Drought-Responsive NAC Transcription Factor RcNAC72 Is Recognized by RcABF4, Interacts with RcDREB2A to Enhance Drought Tolerance in Arabidopsis
Abstract
:1. Introduction
2. Results
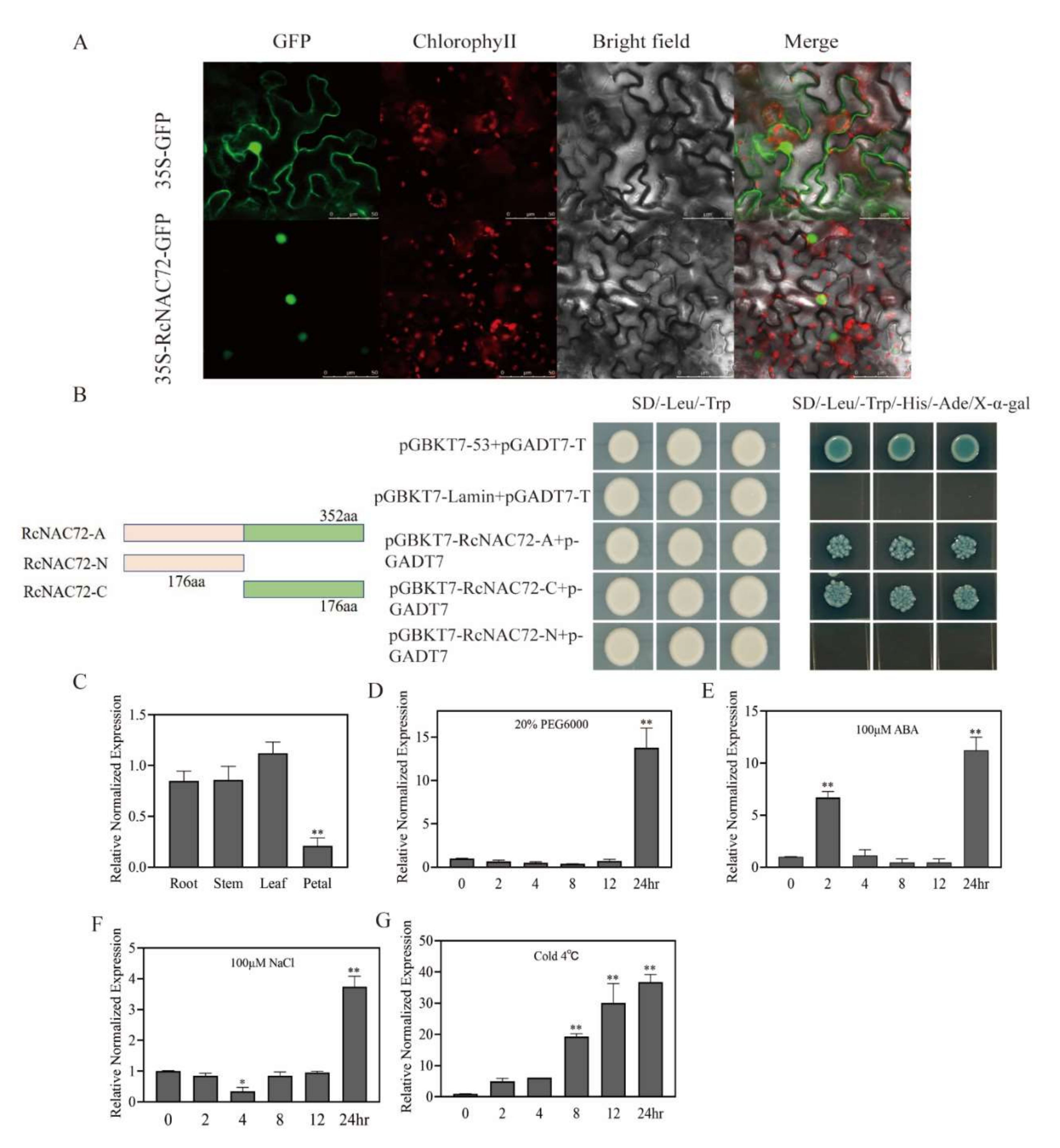
2.1. Bioinformatics Analysis of RcNAC72
2.2. RcNAC72 Tissue Specificity and Expression Analysis under Abiotic Stress
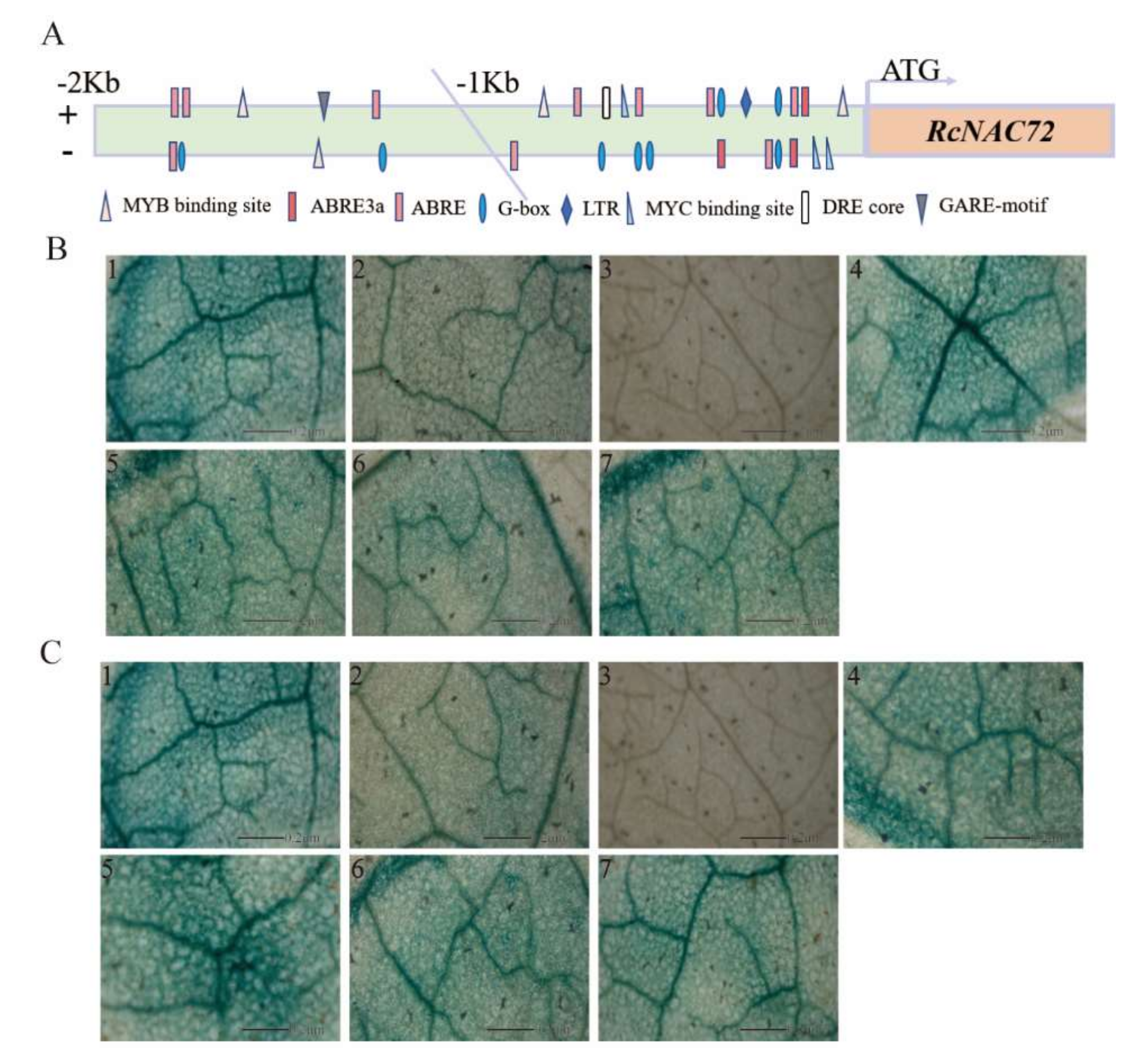
2.3. Promoter Analysis of RcNAC72
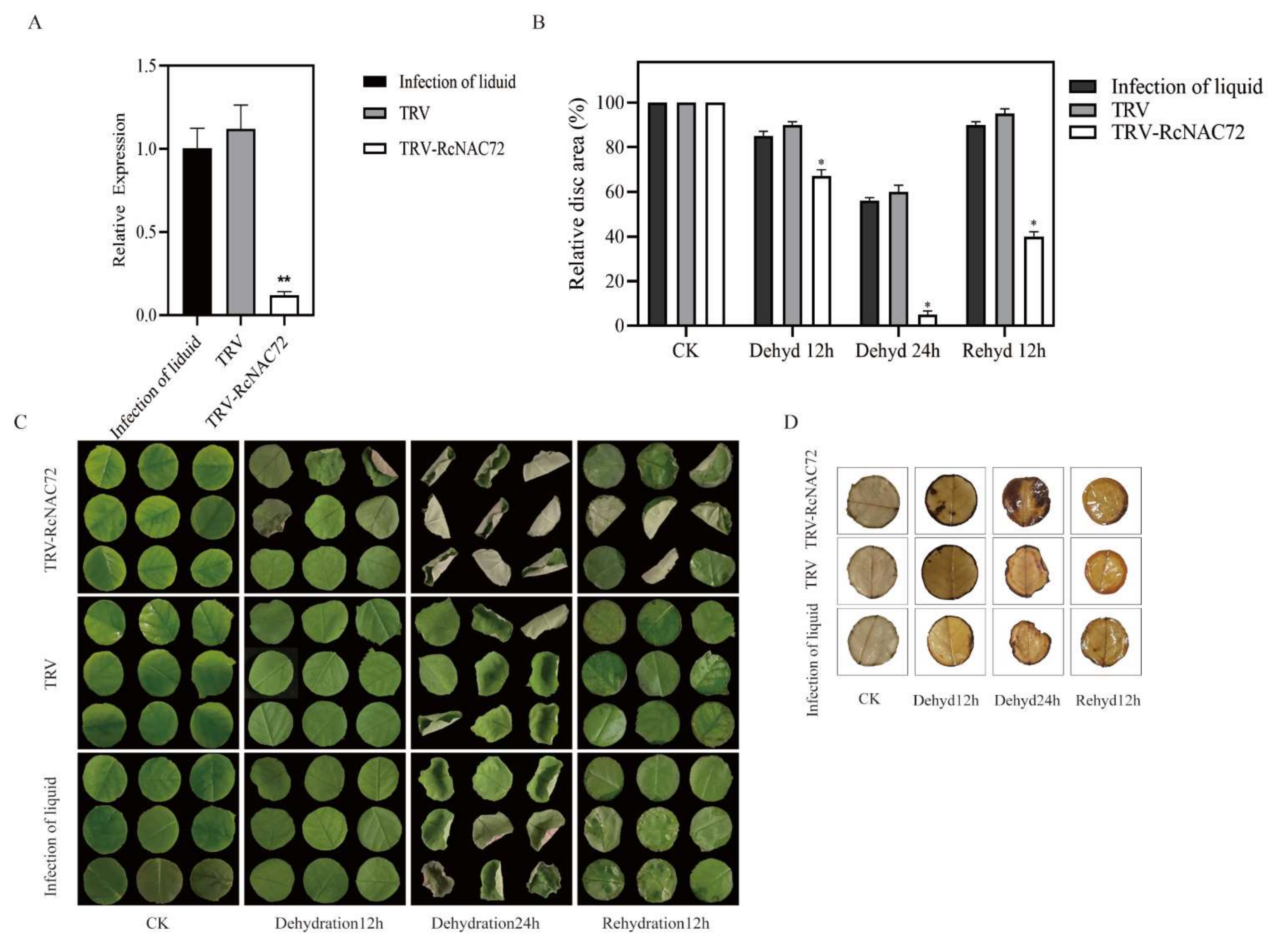
2.4. Silencing RcNAC72 by VIGS Reduced Dehydration Tolerance in Rose Leaf
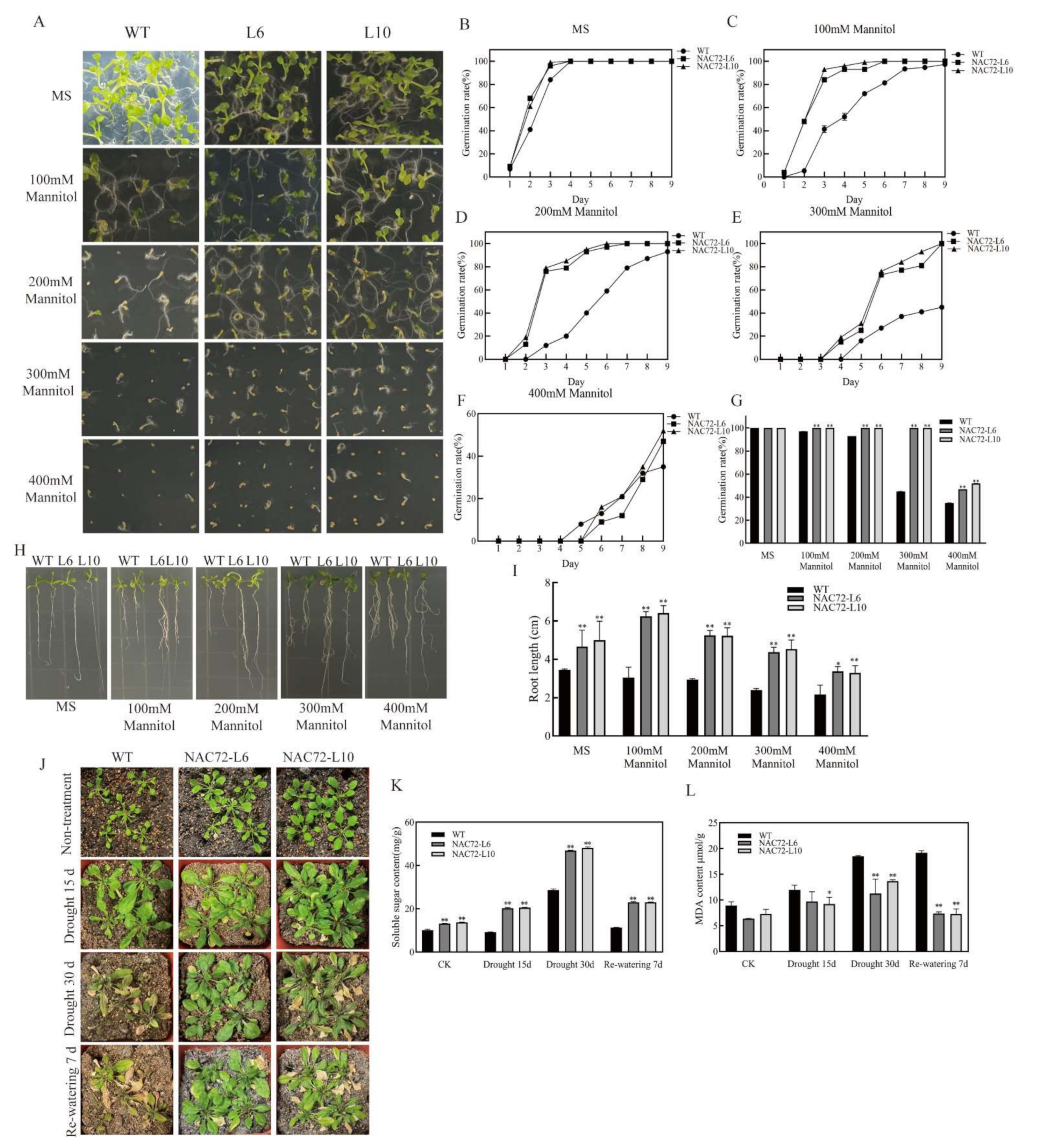
2.5. Overexpression of RcNAC72 in Arabidopsis Enhance Tolerance to Drought Stress
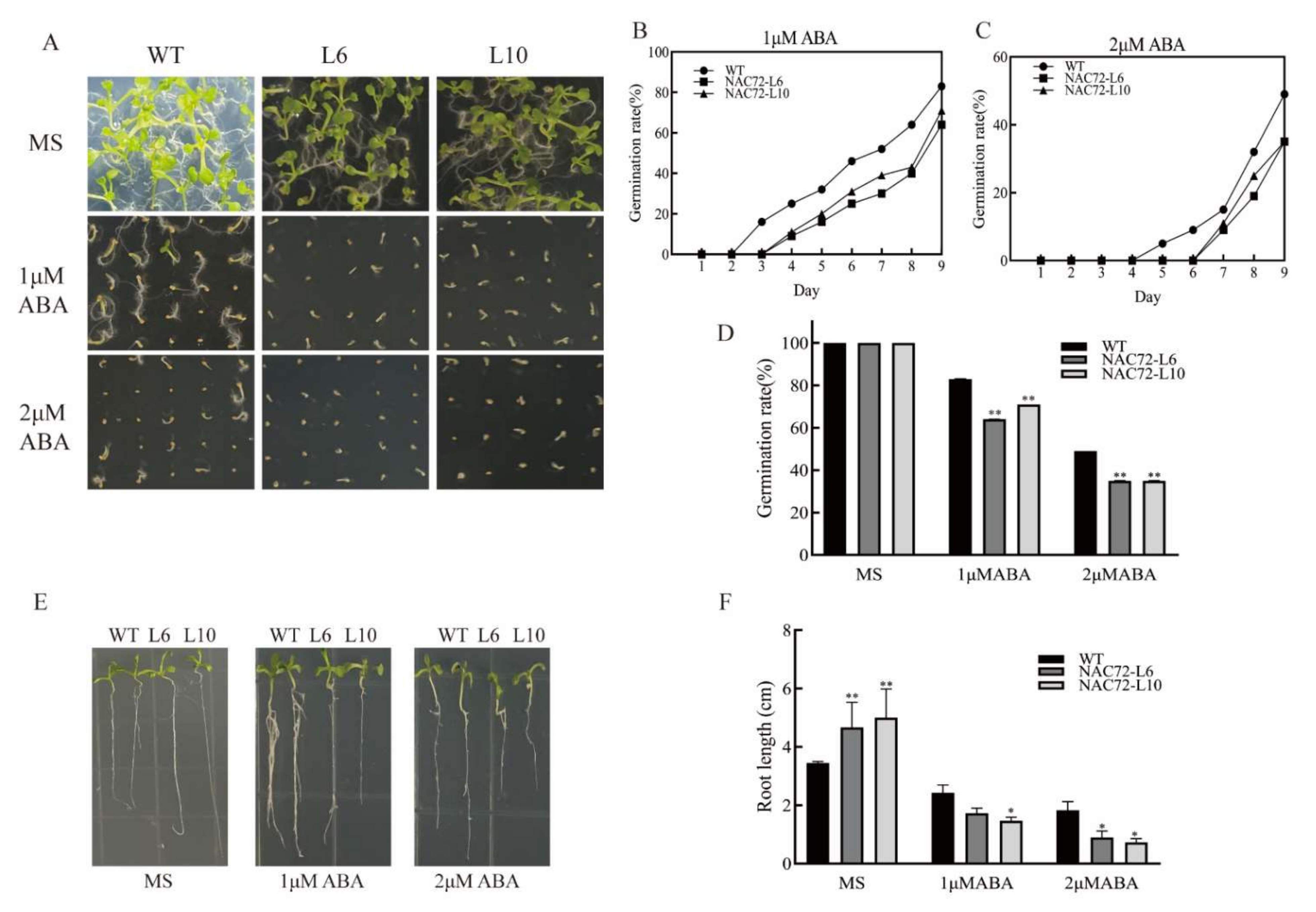
2.6. Overexpression of RcNAC72 in Arabidopsis Enhanced Sensitivity to ABA
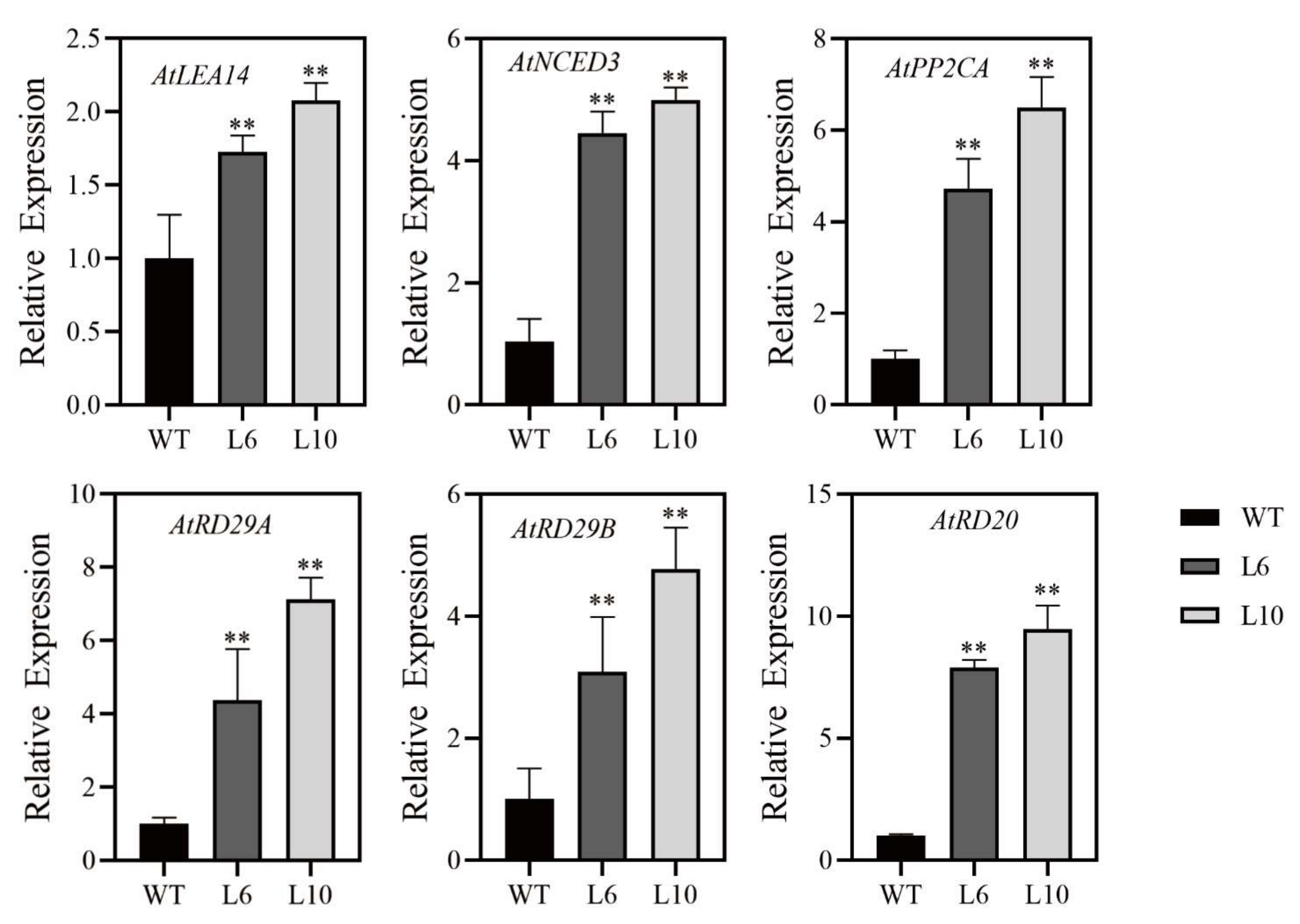
2.7. Altered Expression of Stress-Related Genes in Overexpression RcNAC72 Arabidopsis Plants
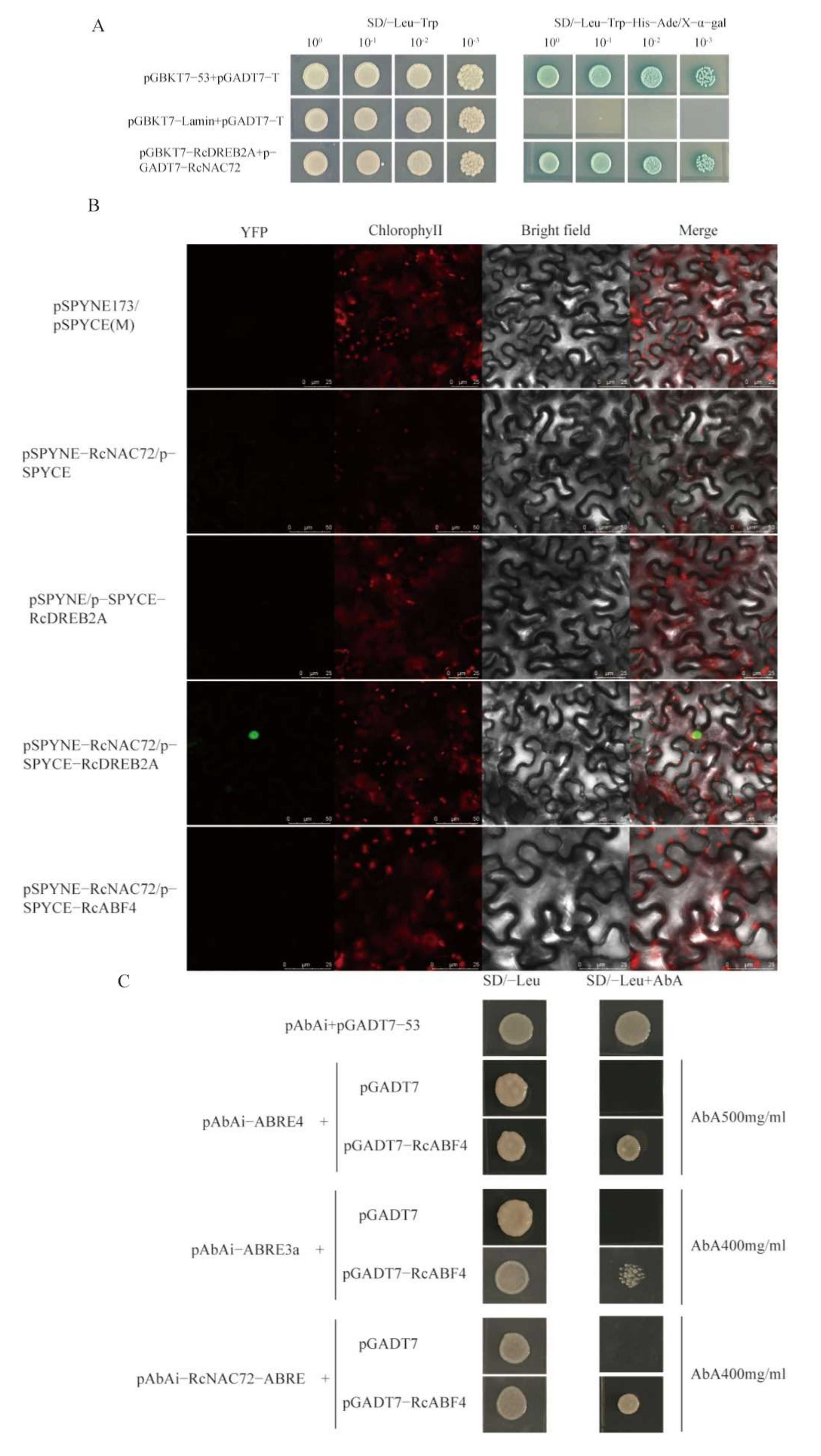
2.8. RcABF4 Combines with the Promoter Region of RcNAC72 and RcNAC72 Interacts with RcDREB2A
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Cloning and Sequencing Analysis of RcNAC72
4.3. RNA Extraction and Quantitative Real-Time PCR
4.4. RcNAC72 Promoter Cloning, Cis-Acting Elements and Promoter Activity Analysis
4.5. Silencing of RcNAC72 in Rose Leave Discs by Virus-Induced Gene Silencing (VIGS)
4.6. Obtainment of Transgenic Arabidopsis with RcNAC72 and Functional Verification
