, M.; Ma, S.; Sonavane, R.; Pratap, U.P.; Altwegg, K.A.; et al. EC359: A First-in-Class Small-Molecule Inhibitor for Targeting Oncogenic LIFR Signaling in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2019, 18, 1341–1354. [Google Scholar] [CrossRef]
- Ghanei, Z.; Mehri, N.; Jamshidizad, A.; Joupari, M.D.; Shamsara, M. Immunization against leukemia inhibitory factor and its receptor suppresses tumor formation of breast cancer initiating cells in BALB/c mouse. Sci. Rep. 2020, 10, 11465. [Google Scholar] [CrossRef]
- Lapeire, L.; Hendrix, A.; Lambein, K.; Van Bockstal, M.; Braems, G.; Van Den Broecke, R.; Limame, R.; Mestdagh, P.; Vandesompele, J.; Vanhove, C.; et al. Cancer-associated adipose tissue promotes breast cancer progression by paracrine oncostatin M and Jak/STAT3 signaling. Cancer Res. 2014, 74, 6806–6819. [Google Scholar] [CrossRef] [Green Version]
- Holzer, R.G.; Ryan, R.E.; Tommack, M.; Schlekeway, E.; Jorcyk, C.L. Oncostatin M stimulates the detachment of a reservoir of invasive mammary carcinoma cells: Role of cyclooxygenase-2. Clin. Exp. Metastasis 2004, 21, 167–176. [Google Scholar] [CrossRef]
- West, N.R.; Murphy, L.C.; Watson, P.H. Oncostatin M suppresses oestrogen receptor-α expression and is associated with poor outcome in human breast cancer. Endocr.-Relat. Cancer 2012, 19, 181–195. [Google Scholar] [CrossRef] [Green Version]
- Tawara, K.; Scott, H.; Emathinger, J.; Wolf, C.; LaJoie, D.; Hedeen, D.; Bond, L.; Montgomery, P.; Jorcyk, C. High expression of OSM and IL-6 are associated with decreased breast cancer survival: Synergistic induction of IL-6 secretion by OSM and IL-1β. Oncotarget 2019, 10, 2068–2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, N.R.; Murray, J.I.; Watson, P.H. Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene 2014, 33, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Junk, D.J.; Bryson, B.L.; Smigiel, J.M.; Parameswaran, N.; Bartel, C.A.; Jackson, M.W. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 2017, 36, 4001–4013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doherty, M.R.; Parvani, J.G.; Tamagno, I.; Junk, D.J.; Bryson, B.L.; Cheon, H.J.; Stark, G.R.; Jackson, M.W. The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer. Breast Cancer Res. 2019, 21, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolin, C.; Tawara, K.; Sutherland, C.; Redshaw, J.; Aranda, P.; Moselhy, J.; Anderson, R.; Jorcyk, C.L. Oncostatin M promotes mammary tumor metastasis to bone and osteolytic bone degradation. Genes Cancer 2012, 3, 117–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tawara, K.; Bolin, C.; Koncinsky, J.; Kadaba, S.; Covert, H.; Sutherland, C.; Bond, L.; Kronz, J.; Garbow, J.R.; Jorcyk, C.L. OSM potentiates preintravasation events, increases CTC counts, and promotes breast cancer metastasis to the lung. Breast Cancer Res. 2018, 20, 1–18. [Google Scholar] [CrossRef]
- Reid, J.; Zamuner, S.; Edwards, K.; Rumley, S.A.; Nevin, K.; Feeney, M.; Zecchin, C.; Fernando, D.; Wisniacki, N. In vivo affinity and target engagement in skin and blood in a first-time-in-human study of an anti-oncostatin M monoclonal antibody. Br. J. Clin. Pharmacol. 2018, 84, 2280–2291. [Google Scholar] [CrossRef] [Green Version]
- Hanavadi, S.; Martin, T.A.; Watkins, G.; Mansel, R.E.; Jiang, W.G. Expression of Interleukin 11 and Its Receptor and Their Prognostic Value in Human Breast Cancer. Ann. Surg. Oncol. 2006, 13, 802–808. [Google Scholar] [CrossRef]
