Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants
Abstract
:1. Introduction
2. An Overview of the Mechanism of Ubiquitylation in Plants
3. NO-Mediated Ubiquitylation Process in Plant Development and Stress Management
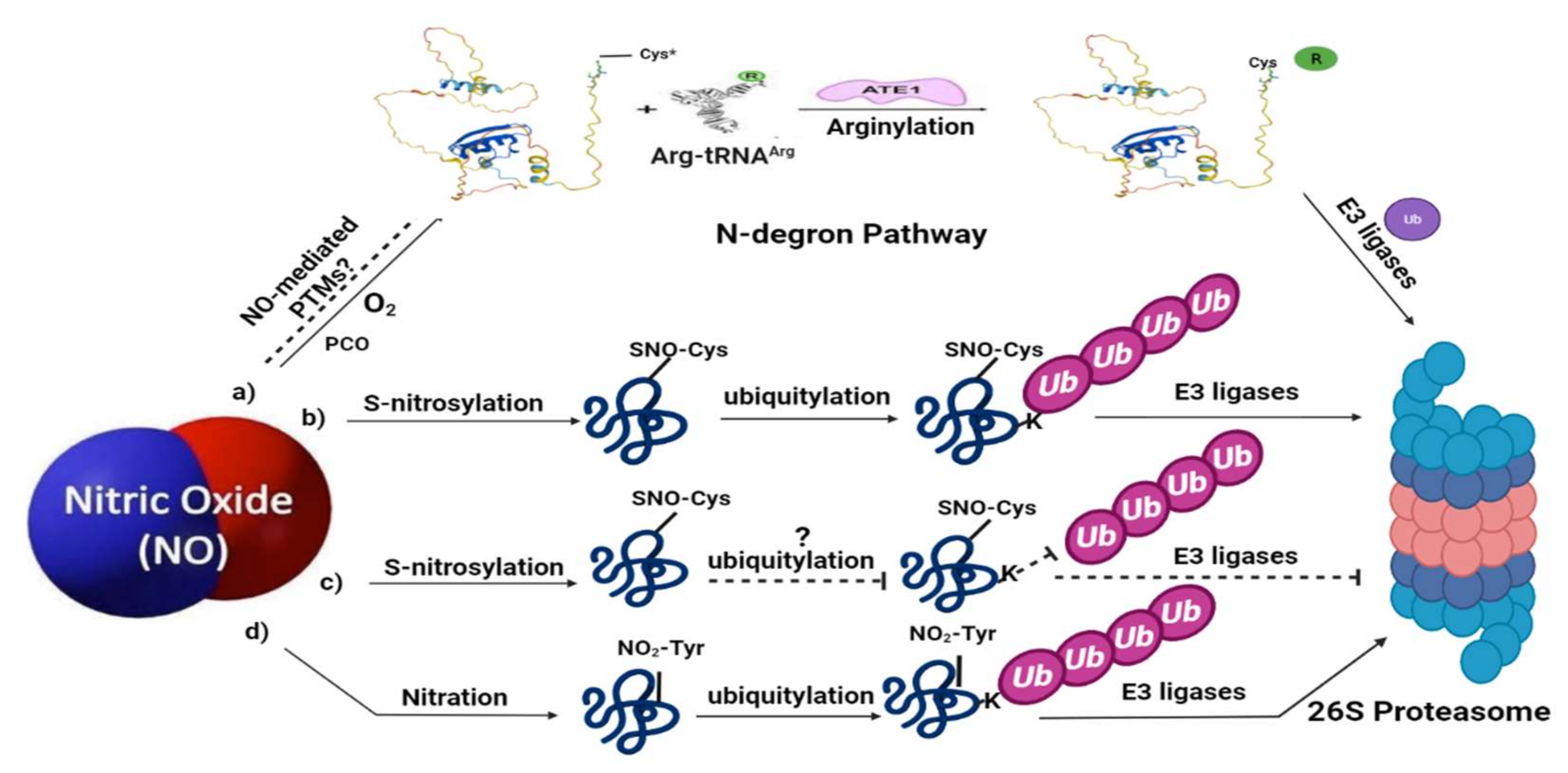
4. Ubiquitylation as Part of NO Sensing in Plants: The N-Degron Pathway
5. NO-Mediated PTMs Associated with Ubiquitin-Mediated Proteolysis
6. NO-Mediated PTMs Regulate the Components of Ubiquitin-Mediated Proteolysis
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, R.M.; Ferrige, A.; Moncada, S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987, 327, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Beligni, M.V.; Lamattina, L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 2000, 210, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.C.; Simontacchi, M.; Puntarulo, S.; Lamattina, L. Nitric oxide is required for root organogenesis. Plant Physiol. 2002, 129, 954–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpas, F.J.; Leterrier, M.; Valderrama, R.; Airaki, M.; Chaki, M.; Palma, J.M.; Barroso, J.B. Nitric oxide imbalance provokes a nitrosative response in plants under abiotic stress. Plant Sci. 2011, 181, 604–611. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Paradiso, A.; Leonetti, P.; de Gara, L. Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. Plant J. 2006, 48, 784–795. [Google Scholar] [CrossRef]
- De Michele, R.; Vurro, E.; Rigo, C.; Costa, A.; Elviri, L.; di Valentin, M.; Careri, M.; Zottini, M.; Sanita di Toppi, L.; lo Schiavo, F. Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol. 2009, 150, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.J.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012, 158, 451–464. [Google Scholar] [CrossRef] [Green Version]
- Serrano, I.; Romero-Puertas, M.C.; Rodríguez-Serrano, M.; Sandalio, L.M.; Olmedilla, A. Peroxynitrite mediates programmed cell death both in papillar cells and in self-incompatible pollen in the olive (Olea europaea L.). J. Exp. Bot. 2012, 63, 1479–1493. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.J.; Kolbert, Z.; Durner, J.; Lindermayr, C.; Corpas, F.J.; Brouquisse, R.; Barroso, J.B.; Umbreen, S.; Palma, J.M.; Hancock, J.T. Regulating the regulator: Nitric oxide control of post-translational modifications. New Phytol. 2020, 227, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Lindermayr, C. Nitric oxide-dependent posttranslational modification in plants: An update. Int. J. Mol. Sci. 2012, 13, 15193–15208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astier, J.; Rasul, S.; Koen, E.; Manzoor, H.; Besson-Bard, A.; Lamotte, O.; Jeandroz, S.; Durner, J.; Lindermayr, C.; Wendehenne, D. S-nitrosylation: An emerging post-translational protein modification in plants. Plant Sci. 2011, 181, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Bacardit, J.; Bachmair, A.; Holdsworth, M.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014, 24, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Isa, N.M.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marín-de la Rosa, N.; Conde, J.V.; Correia, C.S.; Pearce, S.P. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef] [Green Version]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Doroodian, P.; Hua, Z. The ubiquitin switch in plant stress response. Plants 2021, 10, 246. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, C.; Kim, D.Y.; Huang, Y.; Chatt, E.; He, P.; Vierstra, R.D.; Shan, L. Ubiquitylome analysis reveals a central role for the ubiquitin-proteasome system in plant innate immunity. Plant Physiol. 2021, 185, 1943–1965. [Google Scholar] [CrossRef]