4.7. Subcellular Localization of RcNAC72
4.8. Transcription Activation Activity Analysis and Yeast Two-Hybrid Assay
4.9. Bimolecular Fluorescent Complimentary (BiFC) Assay
4.10. Yeast One-Hybrid Assay
4.11. Physiological Parameters Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Razi, K.; Muneer, S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 2021, 41, 669–691. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef] [Green Version]
- Piao, W.; Sakuraba, Y.; Paek, N.-C. Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thaliana. BMB Rep. 2019, 52, 653–658. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Zhang, W.J.; Zhang, Q.; Xu, Z.Q.; Zhu, Z.G.; Duan, F.P.; Wu, R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 2011, 49, 1384–1391. [Google Scholar] [CrossRef]
- Garcia, M.N.M.; Cortelezzi, J.I.; Fumagalli, M.; Capiati, D.A. Expression of the Arabidopsis ABF4 gene in potato increases tuber yield, improves tuber quality and enhances salt and drought tolerance. Plant Mol. Biol. 2018, 98, 137–152. [Google Scholar] [CrossRef]
- Hong, Y.B.; Zhang, H.J.; Huang, L.; Li, D.Y.; Song, F.M. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONACO22 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Dane, F. NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway. Acta Physiol. Plant. 2013, 35, 1397–1408. [Google Scholar] [CrossRef]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant. 2018, 164, 279–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Kim, Y.-S.; Han, S.-H.; Lee, B.-D.; Paek, N.-C. The Arabidopsis Transcription Factor NAC016 Promotes Drought Stress Responses by Repressing AREB1 Transcription through a Trifurcate Feed-Forward Regulatory Loop Involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.-Y.; Kim, S.Y.; Hyeon, D.Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.-H.; Kim, S.-K.; Hong, J.C.; et al. The Arabidopsis NAC Transcription Factor ANAC096 Cooperates with bZIP-Type Transcription Factors in Dehydration and Osmotic Stress Responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Guo, C.; Li, Z.Y.; Sun, J.H.; Deng, Z.C.; Wen, L.C.; Li, X.X.; Guo, Y.F. Potato NAC Transcription Factor StNAC053 Enhances Salt and Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 2568. [Google Scholar] [CrossRef] [PubMed]
- Mergby, D.; Hanin, M.; Saidi, M.N. The durum wheat NAC transcription factor TtNAC2A enhances drought stress tolerance in Arabidopsis. Environ. Exp. Bot. 2021, 186, 104439. [Google Scholar] [CrossRef]
- Ma, J.; Wang, L.Y.; Dai, J.X.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 1–11. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.T.; Bi, Y.; Li, D.Y.; Song, F.M. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef]
