Optogenetic and Chemical Induction Systems for Regulation of Transgene Expression in Plants: Use in Basic and Applied Research
Abstract
1. Introduction
- -
- without specific induction, the system is off and the gene of interest is not expressed;
- -
- upon induction, the promoter is activated, enabling the target gene transcription in specific plant growth periods and tissues;
- -
- the induction can be switched-off if it is needed [33].
2. Optogenetic and Chemical Induction Systems
2.1. Optogenetics and Photoreceptors
2.2. Optogenetic Systems in Plants
2.3. Chemical Induction Systems in Plants
3. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Khan, S.; Anwar, S.; Yu, S.; Sun, M.; Yang, Z.; Gao, Z.-Q. Development of drought-tolerant transgenic wheat: Achievements and limitations. Int. J. Mol. Sci. 2019, 20, 3350. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, H.; Fang, Y.; Guo, W.; Chen, H.; Zhang, X.; Dai, W.; Chen, S.; Hao, Q.; Yuan, S.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.M.; Ali, M.; Feng, X.; Li, X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 2017, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.-Y.; Du, Y.-T.; Ma, J.; Min, D.-H.; Jin, L.-G.; Chen, J.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; et al. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- Fuganti-Pagliarini, R.; Ferreira, L.C.; Rodrigues, F.A.; Molinari, H.B.C.; Marin, S.R.R.; Molinari, M.D.C.; Marcolino-Gomes, J.; Mertz-Henning, L.M.; Farias, J.R.B.; de Oliveira, M.C.N.; et al. Characterization of soybean genetically modified for drought tolerance in field conditions. Front. Plant Sci. 2017, 8, 448. [Google Scholar] [CrossRef]
- Ma, X.-J.; Yu, T.-F.; Li, X.-H.; Cao, X.-Y.; Ma, J.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Zhang, J.-H.; et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 2020, 20, 123. [Google Scholar] [CrossRef]
- Zhang, Z.; Ali, S.; Zhang, T.; Wang, W.; Xie, L. Identification, evolutionary and expression analysis of PYL-PP2C-SnRK2s gene families in soybean. Plants 2020, 9, 1356. [Google Scholar] [CrossRef]
- Li, L.; Du, Y.; He, C.; Dietrich, C.R.; Li, J.; Ma, X.; Wang, R.; Liu, Q.; Liu, S.; Wang, G.; et al. Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 2019, 70, 3089–3099. [Google Scholar] [CrossRef]
- Muppala, S.; Gudlavalleti, P.K.; Malireddy, K.R.; Puligundla, S.K.; Dasari, P. Development of stable transgenic maize plants tolerant for drought by manipulating ABA signaling through Agrobacterium-mediated transformation. J. Genet. Eng. Biotechnol. 2021, 19, 96. [Google Scholar] [CrossRef]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Li, M.; Zhao, D.; Yang, J.-F.; Fu, J.-D.; Xu, Z.-S. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- González, F.G.; Capella, M.; Ribichich, K.F.; Curín, F.; Giacomelli, J.I.; Ayala, F.; Watson, G.; Otegui, M.E.; Chan, R.L. Field-grown transgenic wheat expressing the sunflower gene HaHB4 significantly outyields the wild type. J. Exp. Bot. 2019, 70, 1669–1681. [Google Scholar] [CrossRef]
- Yu, T.-F.; Xu, Z.-S.; Guo, J.-K.; Wang, Y.-X.; Abernathy, B.; Fu, J.-D.; Chen, X.; Zhou, Y.-B.; Chen, M.; Ye, X.-G.; et al. Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 2017, 7, 44050. [Google Scholar] [CrossRef]
- Bi, H.; Shi, J.; Kovalchuk, N.; Luang, S.; Bazanova, N.; Chirkova, L.; Zhang, D.; Shavrukov, Y.; Stepanenko, A.; Tricker, P.; et al. Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance, and no yield penalty under controlled growth conditions. Plant Cell Environ. 2018, 41, 2549–2566. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY transcription tactor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Le Roux, M.L.; Kunert, K.J.; van der Vyver, C.; Cullis, C.A.; Botha, A.-M. Expression of a small ubiquitin-like modifier protease increases drought tolerance in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 266. [Google Scholar] [CrossRef]