- Miricescu, A.; Goslin, K.; Graciet, E. Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J. Exp. Bot. 2018, 69, 4511–4527. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Damaris, R.N.; Li, M.; Khan, I.; Yang, P. Advances on Plant Ubiquitylome—From Mechanism to Application. Int. J. Mol. Sci. 2020, 21, 7909. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Yang, M.; Wang, P.; Zhao, Y.; Ma, C. Interplay between the ubiquitin proteasome system and ubiquitin-mediated autophagy in plants. Cells 2020, 9, 2219. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Sadanandom, A. An insight into the factors influencing specificity of the sumo system in plants. Plants 2020, 9, 1788. [Google Scholar] [CrossRef]
- Denuc, A.; Marfany, G. SUMO and ubiquitin paths converge. Biochem. Soc. Trans. 2010, 38, 34–39. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z.; Yu, P. Diversifying evolution of the ubiquitin-26S proteasome system in Brassicaceae and Poaceae. Int. J. Mol. Sci. 2019, 20, 3226. [Google Scholar] [CrossRef] [Green Version]
- Sadhu, A.; Moriyasu, Y.; Acharya, K.; Bandyopadhyay, M. Nitric oxide and ROS mediate autophagy and regulate Alternaria alternata toxin-induced cell death in tobacco BY-2 cells. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dmitrieva, S.; Ponomareva, A.; Gurjanov, O.; Mazina, A.; Andrianov, V.; Iyudin, V.; Minibayeva, F. Spermine induces autophagy in plants: Possible role of no and reactive oxygen species. In Doklady Biochemistry and Biophysics; Pleiades Publishing: New York, NY, USA, 2018; pp. 341–343. [Google Scholar]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 2018, 71, 142–154.e146. [Google Scholar] [CrossRef] [Green Version]
- Skelly, M.J.; Malik, S.I.; Le Bihan, T.; Bo, Y.; Jiang, J.; Spoel, S.H.; Loake, G.J. A role for S-nitrosylation of the SUMO-conjugating enzyme SCE1 in plant immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17090–17095. [Google Scholar] [CrossRef] [Green Version]
- Park, B.S.; Song, J.T.; Seo, H.S. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2011, 2, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Haas, A.L.; Warms, J.; Hershko, A.; Rose, I.A. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 1982, 257, 2543–2548. [Google Scholar] [CrossRef]
- Hatfield, P.M.; Gosink, M.M.; Carpenter, T.B.; Vierstra, R.D. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 1997, 11, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.-S.; Deng, X.-W.; Callis, J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-H.; Kim, W.T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 2011, 31, 201–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Minaker, S.; Roth, C.; Huang, S.; Hieter, P.; Lipka, V.; Wiermer, M.; Li, X. An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 2014, 26, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Reits, E.A.; Benham, A.M.; Plougastel, B.; Neefjes, J.; Trowsdale, J. Dynamics of proteasome distribution in living cells. EMBO J. 1997, 16, 6087–6094. [Google Scholar] [CrossRef]