- Bockhorn, J.; Dalton, R.; Nwachukwu, C.; Huang, S.; Prat, A.; Yee, K.; Chang, Y.F.; Huo, D.; Wen, Y.; Swanson, K.E.; et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013, 4, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Samaeekia, R.; Adorno-Cruz, V.; Bockhorn, J.; Chang, Y.F.; Huang, S.; Prat, A.; Ha, N.; Kibria, G.; Huo, D.; Zheng, H.; et al. miR-206 Inhibits Stemness and Metastasis of Breast Cancer by Targeting MKL1/IL11 Pathway. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Wang, X.; Dong, Z.; Liu, J.; Zhang, S. Bone metastasis from breast cancer involves elevated IL-11 expression and the gp130/STAT3 pathway. Med. Oncol. 2013, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cosphiadi, I.; Atmakusumah, T.D.; Siregar, N.C.; Muthalib, A.; Harahap, A.; Mansyur, M. Bone Metastasis in Advanced Breast Cancer: Analysis of Gene Expression Microarray. Clin. Breast Cancer 2018, 18, e1117–e1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotiriou, C.; Lacroix, M.; Lespagnard, L.; Larsimont, D.; Paesmans, M.; Body, J.J. Interleukins-6 and -11 expression in primary breast cancer and subsequent development of bone metastases. Cancer Lett. 2001, 169, 87–95. [Google Scholar] [CrossRef]
- Morgan, H.; Tumber, A.; Hill, P.A. Breast cancer cells induce osteoclast formation by stimulating host IL-11 production and downregulating granulocyte/macrophage colony-stimulating factor. Int. J. Cancer 2004, 109, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, H.; Onodera, Y.; Murakami, S.; Suzuki, T.; Motohashi, H. IL-11 contribution to tumorigenesis in an NRF2 addiction cancer model. Oncogene 2017, 36, 6315–6324. [Google Scholar] [CrossRef]
- Callihan, E.B.; Gao, D.; Jindal, S.; Lyons, T.R.; Manthey, E.; Edgerton, S.; Urquhart, A.; Schedin, P.; Borges, V.F. Postpartum diagnosis demonstrates a high risk for metastasis and merits an expanded definition of pregnancy-associated breast cancer. Breast Cancer Res. Treat. 2013, 138, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Goddard, E.T.; Bassale, S.; Schedin, T.; Jindal, S.; Johnston, J.; Cabral, E.; Latour, E.; Lyons, T.R.; Mori, M.; Schedin, P.J.; et al. Association Between Postpartum Breast Cancer Diagnosis and Metastasis and the Clinical Features Underlying Risk. JAMA Netw. Open 2019, 2, e186997. [Google Scholar] [CrossRef] [Green Version]
- Jindal, S.; Gao, D.; Bell, P.; Albrektsen, G.; Edgerton, S.M.; Ambrosone, C.B.; Thor, A.D.; Borges, V.F.; Schedin, P. Postpartum breast involution reveals regression of secretory lobules mediated by tissue-remodeling. Breast Cancer Res. 2014, 16, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lund, L.R.; Rømer, J.; Thomasset, N.; Solberg, H.; Pyke, C.; Bissell, M.J.; Danø, K.; Werb, Z. Two distinct phases of apoptosis in mammary gland involution: Proteinase-independent and -dependent pathways. Development 1996, 122, 181. [Google Scholar] [CrossRef]
- Kritikou, E.A.; Sharkey, A.; Abell, K.; Came, P.J.; Anderson, E.; Clarkson, R.W.; Watson, C.J. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development 2003, 130, 3459–3468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schere-Levy, C.; Buggiano, V.; Quaglino, A.; Gattelli, A.; Cirio, M.C.; Piazzon, I.; Vanzulli, S.; Kordon, E.C. Leukemia inhibitory factor induces apoptosis of the mammary epithelial cells and participates in mouse mammary gland involution. Exp. Cell Res. 2003, 282, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Melenhorst, J.J.; Hennighausen, L. Loss of Interleukin 6 Results in Delayed Mammary Gland Involution: A Possible Role for Mitogen-Activated Protein Kinase and Not Signal Transducer and Activator of Transcription 3. Mol. Endocrinol. 2002, 16, 2902–2912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiffen, P.G.; Omidvar, N.; Marquez-Almuina, N.; Croston, D.; Watson, C.J.; Clarkson, R.W.E. A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo. Mol. Endocrinol. 2008, 22, 2677–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyszynski, A.; Hong, C.C.; Lam, K.; Michailidou, K.; Lytle, C.; Yao, S.; Zhang, Y.; Bolla, M.K.; Wang, Q.; Dennis, J.; et al. An intergenic risk locus containing an enhancer deletion in 2q35 modulates breast cancer risk by deregulating IGFBP5 expression. Hum. Mol. Genet. 2016, 25, 3863–3876. [Google Scholar] [CrossRef] [Green Version]