- Yang, Y.; Luang, S.; Harris, J.; Riboni, M.; Li, Y.; Bazanova, N.; Hrmova, M.; Haefele, S.; Kovalchuk, N.; Lopato, S. Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol. J. 2018, 16, 1227–1240. [Google Scholar] [CrossRef]
- Beznec, A.; Faccio, P.; Miralles, D.J.; Abeledo, L.G.; Oneto, C.D.; Garibotto, M.d.B.; Gonzalez, G.; Moreyra, F.; Elizondo, M.; Ruíz, M.; et al. Stress-induced expression of IPT gene in transgenic wheat reduces grain yield penalty under drought. J. Genet. Eng. Biotechnol. 2021, 19, 67. [Google Scholar] [CrossRef]
- Chen, Q.; Bao, C.; Xu, F.; Ma, C.; Huang, L.; Guo, Q.; Luo, M. Silencing GhJUB1L1 (JUB1-like 1) reduces cotton (Gossypium hirsutum) drought tolerance. PLoS ONE 2021, 16, e0259382. [Google Scholar] [CrossRef]
- Hao, Y.-Q.; Lu, G.-Q.; Wang, L.-H.; Wang, C.-L.; Guo, H.-M.; Li, Y.-F.; Cheng, H.-M. Overexpression of AmDUF1517 enhanced tolerance to salinity, drought, and cold stress in transgenic cotton. J. Integr. Agric. 2018, 17, 2204–2214. [Google Scholar] [CrossRef]
- Crickmore, N.; Berry, C.; Panneerselvam, S.; Mishra, R.; Connor, T.R.; Bonning, B.C. A structure-based nomenclature for Bacillus thuringiensis and other bacteria-derived pesticidal proteins. J. Invertebr. Pathol. 2021, 186, 107438. [Google Scholar] [CrossRef]
- Raspor, M.; Cingel, A. Genetically modified potato for pest resistance: Thrift or threat? In Solanum Tuberosum-A Promising Crop for Starvation Problem; Yildiz, M., Ozgen, Y., Eds.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Kumar, A.M.; Sundaresha, S.; Sreevathsa, R. Transgenic sunflower (Helianthus annuus L.) with enhanced resistance to a fungal pathogen Alternaria helianthi. Transgenic Plant J. 2011, 5, 50–56. [Google Scholar]
- Shen, F.; Ying, J.; Xu, L.; Sun, X.; Wang, J.; Wang, Y.; Mei, Y.; Zhu, Y.; Liu, L. Characterization of of Annexin gene family and functional analysis of RsANN1a involved in heat tolerance in radish (Raphanus sativus L.). Physiol. Mol. Biol. Plants 2021, 27, 2027–2041. [Google Scholar] [CrossRef]
- Czemplik, M.; Kulma, A.; Bazela, K.; Szopa, J. The biomedical potential of genetically modified flax seeds overexpressing the glucosyltransferase gene. BMC Complement. Altern. Med. 2012, 12, 251. [Google Scholar] [CrossRef]
- Da Silva, A.C.; de Freitas Lima, M.; Eloy, N.B.; Thiebaut, F.; Montessoro, P.; Hemerly, A.S.; Ferreira, P.C.G. The Yin and Yang in plant breeding: The trade-off between plant growth yield and tolerance to stresses. Biotechnol. Res. Innov. 2020, 3, 73–79. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.-A.; Yee, K.M.; West, M.A.L.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef]
- Tian, D.; Traw, M.B.; Chen, J.Q.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef]
- Hieno, A.; Naznin, H.A.; Hyakumachi, M.; Sakurai, T.; Tokizawa, M.; Koyama, H.; Sato, N.; Nishiyama, T.; Hasebe, M.; Zimmer, A.D.; et al. ppdb: Plant promoter database version 3.0. Nucleic Acids Res. 2014, 42, D1188–D1192. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Belcher, M.S.; Vuu, K.M.; Zhou, A.; Mansoori, N.; Agosto Ramos, A.; Thompson, M.G.; Scheller, H.V.; Loqué, D.; Shih, P.M. Design of orthogonal regulatory systems for modulating gene expression in plants. Nat. Chem. Biol. 2020, 16, 857–865. [Google Scholar] [CrossRef]
- Misra, S.; Ganesan, M. The impact of inducible promoters in transgenic plant production and crop improvement. Plant Gene 2021, 27, 100300. [Google Scholar] [CrossRef]
- Borghi, L. Inducible gene expression systems for plants. Methods Mol. Biol. 2010, 655, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M. Chemically regulated gene expression in plants. Curr. Opin. Plant Biol. 2003, 6, 169–177. [Google Scholar] [CrossRef]
- Braguy, J.; Zurbriggen, M.D. Synthetic strategies for plant signalling studies: Molecular toolbox and orthogonal platforms. Plant J. 2016, 87, 118–138. [Google Scholar] [CrossRef]
- Andres, J.; Blomeier, T.; Zurbriggen, M.D. Synthetic switches and regulatory circuits in plants. Plant Physiol. 2019, 179, 862–884. [Google Scholar] [CrossRef]
- Herrou, J.; Crosson, S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 2011, 9, 713–723. [Google Scholar] [CrossRef]
