New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways
Abstract
1. Introduction
2. The Chloroplast Outer Membrane
2.1. Composition and Function of the Chloroplast Outer Membrane
2.2. Topologies of Chloroplast Outer Membrane Proteins
3. Protein Entry into Chloroplasts: Structure and Function of the TOC Complex
4. Biogenesis of α-Helical Chloroplast Outer Membrane Proteins
4.1. Signal Anchored (SA) Proteins
4.2. Tail Anchored (TA) Proteins
5. Biogenesis of β-Barrel Chloroplast Outer Membrane Proteins
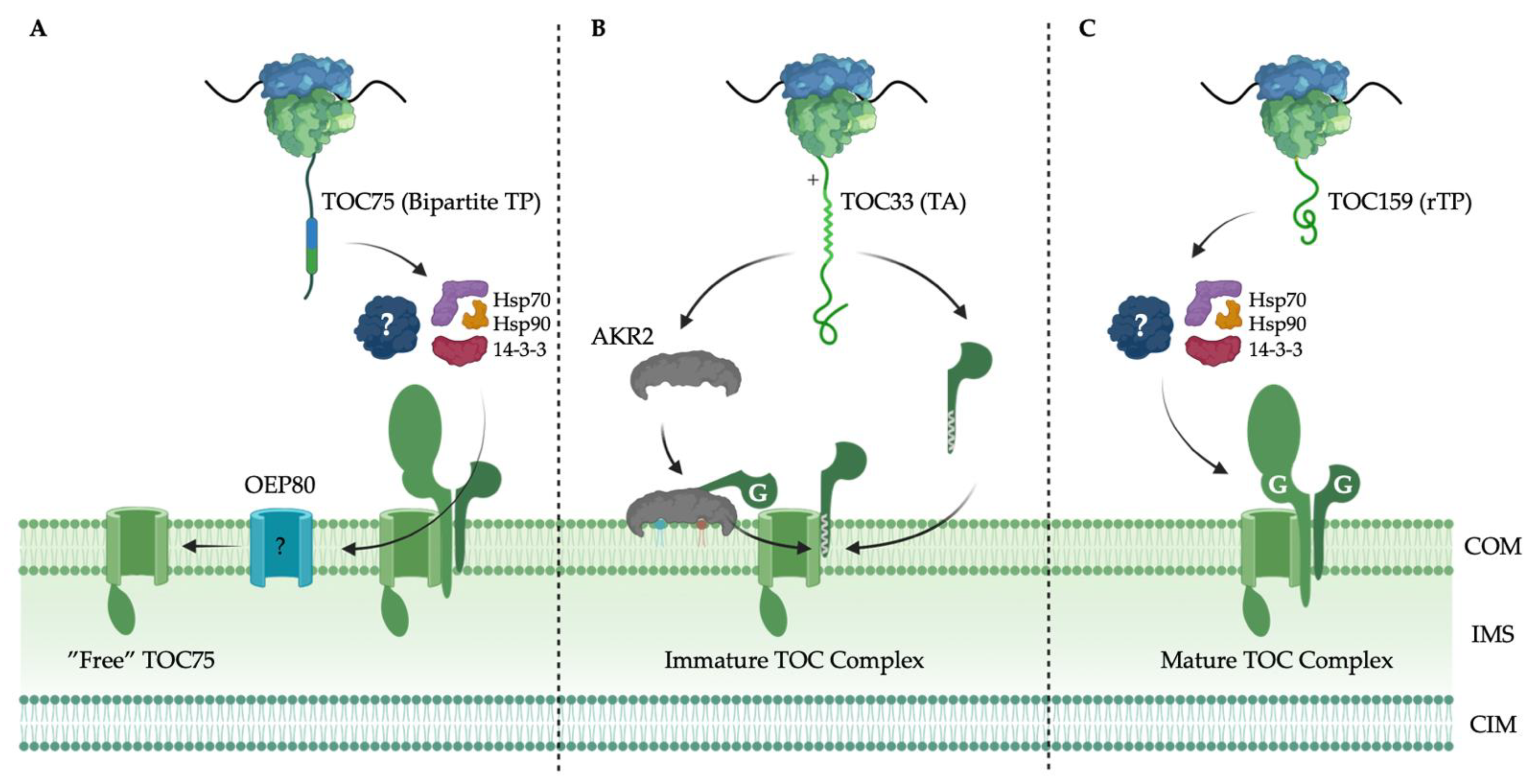
6. Targeting and Assembly of the TOC Complex
6.1. TOC75 Translocation Channel Targeting
6.2. TOC34 GTPase Receptor Family Targeting
6.3. TOC159 GTPase Receptor Family Targeting
7. A Bioinformatic Approach to Identifying Novel Chloroplast Outer Membrane Targeting Signals and Pathways
8. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef]
- Bölter, B. En route into chloroplasts: Preproteins’ way home. Photosynth. Res. 2018, 138, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Day, P.M.; Theg, S.M. Evolution of protein transport to the chloroplast envelope membranes. Photosynth. Res. 2018, 138, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Kessler, F.; Schnell, D. Chloroplast biogenesis: Diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 2009, 21, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Paila, Y.D.; Richardson, L.G.L.; Schnell, D.J. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 2015, 427, 1038–1060. [Google Scholar] [CrossRef]
- Choi, H.; Yi, T.; Ha, S.H. Diversity of plastid types and their interconversions. Front. Plant Sci. 2021, 12, 692024. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.C.; Swamy, K.; Li, H.M. Tissue-specific regulation of plastid protein import via transit-peptide motifs. Plant Cell 2020, 32, 1204–1217. [Google Scholar] [CrossRef]
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta-Bioenerg. 2015, 1847, 1017–1024. [Google Scholar] [CrossRef]
- Chu, C.C.; Li, H.M. Developmental regulation of protein import into plastids. Photosynth. Res. 2018, 138, 327–334. [Google Scholar] [CrossRef]
- Shanmugabalaji, V.; Chahtane, H.; Accossato, S.; Rahire, M.; Gouzerh, G.; Lopez-Molina, L.; Kessler, F. Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr. Biol. 2018, 28, 2616–2623. [Google Scholar] [CrossRef]
- Yan, J.; Smith, M.D.; Glick, B.R.; Liang, Y. Effects of ACC deaminase containing rhizobacteria on plant growth and expression of Toc GTPases in tomato (Solanum lycopersicum) under salt stress. Botany 2014, 92, 775–781. [Google Scholar] [CrossRef]
- Doroodian, P.; Hua, Z. The ubiquitin switch in plant stress response. Plants 2021, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.G.L.; Singhal, R.; Schnell, D.J. The integration of chloroplast protein targeting with plant developmental and stress responses. BMC Biol. 2017, 15, 118. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, Y.; Qi, M.; Liu, Y.; Li, T. Targeted control of chloroplast quality to improve plant acclimation: From protein import to degradation. Front. Plant Sci. 2019, 10, 958. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, H.; Tada, A.; Richardson, L.G.L.; Kakizaki, T.; Uehara, S.; Ito-Inaba, Y.; Inaba, T. Induction of TOC and TIC genes during photomorphogenesis is mediated primarily by cryptochrome 1 in Arabidopsis. Sci. Rep. 2020, 10, 20255. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Rounds, C.; Schnell, D.J. The molecular basis for distinct pathways for protein import into arabidopsis chloroplasts. Plant Cell 2010, 22, 1947–1960. [Google Scholar] [CrossRef]