- Enenkel, C.; Lehmann, A.; Kloetzel, P.M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope–ER network in yeast. EMBO J. 1998, 17, 6144–6154. [Google Scholar] [CrossRef] [Green Version]
- Russell, S.J.; Steger, K.A.; Johnston, S.A. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 1999, 274, 21943–21952. [Google Scholar] [CrossRef] [Green Version]
- Pack, C.-G.; Yukii, H.; Toh-e, A.; Kudo, T.; Tsuchiya, H.; Kaiho, A.; Sakata, E.; Murata, S.; Yokosawa, H.; Sako, Y. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Fu, H.; Walker, J.; Papa, C.M.; Smalle, J.; Ju, Y.-M.; Vierstra, R.D. Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 2004, 279, 6401–6413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedford, L.; Paine, S.; Sheppard, P.W.; Mayer, R.J.; Roelofs, J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010, 20, 391–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finley, D.; Prado, M.A. The proteasome and its network: Engineering for adaptability. Cold Spring Harb. Perspect. Biol. 2020, 12, a033985. [Google Scholar] [CrossRef]
- Ewan, R.; Pangestuti, R.; Thornber, S.; Craig, A.; Carr, C.; O’Donnell, L.; Zhang, C.; Sadanandom, A. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011, 191, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Jung, C.; Seo, J.S.; Kim, J.-K.; Chua, N.-H. The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 2017, 29, 1406–1424. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, D.J.; Tedds, H.M.; Labandera, A.-M.; Bailey, M.; White, M.D.; Hartman, S.; Sprigg, C.; Mogg, S.L.; Osborne, R.; Dambire, C. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Weits, D.A.; Kunkowska, A.B.; Kamps, N.C.; Portz, K.M.; Packbier, N.K.; Venza, Z.N.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; van Dongen, J.T. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 2019, 569, 714–717. [Google Scholar] [CrossRef]
- Holman, T.J.; Jones, P.D.; Russell, L.; Medhurst, A.; Tomás, S.Ú.; Talloji, P.; Marquez, J.; Schmuths, H.; Tung, S.-A.; Taylor, I. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4549–4554. [Google Scholar] [CrossRef] [Green Version]
- Graciet, E.; Walter, F.; Maoiléidigh, D.Ó.; Pollmann, S.; Meyerowitz, E.M.; Varshavsky, A.; Wellmer, F. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. USA 2009, 106, 13618–13623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuessele, C.; Hoernstein, S.N.; Mueller, S.J.; Rodriguez-Franco, M.; Lorenz, T.; Lang, D.; Igloi, G.L.; Reski, R. Spatio-temporal patterning of arginyl-tRNA protein transferase (ATE) contributes to gametophytic development in a moss. New Phytol. 2016, 209, 1014–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicente, J.; Mendiondo, G.M.; Movahedi, M.; Peirats-Llobet, M.; Juan, Y.-T.; Shen, Y.-Y.; Dambire, C.; Smart, K.; Rodriguez, P.L.; Charng, Y.-Y. The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 2017, 27, 3183–3190.e3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riber, W.; Müller, J.T.; Visser, E.J.; Sasidharan, R.; Voesenek, L.A.; Mustroph, A. The greening after extended darkness1 is an N-end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiol. 2015, 167, 1616–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Marchi, R.; Sorel, M.; Mooney, B.; Fudal, I.; Goslin, K.; Kwaśniewska, K.; Ryan, P.T.; Pfalz, M.; Kroymann, J.; Pollmann, S. The N-end rule pathway regulates pathogen responses in plants. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dissmeyer, N. Conditional protein function via N-degron pathway–mediated proteostasis in stress physiology. Annu. Rev. Plant Biol. 2019, 70, 83–117. [Google Scholar] [CrossRef] [Green Version]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr. Opin. Cell Biol. 2009, 21, 894–899. [Google Scholar] [CrossRef] [Green Version]
- Holdsworth, M.J.; Gibbs, D.J. Comparative biology of oxygen sensing in plants and animals. Curr. Biol. 2020, 30, R362–R369. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.W.; Vierstra, R.D. UPL1 and 2, two 405 kDa ubiquitin–protein ligases from Arabidopsis thaliana related to the HECT-domain protein family. Plant J. 1999, 20, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias, M.J.; Casalongué, C.A.; Terrile, M.C. Ubiquitin-proteasome system as part of nitric oxide sensing in plants. In Nitric Oxide in Plant Biology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 653–687. [Google Scholar]
- Van, V.; Smith, A.T. ATE1-mediated post-translational arginylation is an essential regulator of eukaryotic cellular homeostasis. ACS Chem. Biol. 2020, 15, 3073–3085. [Google Scholar] [CrossRef] [PubMed]
- Labandera, A.M.; Tedds, H.M.; Bailey, M.; Sprigg, C.; Etherington, R.D.; Akintewe, O.; Kalleechurn, G.; Holdsworth, M.J.; Gibbs, D.J. The PRT6 N-degron pathway restricts VERNALIZATION 2 to endogenous hypoxic niches to modulate plant development. New Phytol. 2021, 229, 126–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yun, B.-W.; Kwon, E.; Hong, J.K.; Yoon, J.; Loake, G.J. S-nitrosylation: An emerging redox-based post-translational modification in plants. J. Exp. Bot. 2006, 57, 1777–1784. [Google Scholar] [CrossRef]
- Lubega, J.; Umbreen, S.; Loake, G.J. Recent advances in the regulation of plant immunity by S-nitrosylation. J. Exp. Bot. 2021, 72, 864–872. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J.r. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [Green Version]
- Yun, B.W.; Skelly, M.J.; Yin, M.; Yu, M.; Mun, B.G.; Lee, S.U.; Hussain, A.; Spoel, S.H.; Loake, G.J. Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 2016, 211, 516–526. [Google Scholar] [CrossRef] [Green Version]
- Yun, B.-W.; Feechan, A.; Yin, M.; Saidi, N.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.-G.; Kwon, E.; Spoel, S.H. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Feechan, A.; Yun, B.-W.; Shafiei, R.; Hofmann, A.; Taylor, P.; Xue, P.; Yang, F.-Q.; Xie, Z.-S.; Pallas, J.A. S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 2009, 284, 2131–2137. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Loake, G.J. Redox-based protein modifications: The missing link in plant immune signalling. Curr. Opin. Plant Biol. 2011, 14, 358–364. [Google Scholar] [CrossRef]