- Christie, J.M.; Gawthorne, J.; Young, G.; Fraser, N.J.; Roe, A.J. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol. Plant 2012, 5, 533–544. [Google Scholar] [CrossRef]
- Peter, E.; Dick, B.; Baeurle, S.A. Mechanism of signal transduction of the LOV2-Jα photosensor from Avena sativa. Nat. Commun. 2010, 1, 122. [Google Scholar] [CrossRef]
- Yumerefendi, H.; Dickinson, D.J.; Wang, H.; Zimmerman, S.P.; Bear, J.E.; Goldstein, B.; Hahn, K.; Kuhlman, B. Control of protein activity and cell fate specification via light-mediated nuclear translocation. PLoS ONE 2015, 10, e0128443. [Google Scholar] [CrossRef]
- Niopek, D.; Wehler, P.; Roensch, J.; Eils, R.; Di Ventura, B. Optogenetic control of nuclear protein export. Nat. Commun. 2016, 7, 10624. [Google Scholar] [CrossRef]
- Redchuk, T.A.; Omelina, E.S.; Chernov, K.G.; Verkhusha, V.V. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat. Chem. Biol. 2017, 13, 633–639. [Google Scholar] [CrossRef]
- Spiltoir, J.I.; Strickland, D.; Glotzer, M.; Tucker, C.L. Optical control of peroxisomal trafficking. ACS Synth. Biol. 2016, 5, 554–560. [Google Scholar] [CrossRef]
- Renicke, C.; Schuster, D.; Usherenko, S.; Essen, L.-O.; Taxis, C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 2013, 20, 619–626. [Google Scholar] [CrossRef]
- Wu, Y.I.; Frey, D.; Lungu, O.I.; Jaehrig, A.; Schlichting, I.; Kuhlman, B.; Hahn, K.M. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009, 461, 104–108. [Google Scholar] [CrossRef]
- Lee, J.; Natarajan, M.; Nashine, V.C.; Socolich, M.; Vo, T.; Russ, W.P.; Benkovic, S.J.; Ranganathan, R. Surface sites for engineering allosteric control in proteins. Science 2008, 322, 438–442. [Google Scholar] [CrossRef]
- Nash, A.I.; McNulty, R.; Shillito, M.E.; Swartz, T.E.; Bogomolni, R.A.; Luecke, H.; Gardner, K.H. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. USA 2011, 108, 9449–9454. [Google Scholar] [CrossRef]
- Rivera-Cancel, G.; Motta-Mena, L.B.; Gardner, K.H. Identification of natural and artificial DNA substrates for light-activated LOV-HTH transcription factor EL222. Biochemistry 2012, 51, 10024–10034. [Google Scholar] [CrossRef]
- Zoltowski, B.D.; Motta-Mena, L.B.; Gardner, K.H. Blue light-induced dimerization of a bacterial LOV-HTH DNA-binding protein. Biochemistry 2013, 52, 6653–6661. [Google Scholar] [CrossRef]
- Selby, C.P.; Sancar, A. The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry 2012, 51, 167–171. [Google Scholar] [CrossRef]
- Palayam, M.; Ganapathy, J.; Guercio, A.M.; Tal, L.; Deck, S.L.; Shabek, N. Structural insights into photoactivation of plant Cryptochrome-2. Commun. Biol. 2021, 4, 28. [Google Scholar] [CrossRef]
- Kennedy, M.J.; Hughes, R.M.; Peteya, L.A.; Schwartz, J.W.; Ehlers, M.D.; Tucker, C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 2010, 7, 973–975. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Bellini, D.; Papiz, M.Z. Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 2012, 20, 1436–1446. [Google Scholar] [CrossRef]
- Tarutina, M.; Ryjenkov, D.A.; Gomelsky, M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 2006, 281, 34751–34758. [Google Scholar] [CrossRef]
- Ni, M.; Tepperman, J.M.; Quail, P.H. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 1999, 400, 781–784. [Google Scholar] [CrossRef]
- Zhu, Y.; Tepperman, J.M.; Fairchild, C.D.; Quail, P.H. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 2000, 97, 13419–13424. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Huq, E.; Tepperman, J.M.; Quail, P.H. A light-switchable gene promoter system. Nat. Biotechnol. 2002, 20, 1041–1044. [Google Scholar] [CrossRef]
- Müller, K.; Engesser, R.; Metzger, S.; Schulz, S.; Kämpf, M.M.; Busacker, M.; Steinberg, T.; Tomakidi, P.; Ehrbar, M.; Nagy, F.; et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013, 41, e77. [Google Scholar] [CrossRef]
- Müller, K.; Siegel, D.; Rodriguez Jahnke, F.; Gerrer, K.; Wend, S.; Decker, E.L.; Reski, R.; Weber, W.; Zurbriggen, M.D. A red light-controlled synthetic gene expression switch for plant systems. Mol. Biosyst. 2014, 10, 1679–1688. [Google Scholar] [CrossRef]