- Li, H.; Teng, Y.S. Transit peptide design and plastid import regulation. Trends Plant Sci. 2013, 18, 360–366. [Google Scholar] [CrossRef]
- Chu, C.C.; Li, H.M. Protein import into isolated pea root leucoplasts. Front. Plant Sci. 2015, 6, 690. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Chloroplast research in the genomic age. Trends Genet. 2003, 19, 47–56. [Google Scholar] [CrossRef]
- Kleffmann, T.; Russenberger, D.; Von Zychlinski, A.; Christopher, W.; Sjölander, K.; Gruissem, W.; Baginsky, S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 2004, 14, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Zybailov, B.; Rutschow, H.; Friso, G.; Rudella, A.; Emanuelsson, O.; Sun, Q.; Van Wijk, K.J. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 2008, 3, e1994. [Google Scholar] [CrossRef] [PubMed]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, P.; Paul, J. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008, 179, 257–285. [Google Scholar] [CrossRef]
- Schnell, D.J. The TOC GTPase receptors: Regulators of the fidelity, specificity and substrate profiles of the general protein import machinery of chloroplasts. Protein J. 2019, 38, 343–350. [Google Scholar] [CrossRef]
- Richardson, L.G.L.; Paila, Y.D.; Siman, S.R.; Chen, Y.; Smith, M.D.; Schnell, D.J. Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 2014, 5, 269. [Google Scholar] [CrossRef]
- Kim, J.; Na, Y.J.; Park, S.J.; Baek, S.H.; Kim, D.H. Biogenesis of chloroplast outer envelope membrane proteins. Plant Cell Rep. 2019, 38, 783–792. [Google Scholar] [CrossRef]
- Sjuts, I.; Soll, J.; Bölter, B. Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front. Plant Sci. 2017, 8, 168. [Google Scholar] [CrossRef]
- Bouchnak, I.; Brugière, S.; Moyet, L.; Le Gall, S.; Salvi, D.; Kuntz, M.; Tardif, M.; Rolland, N. Unraveling hidden components of the chloroplast envelope proteome: Opportunities and limits of better MS sensitivity. Mol. Cell. Proteom. 2019, 18, 1285–1306. [Google Scholar] [CrossRef]
- Inoue, K. Emerging knowledge of the organelle outer membranes—Research snapshots and an updated list of the chloroplast outer envelope proteins. Front. Plant Sci. 2015, 6, 278. [Google Scholar] [CrossRef][Green Version]
- Inoue, K. Emerging roles of the chloroplast outer envelope membrane. Trends Plant Sci. 2011, 16, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Breuers, F.K.H.; Bräutigam, A.; Weber, A.P.M. The plastid outer envelope—A highly dynamic interface between plastid and cytoplasm. Front. Plant Sci. 2011, 2, 97. [Google Scholar] [CrossRef]
- Schleiff, E.; Tien, R.; Salomon, M.; Soll, J. Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell 2001, 12, 4090–4102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cline, K.; Keegstra, K. Galactosyltransferases involved in galactolipid biosynthesis are located in the outer membrane of pea chloroplast envelopes. Plant Physiol. 1983, 71, 366–372. [Google Scholar] [CrossRef]