- Takabatake, Y.; Oxvig, C.; Nagi, C.; Adelson, K.; Jaffer, S.; Schmidt, H.; Keely, P.J.; Eliceiri, K.W.; Mandeli, J.; Germain, D. Lactation opposes pappalysin-1-driven pregnancy-associated breast cancer. EMBO Mol. Med. 2016, 8, 388–406. [Google Scholar] [CrossRef]
- Basree, M.M.; Shinde, N.; Koivisto, C.; Cuitino, M.; Kladney, R.; Zhang, J.; Stephens, J.; Palettas, M.; Zhang, A.; Kim, H.K.; et al. Abrupt involution induces inflammation, estrogenic signaling, and hyperplasia linking lack of breastfeeding with increased risk of breast cancer. Breast Cancer Res. 2019, 21, 80. [Google Scholar] [CrossRef] [Green Version]
- Borges, V.F.; Lyons, T.R.; Germain, D.; Schedin, P. Postpartum Involution and Cancer: An Opportunity for Targeted Breast Cancer Prevention and Treatments? Cancer Res. 2020, 80, 1790–1798. [Google Scholar] [CrossRef] [Green Version]
- Wambach, K.A. Lactation mastitis: A descriptive study of the experience experiencia. J. Hum. Lact. 2003, 19, 24–34. [Google Scholar] [CrossRef]
- Winder, C.B.; Sargeant, J.M.; Hu, D.; Wang, C.; Kelton, D.F.; Godkin, M.A.; Churchill, K.J.; O’Connor, A.M. Comparative efficacy of antimicrobials for treatment of clinical mastitis in lactating dairy cattle: A systematic review and network meta-analysis. Anim. Health Res. Rev. 2019, 20, 229–246. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, Y.; Dong, X.; Chen, J.; Wang, Z.; Chen, J.; Dong, G. The synergism of PGN, LTA and LPS in inducing transcriptome changes, inflammatory responses and a decrease in lactation aswell as the associated epigenetic mechanisms in bovine mammary epithelial cells. Toxins 2020, 12, 387. [Google Scholar] [CrossRef] [PubMed]
- Vangroenweghe, F.; Lamote, I.; Burvenich, C. Physiology of the periparturient period and its relation to severity of clinical mastitis. Domest. Anim. Endocrinol. 2005, 29, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Taponen, S.; Pyorala, S. Coagulase-negative staphylococci as cause of bovine mastitis—Not so different from Staphylococcus aureus? Vet. Microbiol. 2009, 134, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Günther, J.; Esch, K.; Poschadel, N.; Petzl, W.; Zerbe, H.; Mitterhuemer, S.; Blum, H.; Seyfert, H.M. Comparative kinetics of Escherichia coli-And Staphylococcus aureus-Specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha. Infect. Immun. 2011, 79, 695–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa-Cesnik, C. Lactation Mastitis. JAMA 2003, 289, 1609. [Google Scholar] [CrossRef] [PubMed]
- Sakemi, Y.; Tamura, Y.; Hagiwara, K. Interleukin-6 in quarter milk as a further prediction marker for bovine subclinical mastitis. J. Dairy Res. 2011, 78, 118–121. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Jiang, Q.; Hao, H.; Ju, Z.; Yang, C.; Sun, Y.; Wang, C.; Zhong, J.; Huang, J.; et al. DNA methylation rather than single nucleotide polymorphisms regulates the production of an aberrant splice variant of IL6R in mastitic cows. Cell Stress Chaperones 2018, 23, 617–628. [Google Scholar] [CrossRef]
- Zhu, Y.; Berg, M.; Fossum, C.; Magnusson, U. Proinflammatory cytokine mRNA expression in mammary tissue of sows following intramammary inoculation with Escherichia coli. Vet. Immunol. Immunopathol. 2007, 116, 98–103. [Google Scholar] [CrossRef]
- Mizuno, K.; Hatsuno, M.; Aikawa, K.; Takeichi, H.; Himi, T.; Kaneko, A.; Kodaira, K.; Takahashi, H.; Itabashi, K. Mastitis is associated with IL-6 levels and milk fat globule size in breast milk. J. Hum. Lact. 2012, 28, 529–534. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Zhou, Y.H.; Jiang, Y.N.; Zhang, W.; Tang, X.J.; Ren, Y.; Han, S.P.; Liu, P.J.; Xu, J.; et al. IL-6/STAT3 signaling pathway is activated in plasma cell mastitis. Int. J. Clin. Exp. Pathol. 2015, 8, 12541. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4680387 (accessed on 30 December 2021).