- Chatelle, C.; Ochoa-Fernandez, R.; Engesser, R.; Schneider, N.; Beyer, H.M.; Jones, A.R.; Timmer, J.; Zurbriggen, M.D.; Weber, W. A Green-light-responsive system for the control of transgene expression in mammalian and plant cells. ACS Synth. Biol. 2018, 7, 1349–1358. [Google Scholar] [CrossRef]
- Wang, Y.; Folta, K.M. Contributions of green light to plant growth and development. Am. J. Bot. 2013, 100, 70–78. [Google Scholar] [CrossRef]
- Battle, M.W.; Jones, M.A. Cryptochromes integrate green light signals into the circadian system. Plant Cell Environ. 2020, 43, 16–27. [Google Scholar] [CrossRef]
- Jost, M.; Fernández-Zapata, J.; Polanco, M.C.; Ortiz-Guerrero, J.M.; Chen, P.Y.-T.; Kang, G.; Padmanabhan, S.; Elías-Arnanz, M.; Drennan, C.L. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 2015, 526, 536–541. [Google Scholar] [CrossRef]
- Schneider, N.; Chatelle, C.V.; Ochoa-Fernandez, R.; Zurbriggen, M.D.; Weber, W. Green light-controlled gene switch for mammalian and plant cells. Methods Mol. Biol. 2021, 2312, 89–107. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Hu, C.; Sun, T.; Zeng, Z.; Cai, X.; Li, H.; Hu, Z. Optogenetic regulation of artificial microRNA improves H2 production in green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2017, 10, 257. [Google Scholar] [CrossRef]
- Ochoa-Fernandez, R.; Abel, N.B.; Wieland, F.-G.; Schlegel, J.; Koch, L.-A.; Miller, J.B.; Engesser, R.; Giuriani, G.; Brandl, S.M.; Timmer, J.; et al. Optogenetic control of gene expression in plants in the presence of ambient white light. Nat. Methods 2020, 17, 717–725. [Google Scholar] [CrossRef]
- Ikeda, M.; Ohme-Takagi, M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 2009, 50, 970–975. [Google Scholar] [CrossRef]
- Papanatsiou, M.; Petersen, J.; Henderson, L.; Wang, Y.; Christie, J.M.; Blatt, M.R. Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 2019, 363, 1456–1459. [Google Scholar] [CrossRef]
- Cosentino, C.; Alberio, L.; Gazzarrini, S.; Aquila, M.; Romano, E.; Cermenati, S.; Zuccolini, P.; Petersen, J.; Beltrame, M.; Van Etten, J.L.; et al. Optogenetics. Engineering of a light-gated potassium channel. Science 2015, 348, 707–710. [Google Scholar] [CrossRef]
- Zhou, Y.; Ding, M.; Gao, S.; Yu-Strzelczyk, J.; Krischke, M.; Duan, X.; Leide, J.; Riederer, M.; Mueller, M.J.; Hedrich, R.; et al. Optogenetic control of plant growth by a microbial rhodopsin. Nat. Plants 2021, 7, 144–151. [Google Scholar] [CrossRef]
- Huang, S.; Ding, M.; Roelfsema, M.R.G.; Dreyer, I.; Scherzer, S.; Al-Rasheid, K.A.S.; Gao, S.; Nagel, G.; Hedrich, R.; Konrad, K.R. Optogenetic control of the guard cell membrane potential and stomatal movement by the light-gated anion channel GtACR1. Sci. Adv. 2021, 7, eabg4619. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Janz, R.; Liu, X.; Spudich, J.L. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 2015, 349, 647–650. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Rodarte, E.M.; Janz, R.; Morelle, O.; Melkonian, M.; Wong, G.K.-S.; Spudich, J.L. The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 2017, 7, 43358. [Google Scholar] [CrossRef]
- Zhou, Y.; Ding, M.; Duan, X.; Konrad, K.R.; Nagel, G.; Gao, S. Extending the anion channelrhodopsin-based toolbox for plant optogenetics. Membranes 2021, 11, 287. [Google Scholar] [CrossRef]
- Dawydow, A.; Gueta, R.; Ljaschenko, D.; Ullrich, S.; Hermann, M.; Ehmann, N.; Gao, S.; Fiala, A.; Langenhan, T.; Nagel, G.; et al. Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl. Acad. Sci. USA 2014, 111, 13972–13977. [Google Scholar] [CrossRef]
- Reyer, A.; Häßler, M.; Scherzer, S.; Huang, S.; Pedersen, J.T.; Al-Rascheid, K.A.S.; Bamberg, E.; Palmgren, M.; Dreyer, I.; Nagel, G.; et al. Channelrhodopsin-mediated optogenetics highlights a central role of depolarization-dependent plant proton pumps. Proc. Natl. Acad. Sci. USA 2020, 117, 20920–20925. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- De Veylder, L.; Beeckman, T.; Van Montagu, M.; Inzé, D. Increased leakiness of the tetracycline-inducible Triple-Op promoter in dividing cells renders it unsuitable for high inducible levels of a dominant negative CDC2aAt gene. J. Exp. Bot. 2000, 51, 1647–1653. [Google Scholar] [CrossRef]