- Sahin, C.; Reid, D.J.; Marty, M.T.; Landreh, M. Scratching the surface: Native mass spectrometry of peripheral membrane protein complexes. Biochem. Soc. Trans. 2020, 48, 547–558. [Google Scholar] [CrossRef]
- Murcha, M.W.; Kmiec, B.; Kubiszewski-Jakubiak, S.; Teixeira, P.F.; Glaser, E.; Whelan, J. Protein import into plant mitochondria: Signals, machinery, processing, and regulation. J. Exp. Bot. 2014, 65, 6301–6335. [Google Scholar] [CrossRef]
- Dhanoa, P.K.; Richardson, L.G.L.; Smith, M.D.; Gidda, S.K.; Henderson, M.P.A.; Andrews, D.W.; Mullen, R.T. Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 2010, 5, e10098. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Lee, J.; Hwang, I. Sorting of nuclear-encoded chloroplast membrane proteins. Curr. Opin. Plant Biol. 2017, 40, 1–7. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.H.; Hwang, I. Specific targeting of proteins to outer envelope membranes of endosymbiotic organelles, chloroplasts, and mitochondria. Front. Plant Sci. 2014, 5, 173. [Google Scholar] [CrossRef]
- Richardson, L.G.L.; Schnell, D.J. Origins, function, and regulation of the TOC-TIC general protein import machinery of plastids. J. Exp. Bot. 2020, 71, 1226–1238. [Google Scholar] [CrossRef]
- Thomson, S.M.; Pulido, P.; Jarvis, R.P. Protein import into chloroplasts and its regulation by the ubiquitin-proteasome system. Biochem. Soc. Trans. 2020, 48, 71–82. [Google Scholar] [CrossRef]
- Andrès, C.; Agne, B.; Kessler, F. The TOC complex: Preprotein gateway to the chloroplast. Biochim. Biophys. Acta-Mol. Cell Res. 2010, 1803, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M. New perspectives on chloroplast protein import. Plant Cell Physiol. 2018, 59, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D. Protein import into chloroplasts: An ever-evolving story. Can. J. Bot. 2006, 84, 531–542. [Google Scholar] [CrossRef]
- Lee, D.W.; Jung, C.; Hwang, I. Cytosolic events involved in chloroplast protein targeting. Biochim. Biophys. Acta-Mol. Cell Res. 2013, 1833, 245–252. [Google Scholar] [CrossRef]
- Patron, N.J.; Waller, R.F. Transit peptide diversity and divergence: A global analysis of plastid targeting signals. BioEssays 2007, 29, 1048–1058. [Google Scholar] [CrossRef]
- Lee, D.W.; Hwang, I. Evolution and design principles of the diverse chloroplast transit peptides. Mol. Cells 2018, 41, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Yoo, Y.J.; Razzak, M.A.; Hwang, I. Prolines in transit peptides are crucial for efficient preprotein translocation into chloroplasts. Plant Physiol. 2018, 176, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Bruce, B.D. The paradox of plastid transit peptides: Conservation of function despite divergence in primary structure. Biochim. Biophys. Acta-Mol. Cell Res. 2001, 1541, 2–21. [Google Scholar] [CrossRef]
- Dong, W.L.; Jong, K.K.; Lee, S.; Choi, S.; Kim, S.; Hwang, I. Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 2008, 20, 1603–1622. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, S.; Lee, J.; Woo, S.; Razzak, M.A.; Vitale, A.; Hwang, I. Molecular mechanism of the specificity of protein import into chloroplasts and mitochondria in plant cells. Mol. Plant 2019, 12, 951–966. [Google Scholar] [CrossRef]