- Guan, H.; Liu, X.; Niu, F.; Zhao, Q.; Fan, N.; Cao, D.; Meng, D.; He, W.; Guo, B.; Wei, Y.; et al. OoNAC72, a NAC-Type Oxytropis ochrocephala Transcription Factor, Conferring Enhanced Drought and Salt Stress Tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 890. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.J.; Wang, Y.; Li, B.; Chang, J.L.; Chen, M.J.; Li, K.X.; Yang, G.X.; He, G.Y. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Gong, Y.; Gao, Y.; Zhou, Y.B.; Chen, M.; Xu, Z.S.; Guo, C.H.; Ma, Y.Z. TaNAC48 positively regulates drought tolerance and ABA responses in wheat (Triticum aestivum L.). Crop J. 2021, 9, 785–793. [Google Scholar] [CrossRef]
- Wang, L.; Hu, Z.; Zhu, M.; Zhu, Z.; Hu, J.; Qanmber, G.; Chen, G. The abiotic stress-responsive NAC transcription factor SlNAC11 is involved in drought and salt response in tomato (Solanum lycopersicum L.). Plant Cell Tissue Organ Cult. 2017, 129, 161–174. [Google Scholar] [CrossRef]
- Jian, W.; Zheng, Y.X.; Yu, T.T.; Cao, H.H.; Chen, Y.; Cui, Q.Y.; Xu, C.; Li, Z.G. SlNAC6, A NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J. Plant Physiol. 2021, 264, 153483. [Google Scholar] [CrossRef]
- Li, M.; Wu, Z.Y.; Gu, H.; Cheng, D.W.; Guo, X.Z.; Li, L.; Shi, C.Y.; Xu, G.Y.; Gu, S.C.; Abid, M.; et al. AvNAC030, a NAC Domain Transcription Factor, Enhances Salt Stress Tolerance in Kiwifruit. Int. J. Mol. Sci. 2021, 22, 11897. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Zhang, C.; Jiang, X.; Kang, M.; Yin, X.; Lu, P.; Zhang, X.; Zheng, Y.; Gao, J. RhNAC2 and RhEXPA4 Are Involved in the Regulation of Dehydration Tolerance during the Expansion of Rose Petals. Plant Physiol. 2012, 160, 2064–2082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Zhang, C.; Lu, P.; Jiang, G.; Liu, X.; Dai, F.; Gao, J. RhNAC3, a stress- associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress- related genes in rose petals. Plant Biotechnol. J. 2014, 12, 38–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, A.; Li, S.; Li, W.; Hao, Q.; Wan, X.; Wang, K.; Liu, Q.; Liu, Q.; Jiang, X. RhNAC31, a novel rose NAC transcription factor, enhances tolerance to multiple abiotic stresses in Arabidopsis. Acta Physiol. Plant. 2019, 41, 1–16. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhong, M.; He, L.; Wang, B.; Liu, Q.L.; Pan, Y.Z.; Jiang, B.B.; Zhang, L. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves drought tolerance in chrysanthemum. Plant Cell Tissue Organ Cult. 2018, 135, 119–132. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Long, Y.; Chen, X.X.; Zhang, B.L.; Xin, Y.F.; Li, L.Y.; Cao, S.L.; Liu, F.H.; Wang, Z.G.; Huang, H.; et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 2021, 21, 1–13. [Google Scholar] [CrossRef]