- Das, A.T.; Tenenbaum, L.; Berkhout, B. Tet-On systems for doxycycline-inducible gene expression. Curr. Gene Ther. 2016, 16, 156–167. [Google Scholar] [CrossRef]
- Gatz, C.; Frohberg, C.; Wendenburg, R. Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 1992, 2, 397–404. [Google Scholar] [CrossRef]
- David, K.M.; Perrot-Rechenmann, C. Characterization of a tobacco Bright Yellow 2 cell line expressing the tetracycline repressor at a high level for strict regulation of transgene expression. Plant Physiol. 2001, 125, 1548–1553. [Google Scholar] [CrossRef] [PubMed]
- Love, J.; Scott, A.C.; Thompson, W.F. Stringent control of transgene expression in Arabidopsis thaliana using the Top10 promoter system. Plant J. 2000, 21, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, M.; Gatz, C.; Hartmann, E.; Hughes, J. Tetracycline-regulated reporter gene expression in the moss Physcomitrella patens. Plant Mol. Biol. 1996, 30, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, J.-H.; Pan, Y.; Zheng, Y.; Pan, Y.-l.; Ding, Y.-M.; Su, C.-G.; Zhang, X.-G. Establishment of a tetracycline-off and heat shock-on gene expression system in tobacco. J. Integr. Agric. 2017, 16, 1112–1119. [Google Scholar] [CrossRef]
- Aoyama, T.; Chua, N.-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997, 11, 605–612. [Google Scholar] [CrossRef]
- Mori, M.; Fujihara, N.; Mise, K.; Furusawa, I. Inducible high-level mRNA amplification system by viral replicase in transgenic plants. Plant J. 2001, 27, 79–86. [Google Scholar] [CrossRef]
- Andersen, S.U.; Cvitanich, C.; Hougaard, B.K.; Roussis, A.; Grønlund, M.; Jensen, D.B.; Frøkjaer, L.A.; Jensen, E.O. The glucocorticoid-inducible GVG system causes severe growth defects in both root and shoot of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 2003, 16, 1069–1076. [Google Scholar] [CrossRef]
- Moore, I.; Samalova, M.; Kurup, S. Transactivated and chemically inducible gene expression in plants. Plant J. 2006, 45, 651–683. [Google Scholar] [CrossRef]
- Schürholz, A.-K.; López-Salmerón, V.; Li, Z.; Forner, J.; Wenzl, C.; Gaillochet, C.; Augustin, S.; Barro, A.V.; Fuchs, M.; Gebert, M.; et al. A comprehensive toolkit for inducible, cell type-specific gene expression in Arabidopsis. Plant Physiol. 2018, 178, 40–53. [Google Scholar] [CrossRef]
- Samalova, M.; Brzobohaty, B.; Moore, I. pOp6/LhGR: A stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. Plant J. 2005, 41, 919–935. [Google Scholar] [CrossRef]
- Vlad, D.; Abu-Jamous, B.; Wang, P.; Langdale, J.A. A modular steroid-inducible gene expression system for use in rice. BMC Plant Biol. 2019, 19, 426. [Google Scholar] [CrossRef]
- Rutherford, S.; Brandizzi, F.; Townley, H.; Craft, J.; Wang, Y.; Jepson, I.; Martinez, A.; Moore, I. Improved transcriptional activators and their use in mis-expression traps in Arabidopsis. Plant J. 2005, 43, 769–788. [Google Scholar] [CrossRef]
- Samalova, M.; Kirchhelle, C.; Moore, I. Universal methods for transgene induction using the dexamethasone-inducible transcription activation system pOp6/LhGR in Arabidopsis and other plant species. Curr. Protoc. Plant Biol. 2019, 4, e20089. [Google Scholar] [CrossRef]
- Samalova, M.; Moore, I. The steroid-inducible pOp6/LhGR gene expression system is fast, sensitive and does not cause plant growth defects in rice (Oryza sativa). BMC Plant Biol. 2021, 21, 461. [Google Scholar] [CrossRef]
- Bruce, W.; Folkerts, O.; Garnaat, C.; Crasta, O.; Roth, B.; Bowen, B. Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 2000, 12, 65–79. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.-K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.-W.; Chua, N.-H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000, 24, 265–273. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.-W.; Frugis, G.; Chua, N.-H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef]