- Flores-Pérez, Ú.; Jarvis, P. Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta-Mol. Cell Res. 2013, 1833, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Hristou, A.; Grimmer, J.; Baginsky, S. The secret life of chloroplast precursor proteins in the cytosol. Mol. Plant 2020, 13, 1111–1113. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, R.; Whelan, J.; Vrielink, A. Exploring ligand recognition, selectivity and dynamics of TPR domains of chloroplast Toc64 and mitochondria Om64 from Arabidopsis thaliana. J. Mol. Recognit. 2014, 27, 402–414. [Google Scholar] [CrossRef]
- Qbadou, S.; Becker, T.; Mirus, O.; Tews, I.; Soll, J.; Schleiff, E. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006, 25, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Kouranov, A.; Schnell, D.J. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 1997, 139, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Rounds, C.M.; Wang, F.; Chen, K.; Afitlhile, M.; Schnell, D.J. atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 2004, 165, 323–334. [Google Scholar] [CrossRef]
- Kessler, F.; Blobel, G.; Patel, H.A.; Schnell, D.J. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 1994, 266, 1035–1039. [Google Scholar] [CrossRef]
- Yan, J.; Campbell, J.H.; Glick, B.R.; Smith, M.D.; Liang, Y. Molecular characterization and expression analysis of chloroplast protein import components in tomato (Solanum lycopersicum). PLoS ONE 2014, 9, e95088. [Google Scholar] [CrossRef]
- Ramundo, S.; Asakura, Y.; Salomé, P.A.; Strenkert, D.; Boone, M.; Mackinder, L.C.M.; Takafuji, K.; Dinc, E.; Rahire, M.; Crèvecoeur, M.; et al. Coexpressed subunits of dual genetic origin define a conserved supercomplex mediating essential protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2020, 117, 32739–32749. [Google Scholar] [CrossRef]
- Agne, B.; Kessler, F. Protein transport in organelles: The Toc complex way of preprotein import. FEBS J. 2009, 276, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Schleiff, E.; Soll, J.; Küchler, M.; Kühlbrandt, W.; Harrer, R. Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 2003, 160, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Li, H.M. Stable megadalton TOC–TIC supercomplexes as major mediators of protein import into chloroplasts. Plant J. 2017, 92, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Hirohashi, T.; Nakai, M. Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol. 2006, 47, 363–371. [Google Scholar] [CrossRef]
- Chen, K.Y.; Li, H.M. Precursor binding to an 880-kDa Toc complex as an early step during active import of protein into chloroplasts. Plant J. 2007, 49, 149–158. [Google Scholar] [CrossRef]
- Jackson-Constan, D.; Keegstra, K. Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol. 2001, 125, 1567–1576. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Becker, T. Mechanisms and pathways of mitochondrial outer membrane protein biogenesis. Biochim. Biophys. Acta-Bioenerg. 2021, 1862, 148323. [Google Scholar] [CrossRef]