- Luo, P.; Chen, Y.; Rong, K.; Lu, Y.; Wang, N.; Xu, Z.; Pang, B.; Zhou, D.; Weng, J.; Li, M.; et al. ZmSNAC13, a maize NAC transcription factor conferring enhanced resistance to multiple abiotic stresses in transgenic Arabidopsis. Plant Physiol. Biochem. PPB 2021, 170, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, B.-G.; Chao, Q.; Wang, B.; Zhang, Q.; Zhang, C.; Li, S.; Jin, F.; Yang, D.; Li, X. Function analysis of ZmNAC33, a positive regulator in drought stress response in Arabidopsis. Plant Physiol. Biochem. 2019, 145, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Li, X.L.; Li, M.J.; Yan, Y.C.; Liu, X.; Li, L. Dual Function of NAC072 in ABF3-Mediated ABA-Responsive Gene Regulation in Arabidopsis. Front. Plant Sci. 2016, 7, 1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Liu, J.; Du, B.; Zhang, M.; Wang, A.; Zhang, L. NAC Transcription Factor PwNAC11 Activates ERD1 by Interaction with ABF3 and DREB2A to Enhance Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6952. [Google Scholar] [CrossRef]
- Yong, Y.; Zhang, Y.; Lyu, Y. A Stress-Responsive NAC Transcription Factor from Tiger Lily (LlNAC2) Interacts with LlDREB1 and LlZHFD4 and Enhances Various Abiotic Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 3225. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Feng, H.; Bu, Y.; Ji, N.; Lyu, Y.; Zhao, S. Comparative Transcriptome and Weighted Gene Co-expression Network Analysis Identify Key Transcription Factors of Rosa chinensis ʻOld Blushʼ After Exposure to a Gradual Drought Stress Followed by Recovery. Front. Genet. 2021, 12, 690264. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [Green Version]