- Gonzalez, J.H.; Taylor, J.S.; Reed, K.M.; Wright, R.C.; Bargmann, B.O.R. Temporal control of morphogenic factor expression determines efficacy in enhancing regeneration. Plants 2021, 10, 2271. [Google Scholar] [CrossRef]
- Okuzaki, A.; Konagaya, K.-i.; Nanasato, Y.; Tsuda, M.; Tabei, Y. Estrogen-inducible GFP expression patterns in rice (Oryza sativa L.). Plant Cell Rep. 2011, 30, 529–538. [Google Scholar] [CrossRef]
- Siligato, R.; Wang, X.; Yadav, S.R.; Lehesranta, S.; Ma, G.; Ursache, R.; Sevilem, I.; Zhang, J.; Gorte, M.; Prasad, K.; et al. MultiSite Gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 2016, 170, 627–641. [Google Scholar] [CrossRef]
- Chen, Z.; Cheng, Q.; Hu, C.; Guo, X.; Chen, Z.; Lin, Y.; Hu, T.; Bellizzi, M.; Lu, G.; Wang, G.-L.; et al. A chemical-induced, seed-soaking activation procedure for regulated gene expression in rice. Front. Plant Sci. 2017, 8, 1447. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Ouyang, B.; Lu, Y.; Ye, Z. Chemical-induced autoexcision of selectable markers in elite tomato plants transformed with a gene conferring resistance to lepidopteran insects. Biotechnol. Lett. 2006, 28, 1247–1253. [Google Scholar] [CrossRef]
- Hirose, T.; Mizutani, R.; Mitsui, T.; Terao, T. A chemically inducible gene expression system and its application to inducible gene suppression in rice. Plant Prod. Sci. 2012, 15, 73–78. [Google Scholar] [CrossRef][Green Version]
- Liu, S.; Yoder, J.I. Chemical induction of hairpin RNAi molecules to silence vital genes in plant roots. Sci. Rep. 2016, 6, 37711. [Google Scholar] [CrossRef]
- Böhner, S.; Lenk, I.; Rieping, M.; Herold, M.; Gatz, C. Transcriptional activator TGV mediates dexamethasone-inducible and tetracycline-inactivatable gene expression. Plant J. 1999, 19, 87–95. [Google Scholar] [CrossRef]
- Martinez, A.; Sparks, C.; Hart, C.A.; Thompson, J.; Jepson, I. Ecdysone agonist inducible transcription in transgenic tobacco plants. Plant J. 1999, 19, 97–106. [Google Scholar] [CrossRef]
- Padidam, M.; Gore, M.; Lu, D.L.; Smirnova, O. Chemical-inducible, ecdysone receptor-based gene expression system for plants. Transgenic Res. 2003, 12, 101–109. [Google Scholar] [CrossRef]
- Nonogaki, M.; Sekine, T.; Nonogaki, H. Chemically inducible gene expression in seeds before testa rupture. Seed Sci. Res. 2015, 25, 345–352. [Google Scholar] [CrossRef]
- Saijo, T.; Nagasawa, A. Development of a tightly regulated and highly responsive copper-inducible gene expression system and its application to control of flowering time. Plant Cell Rep. 2014, 33, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Granger, C.L.; Cyr, R.J. Characterization of the yeast copper-inducible promoter system in Arabidopsis thaliana. Plant Cell Rep. 2001, 20, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Felenbok, B. The ethanol utilization regulon of Aspergillus nidulans: The alcA-alcR system as a tool for the expression of recombinant proteins. J. Biotechnol. 1991, 17, 11–17. [Google Scholar] [CrossRef]
- Roslan, H.A.; Salter, M.G.; Wood, C.D.; White, M.R.H.; Croft, K.P.; Robson, F.; Coupland, G.; Doonan, J.; Laufs, P.; Tomsett, A.B.; et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 2001, 28, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Sakvarelidze, L.; Tao, Z.; Bush, M.; Roberts, G.R.; Leader, D.J.; Doonan, J.H.; Rawsthorne, S. Coupling the GAL4 UAS system with alcR for versatile cell type-specific chemically inducible gene expression in Arabidopsis. Plant Biotechnol. J. 2007, 5, 465–476. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.J.; Choi, S.; Park, S.-B.; Tran, Q.-G.; Heo, J.; Kim, H.-S. Development of an alcohol-inducible gene expression system for recombinant protein expression in Chlamydomonas reinhardtii. J. Appl. Phycol. 2018, 30, 2297–2304. [Google Scholar] [CrossRef]