- Lindermayr, C.; Sell, S.; Müller, B.; Leister, D.; Durner, J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Mu, J.; Chen, L.; Feng, J.; Hu, J.; Li, L.; Zhou, J.-M.; Zuo, J. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015, 167, 1604–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pinto, M.C.; Locato, V.; Sgobba, A.; Romero-Puertas, M.d.C.; Gadaleta, C.; Delledonne, M.; de Gara, L. S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol. 2013, 163, 1766–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Dommel, M.; Mou, Z. Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR 1 in Arabidopsis thaliana. Plant J. 2016, 86, 20–34. [Google Scholar] [CrossRef] [Green Version]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Correa-Aragunde, N.; Cejudo, F.J.; Lamattina, L. Nitric oxide is required for the auxin-induced activation of NADPH-dependent thioredoxin reductase and protein denitrosylation during root growth responses in arabidopsis. Ann. Bot. 2015, 116, 695–702. [Google Scholar] [CrossRef] [Green Version]
- Rizza, S.; Montagna, C.; Di Giacomo, G.; Cirotti, C.; Filomeni, G. S-nitrosation and ubiquitin-proteasome system interplay in neuromuscular disorders. Int. J. Cell Biol. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Melcher, K.; Ng, L.-M.; Zhou, X.E.; Soon, F.-F.; Xu, Y.; Suino-Powell, K.M.; Park, S.-Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Juste, J.; Leoݩn, J. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR-and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 2010, 152, 891–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, M.-C.; Lozano-Juste, J.; González-Guzmán, M.; Rodriguez, L.; Rodriguez, P.L.; León, J. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 2015, 8, ra89. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Van Wees, S.C.; Van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; Van Loon, L.C. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Backer, R.; Naidoo, S.; van den Berg, N. The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and related family: Mechanistic insights in plant disease resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef] [Green Version]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef] [Green Version]
- Chung, K.K.; Thomas, B.; Li, X.; Pletnikova, O.; Troncoso, J.C.; Marsh, L.; Dawson, V.L.; Dawson, T.M. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science 2004, 304, 1328–1331. [Google Scholar] [CrossRef]
- Nakamura, T.; Wang, L.; Wong, C.C.; Scott, F.L.; Eckelman, B.P.; Han, X.; Tzitzilonis, C.; Meng, F.; Gu, Z.; Holland, E.A. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell 2010, 39, 184–195. [Google Scholar] [CrossRef] [Green Version]
- Kapadia, M.R.; Eng, J.W.; Jiang, Q.; Stoyanovsky, D.A.; Kibbe, M.R. Nitric oxide regulates the 26S proteasome in vascular smooth muscle cells. Nitric Oxide 2009, 20, 279–288. [Google Scholar] [CrossRef]
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terrile, M.C.; París, R.; Calderón-Villalobos, L.I.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongué, C.A. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, G.H.; Cheng, H.; Choe, V.; Bao, X.; Shao, J.; Luo, S.; Rao, H. Cdc48: A swiss army knife of cell biology. J. Amino Acids 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, C.; Gauss, R.; Horn, S.C.; Neuber, O.; Sommer, T. The ubiquitylation machinery of the endoplasmic reticulum. Nature 2009, 458, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, M.; Takata, T.; Kimura, Y.; Manno, A.; Murakami, K.; Koike, M.; Ohizumi, H.; Hori, S.; Kakizuka, A. ATPase