- Liu, Y.; Sun, Y.; Zhou, Y.; Tang, X.; Wang, K.; Ren, Y.; He, J. Sinomenine hydrochloride inhibits the progression of plasma cell mastitis by regulating IL-6/JAK2/STAT3 pathway. Int. Immunopharmacol. 2020, 81, 106025. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Y.; Yan, Z.; Yang, H.; Sun, W.; Yao, Y.; Chen, Y.; Jiang, R. IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J. ImmunoTher. Cancer 2020, 8, e000285. [Google Scholar] [CrossRef] [PubMed]
| Topic | Cytokines [Refs] |
|---|---|
| Stat-3 phosphorylation induction | IL-6 [41]; LIF [67,69,70]; OSM [83] |
| High expression and/or circulating levels are associated with cancer stage or prognosis | IL-6 [44,45]; LIF [66]; OSM [66,81,82]; IL-11 [88] |
| ER+ breast cancer cells express and/or secrete lower cytokine levels than ER− cells | IL-6 [11,47,48]; LIF [67] |
| CSC maintenance, EMT, cell migration and/or invasion induction | IL-6 [43,49,50,51,52]; LIF [67]; OSM [78,79,82,83,84] |
| Expression by diverse cell types in tumor microenvironment | IL-6 [31,38,47,53,54,55,56,57,59]; IL-11 [94,96] |
| Association with Her2 or endocrine therapy resistance | IL-6 [61,62] |
| Induction of TNBC progression | IL-6 [54,64,65] |
| Use in targeted therapy | IL-6 [54,62]; LIF [74,75,76,77]; OSM [87] |
| Receptor as tumor suppressor | LIF [71,72,73] |
| Tumor miRNA modulation | LIF [67]; IL-11 [89,90] |
| Involvement in metastatic progression | OSM [85,86]; IL-11 [91,92,93,94,95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felcher, C.M.; Bogni, E.S.; Kordon, E.C. IL-6 Cytokine Family: A Putative Target for Breast Cancer Prevention and Treatment. Int. J. Mol. Sci. 2022, 23, 1809. https://doi.org/10.3390/ijms23031809
Felcher CM, Bogni ES, Kordon EC. IL-6 Cytokine Family: A Putative Target for Breast Cancer Prevention and Treatment. International Journal of Molecular Sciences. 2022; 23(3):1809. https://doi.org/10.3390/ijms23031809
Chicago/Turabian StyleFelcher, Carla M., Emilia S. Bogni, and Edith C. Kordon. 2022. "IL-6 Cytokine Family: A Putative Target for Breast Cancer Prevention and Treatment" International Journal of Molecular Sciences 23, no. 3: 1809. https://doi.org/10.3390/ijms23031809
APA StyleFelcher, C. M., Bogni, E. S., & Kordon, E. C. (2022). IL-6 Cytokine Family: A Putative Target for Breast Cancer Prevention and Treatment. International Journal of Molecular Sciences, 23(3), 1809. https://doi.org/10.3390/ijms23031809