- Kinkema, M.; Geijskes, R.J.; Shand, K.; Coleman, H.D.; De Lucca, P.C.; Palupe, A.; Harrison, M.D.; Jepson, I.; Dale, J.L.; Sainz, M.B. An improved chemically inducible gene switch that functions in the monocotyledonous plant sugar cane. Plant Mol. Biol. 2014, 84, 443–454. [Google Scholar] [CrossRef]
- Randall, R.S. The plant AlcR-pAlcA ethanol-inducible system displays gross growth artefacts independently of downstream pAlcA-regulated inducible constructs. Sci. Rep. 2021, 11, 2142. [Google Scholar] [CrossRef]
- Redchuk, T.A.; Karasev, M.M.; Omelina, E.S.; Verkhusha, V.V. Near-infrared light-controlled gene expression and protein targeting in neurons and non-neuronal cells. Chembiochem 2018, 19, 1334–1340. [Google Scholar] [CrossRef]
- Lewis, K.A.; Tzilivakis, J.; Warner, D.J.; Green, A. An international database for pesticide risk assessments and management. Hum. Ecol. Risk Assess. 2016, 22, 1050–1064. [Google Scholar] [CrossRef]
- Yuan, L.-B.; Chen, L.; Zhai, N.; Zhou, Y.; Zhao, S.-S.; Shi, L.-L.; Xiao, S.; Yu, L.-J.; Xie, L.-J. The anaerobic product ethanol promotes autophagy-dependent submergence tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7361. [Google Scholar] [CrossRef]
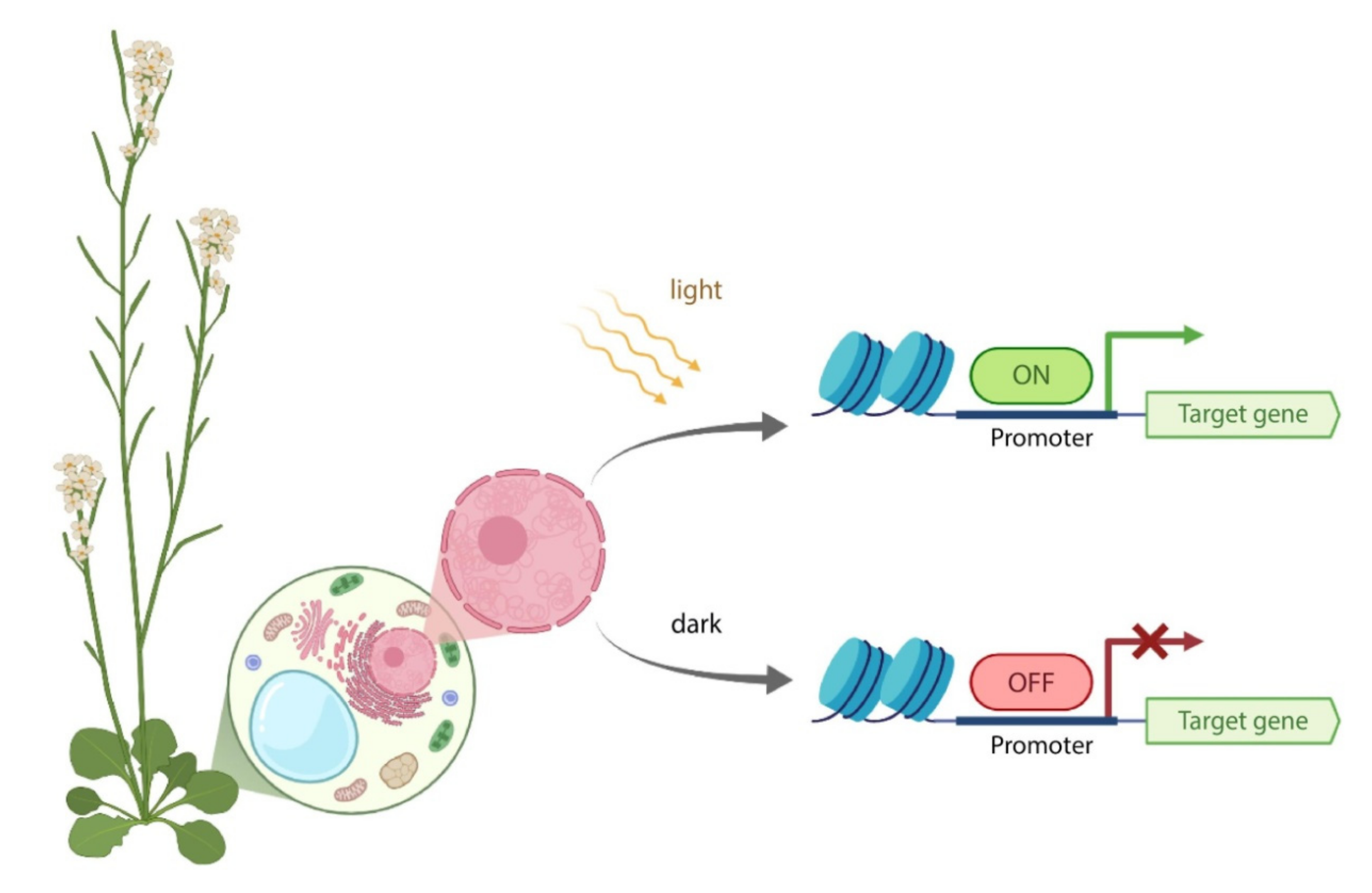
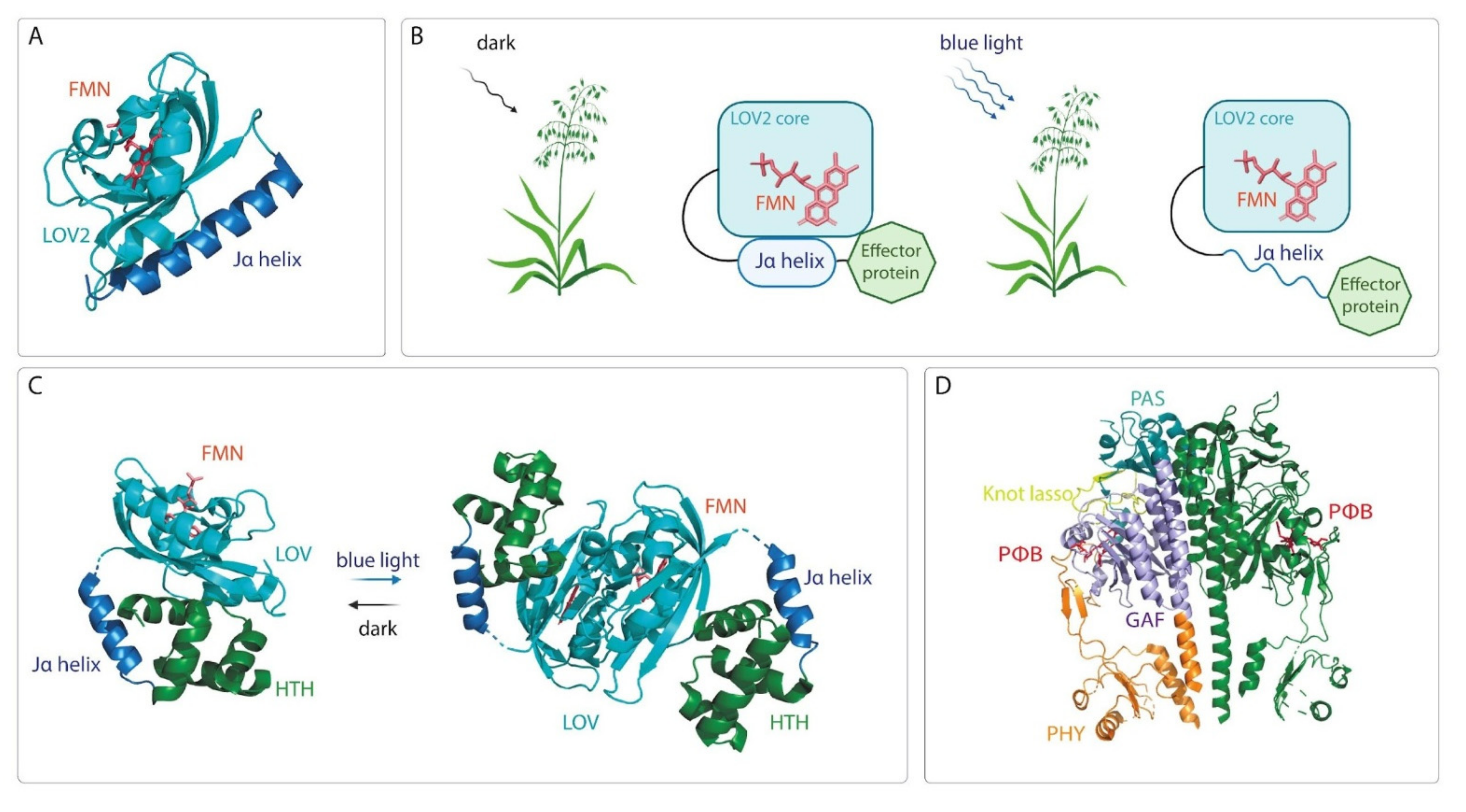
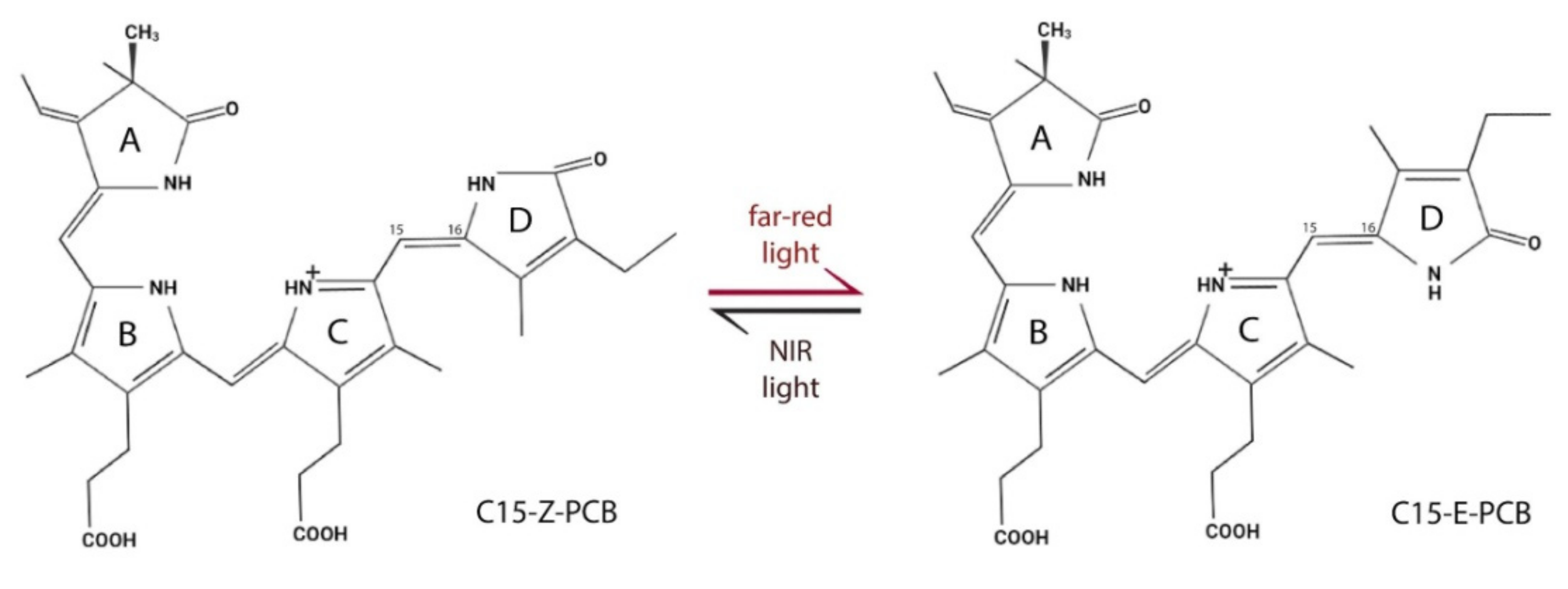
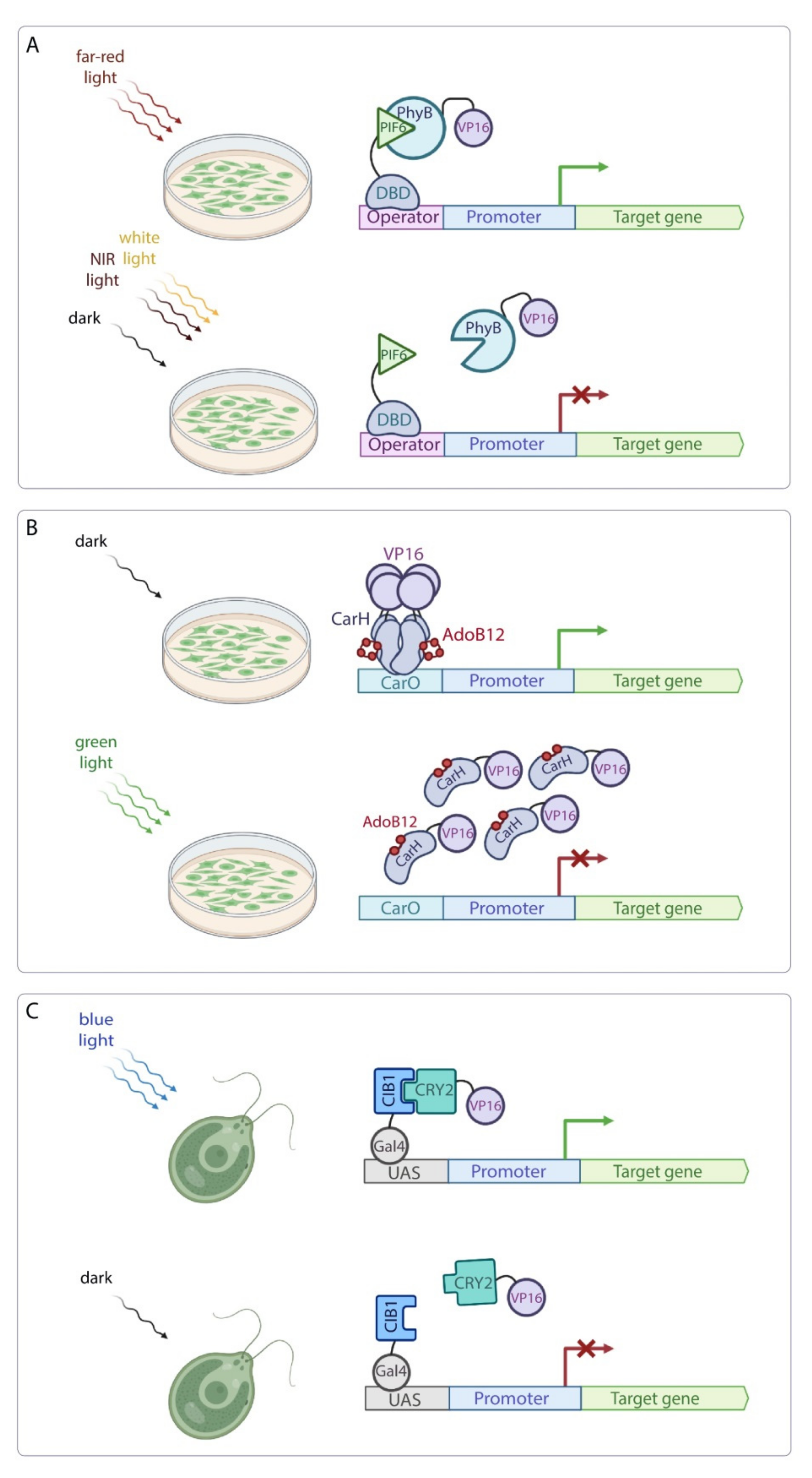

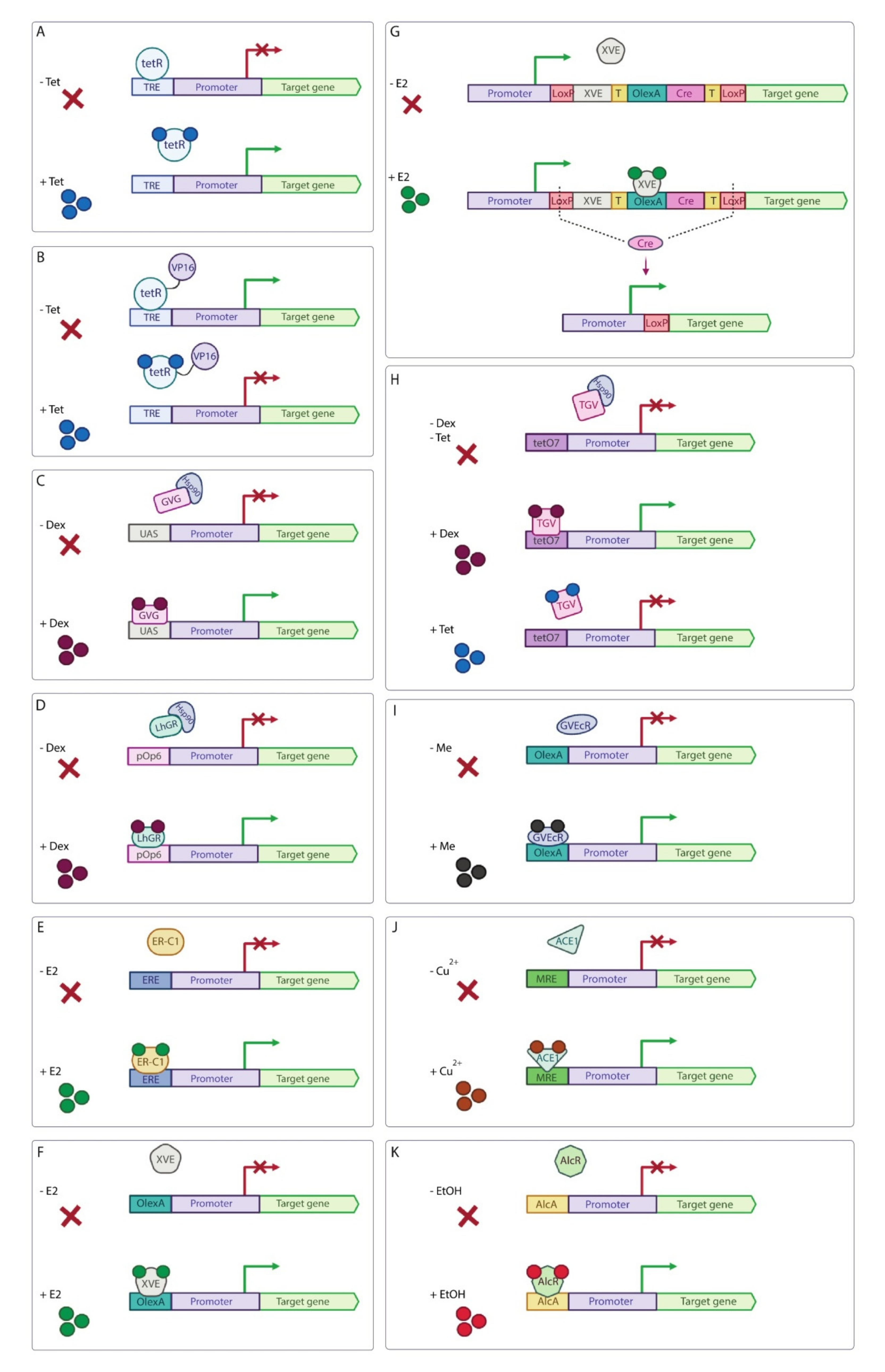
| Inducible System | Use in Basic Science | Potential for Use in Applied Research and Notes | ||||
|---|---|---|---|---|---|---|
| Organism | Purpose | Features | Ref | |||
| Optogenetic induction systems | ||||||
| System based on the PhyB-PIF6 interaction | A. thaliana N. benthamiana | Reversed far-red light-induced transcription activation of the target gene. | The PCB chromophore is produced in plant cells. | [60] | Low | PhyB is a plant phytochrome that can interact with the endogenous proteins of the plant cells, affecting endogenous metabolic pathways. This system can be unintentionally activated under ambient light. |
| System based on the CarH photoreceptor | A. thaliana | Green light-induced transcription inhibition of the target gene. | Plant photoreceptors show reduced activity in green light. | [61] | Low | The AdoB12 chromophore is not produced in plants. This system can be unintentionally activated under ambient light. |
| System based on the CRY2-CIB1 interaction | C. reinhardtii | Blue light-induced hydrogen production in transgenic alga. | The FAD chromophore is produced in all plant cells. | [66] | Low | This system can be unintentionally activated under ambient light. |
| PULSE system | A. thaliana, N. benthamiana | Dual-controlled system for transcription activation of the target gene. | The system is insensitive to ambient light. | [67] | Moderate | This system cannot be accidentally activated under ambient light. However, interaction of PhyB and PIF6 with endogenous proteins could affect plant physiology. |
| BLINK1 system | A. thaliana | Blue light-induced regulation of stomatal kinetics. | The FMN chromophore is produced in all plant cells. BLINK1 improves water use efficiency without penalty in carbon fixation. | [70] | Low | This system can be unintentionally activated under ambient light. |
| Chemical induction systems | ||||||
| Tet-derepressible system | N. benthamiana | Tet-induced transcription derepression of the target gene. | Low background expression level, small amount of Tet to launch the system. | [79,81,82] | Low | Tet is an antibiotic and its usage in field experiments should be avoided. |
| Tet-off system | A. thaliana, P. patens, N. benthamiana | Tet-induced transcription inhibition of the target gene. | Low background expression level, small amount of Tet to launch the system. | [83,84,85] | Low | Tet is an antibiotic and its usage in field experiments should be avoided. |
| GVG/UAS system | A. thaliana, N. benthamiana, L. japonicas | Steroid-induced transcription activation of the target gene. | The system has low background expres-sion level. | [86,87,88] | Low | Dexamethasone is steroid and its usage in field experiments is highly undesirable. Steroid-inducible systems can cause severe growth disturbance in transgenic plants. |