- Hou, L.; Chen, L.J.; Wang, J.Y.; Xu, D.F.; Dai, L.X.; Zhang, H.; Zhao, Y.X. Construction of Stress Responsive Synthetic Promoters and Analysis of Their Activity in Transgenic Arabidopsis thaliana. Plant Mol. Biol. Rep. 2012, 30, 1496–1506. [Google Scholar] [CrossRef]
- Wu, H.; Fu, B.; Sun, P.; Xiao, C.; Liu, J.-H. A NAC Transcription Factor Represses Putrescine Biosynthesis and Affects Drought Tolerance. Plant Physiol. 2016, 172, 1532–1547. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.J.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaheen, T.; Mahmood ur, R.; Abbas, M.F.; Zaib, P.; Mehboob ur, R.; Ullah, I.; ul Qamar, M.T.; Arif, A. Identification, Characterization, Homology Modeling and Protein-Protein Interactions of Cotton (Gossypium arboreum) DREB Gene. Int. J. Agric. Biol. 2018, 20, 1055–1061. [Google Scholar]
- Jin, C.; Li, K.Q.; Xu, X.Y.; Zhang, H.P.; Chen, H.X.; Chen, Y.H.; Hao, J.; Wang, Y.; Huang, X.S.; Zhang, S.L. A Novel NAC Transcription Factor, PbeNAC1, of Pyrus betulifolia Confers Cold and Drought Tolerance via Interacting with PbeDREBs and Activating the Expression of Stress-Responsive Genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE Promoter Sequence