activity of p97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in Walker A motif. J. Biol. Chem. 2005, 280, 41332–41341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astier, J.; Besson-Bard, A.; Lamotte, O.; Bertoldo, J.; Bourque, S.; Terenzi, H.; Wendehenne, D. Nitric oxide inhibits the ATPase activity of the chaperone-like AAA+ ATPase CDC48, a target for S-nitrosylation in cryptogein signalling in tobacco cells. Biochem. J. 2012, 447, 249–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosnoblet, C.; Bègue, H.; Blanchard, C.; Pichereaux, C.; Besson-Bard, A.; Aimé, S.; Wendehenne, D. Functional characterization of the chaperon-like protein Cdc48 in cryptogein-induced immune response in tobacco. Plant Cell Environ. 2017, 40, 491–508. [Google Scholar] [CrossRef]
- Rosnoblet, C.; Chatelain, P.; Klinguer, A.; Bègue, H.; Winckler, P.; Pichereaux, C.; Wendehenne, D. The chaperone-like protein Cdc48 regulates ubiquitin-proteasome system in plants. Plant Cell Environ. 2021. [Google Scholar] [CrossRef]

| Targets for S-Nitrosylation | NO-Mediated PTMs | Target for Ubiquitylation | Ubiquitin–Proteasomal Machinery | Purpose | References |
|---|---|---|---|---|---|
| ABI5 | S-nitrosylation at Cys153 | ABI5 | CULLIN4-based and KEEP ON GOING E3 ligases | Germination and early seed development | [78] |
| NPR1 | S-nitrosylation at Cys156 | NPR1 | ABA-mediated degradation through CULLIN3-RING E3 ligase and E4 ligase | Protection from ABA-mediated proteasomal degradation. | [89] |
| ABA receptors of the PYR/PYL/RCAR family | Tyrosine nitration | PYR1 | Polyubiquitinylation for degradation. | Reduce the activity of ABA receptors and hence limit ABA response. | [86] |
| cAPX1 | S-nitrosylation at Cys32 | cAPX1 | Ubiquitylation for degradation. | H2O2 -mediated programed cell death | [75,76] |
| TIR1 | S-nitrosylation at Cys140 and Cys480 | Aux/IAA repressors | E3-ubiquitin ligase complex, SCFTIR1/AFB | Degradation of Aux/IAA repressors to induce auxin-regulated responses. | [95] |
| CDC48 | S-nitrosylation at Cys526 | CDC48 | Itself has ubiquitin–proteasome activity | Compromised immunity against P. cryptogea | [99,101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pande, A.; Mun, B.-G.; Khan, M.; Rahim, W.; Lee, D.-S.; Lee, G.-M.; Al Azawi, T.N.I.; Hussain, A.; Yun, B.-W. Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants. Int. J. Mol. Sci. 2022, 23, 1657. https://doi.org/10.3390/ijms23031657
Pande A, Mun B-G, Khan M, Rahim W, Lee D-S, Lee G-M, Al Azawi TNI, Hussain A, Yun B-W. Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants. International Journal of Molecular Sciences. 2022; 23(3):1657. https://doi.org/10.3390/ijms23031657
Chicago/Turabian StylePande, Anjali, Bong-Gyu Mun, Murtaza Khan, Waqas Rahim, Da-Sol Lee, Geun-Mo Lee, Tiba Nazar Ibrahim Al Azawi, Adil Hussain, and Byung-Wook Yun. 2022. "Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants" International Journal of Molecular Sciences 23, no. 3: 1657. https://doi.org/10.3390/ijms23031657
APA StylePande, A., Mun, B.-G., Khan, M., Rahim, W., Lee, D.-S., Lee, G.-M., Al Azawi, T. N. I., Hussain, A., & Yun, B.-W. (2022). Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants. International Journal of Molecular Sciences, 23(3), 1657. https://doi.org/10.3390/ijms23031657