| pOp6/LhGR system | A. thaliana, N. benthamiana, O. sativa, Zea mays | Steroid-induced transcription activation of the target gene. | Practically complete inhibition of the LhGR activity in the absence of inducer. | [89,90,91,92,93,94,95] | Low | Dexamethasone is steroid and its usage in field experiments is highly undesirable. Steroid-inducible systems can cause severe growth disturbance in transgenic plants. |
| β-estradiol (E2) induction system | Zea mays | E2-induced transcription activation of the target gene. | Low amount of inducer is needed to activate the system. | [96] | Low | Despite the fact that E2 causes minimal physiological or developmental abnormalities, the usage of steroid-inducible system in field experiments is highly undesirable. |
| XVE system | A. thaliana, N. benthamiana, O. sativa, S. lycopersicum, M. truncatula | E2-induced transcription activation of the target gene. | The system has low background expression level | [99,100,101,102,103,104,107] | Low | Despite the fact that E2 causes minimal physiological or developmental abnormalities, the usage of steroid-inducible system in field experiments is highly undesirable. |
| TGV system | N. benthamiana | Dual-controlled dexamethasone-induced transcription activation and Tet-induced transcription inhibition of the target gene. | The system has low background expression level. | [108] | Low | The usage of steroid dexamethasone and antibiotic Tet in field experiments is highly undesirable. |
| Insecticide-induced systems | A. thaliana, N. benthamiana | Insecticide-induced transcription activation of the target gene. | These systems have low background expression level. | [109,110,111] | Moderate | These systems can be applied in large-scale field experiments and can be used for seed germination recovery and enhancement technologies. |
| Copper-induced system | A. thaliana, N. benthamiana | Copper-induced transcription activation of the target gene. | Copper is already registered as a fungicide for field use in non-toxic concentrations. | [112,113] | High | This system is applicable in large-scale field experiments with copper used in low non-toxic concentrations. |
| Ethanol-induced system | A. thaliana, Saccharum officinarum, C. reinhardtii | Ethanol-induced transcription activation of the target gene. | The system has a fast response. | [115,116,118,119] | High | System is suitable for field application. However, plants produce ethanol under hypoxia that may cause unwanted transgene expression. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omelina, E.S.; Yushkova, A.A.; Motorina, D.M.; Volegov, G.A.; Kozhevnikova, E.N.; Pindyurin, A.V. Optogenetic and Chemical Induction Systems for Regulation of Transgene Expression in Plants: Use in Basic and Applied Research. Int. J. Mol. Sci. 2022, 23, 1737. https://doi.org/10.3390/ijms23031737
Omelina ES, Yushkova AA, Motorina DM, Volegov GA, Kozhevnikova EN, Pindyurin AV. Optogenetic and Chemical Induction Systems for Regulation of Transgene Expression in Plants: Use in Basic and Applied Research. International Journal of Molecular Sciences. 2022; 23(3):1737. https://doi.org/10.3390/ijms23031737
Chicago/Turabian StyleOmelina, Evgeniya S., Anastasiya A. Yushkova, Daria M. Motorina, Grigorii A. Volegov, Elena N. Kozhevnikova, and Alexey V. Pindyurin. 2022. "Optogenetic and Chemical Induction Systems for Regulation of Transgene Expression in Plants: Use in Basic and Applied Research" International Journal of Molecular Sciences 23, no. 3: 1737. https://doi.org/10.3390/ijms23031737
APA StyleOmelina, E. S., Yushkova, A. A., Motorina, D. M., Volegov, G. A., Kozhevnikova, E. N., & Pindyurin, A. V. (2022). Optogenetic and Chemical Induction Systems for Regulation of Transgene Expression in Plants: Use in Basic and Applied Research. International Journal of Molecular Sciences, 23(3), 1737. https://doi.org/10.3390/ijms23031737