is Involved in Osmotic Stress-Responsive Expression of the DREB2A Gene, Which Encodes a Transcription Factor Regulating Drought-Inducible Genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Song, M.; Guo, Y.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Zeng, Z.; Lyu, Y.; Zhao, S. Drought-Responsive NAC Transcription Factor RcNAC72 Is Recognized by RcABF4, Interacts with RcDREB2A to Enhance Drought Tolerance in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1755. https://doi.org/10.3390/ijms23031755
Jia X, Zeng Z, Lyu Y, Zhao S. Drought-Responsive NAC Transcription Factor RcNAC72 Is Recognized by RcABF4, Interacts with RcDREB2A to Enhance Drought Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(3):1755. https://doi.org/10.3390/ijms23031755
Chicago/Turabian StyleJia, Xin, Zhen Zeng, Yingmin Lyu, and Shiwei Zhao. 2022. "Drought-Responsive NAC Transcription Factor RcNAC72 Is Recognized by RcABF4, Interacts with RcDREB2A to Enhance Drought Tolerance in Arabidopsis" International Journal of Molecular Sciences 23, no. 3: 1755. https://doi.org/10.3390/ijms23031755
APA StyleJia, X., Zeng, Z., Lyu, Y., & Zhao, S. (2022). Drought-Responsive NAC Transcription Factor RcNAC72 Is Recognized by RcABF4, Interacts with RcDREB2A to Enhance Drought Tolerance in Arabidopsis. International Journal of Molecular Sciences, 23(3), 1755. https://doi.org/10.3390/ijms23031755