- Lundquist, K.; Billings, E.; Bi, M.; Wellnitz, J.; Noinaj, N. The assembly of β-barrel membrane proteins by BAM and SAM. Mol. Microbiol. 2021, 115, 425–435. [Google Scholar] [CrossRef]
- Teresinski, H.J.; Gidda, S.K.; Nguyen, T.N.D.; Howard, N.J.M.; Porter, B.K.; Grimberg, N.; Smith, M.D.; Andrews, D.W.; Dyer, J.M.; Mullen, R.T. An RK/ST C-terminal motif is required for Ttrgeting of OEP7.2 and a subset of other Arabidopsis tail-anchored proteins to the plastid outer envelope membrane. Plant Cell Physiol. 2019, 60, 516–537. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, H.; Kim, J.; Lee, S.; Kim, D.H.; Kim, S.; Hwang, I. Both the hydrophobicity and a positively charged region flanking the c-terminal region of the transmembrane domain of signal-anchored proteins play critical roles in determining their targeting specificity to the endoplasmic reticulum or endosymbiotic org. Plant Cell 2011, 23, 1588–1607. [Google Scholar] [CrossRef]
- Lee, Y.J.; Sohn, E.J.; Lee, K.H.; Lee, D.W.; Hwang, I. The transmembrane domain of AtTco64 and its C-terminal lysine-rich flanking region are targeting signals to the chloroplast outer envelope membrane. Mol. Cells 2004, 17, 281–291. [Google Scholar]
- Bae, W.; Lee, Y.J.; Kim, D.H.; Lee, J.; Kim, S.; Sohn, E.J.; Hwang, I. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 2008, 10, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lee, J.E.; Xu, Z.Y.; Geem, K.R.; Kwon, Y.; Park, J.W.; Hwang, I. Cytosolic targeting factor AKR2A captures chloroplast outer membrane-localized client proteins at the ribosome during translation. Nat. Commun. 2015, 6, 6843. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, M.J.; Gwon, G.H.; Silkov, A.; Xu, Z.Y.; Yang, E.C.; Song, S.; Song, K.; Kim, Y.; Yoon, H.S.; et al. An ankyrin repeat domain of AKR2 drives chloroplast targeting through coincident binding of two chloroplast lipids. Dev. Cell 2014, 30, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Chung, K.P.; Jiang, L. Targeting tail-anchored proteins into plant organelles. Proc. Natl. Acad. Sci. USA 2017, 114, 1762–1764. [Google Scholar] [CrossRef] [PubMed]
- Moog, D. Higher complexity requires higher accuracy: Tail-anchored protein targeting to the outer envelope membrane of plant plastids via a specific C-terminal motif. Plant Cell Physiol. 2019, 60, 489–491. [Google Scholar] [CrossRef]
- Marty, N.J.; Teresinski, H.J.; Hwang, Y.T.; Clendening, E.A.; Gidda, S.K.; Sliwinska, E.; Zhang, D.; Miernyk, J.A.; Brito, G.C.; Andrews, D.W.; et al. New insights into the targeting of a subset of tail-anchored proteins to the outer mitochondrial membrane. Front. Plant Sci. 2014, 5, 426. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Formighieri, C.; Cazzaniga, S.; Kuras, R.; Bassi, R. Biogenesis of photosynthetic complexes in the chloroplast of Chlamydomonas reinhardtii requires ARSA1, a homolog of prokaryotic arsenite transporter and eukaryotic TRC40 for guided entry of tail-anchored proteins. Plant J. 2013, 73, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.W.; Chen, C.C.; Wu, S.M.; Chang, Y.C.; Li, Y.C.; Su, Y.W.; Hsiao, C.D.; Chang, H.Y. Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1. Plant J. 2019, 99, 128–143. [Google Scholar] [CrossRef]
- Maestre-Reyna, M.; Wu, S.M.; Chang, Y.C.; Chen, C.C.; Maestre-Reyna, A.; Wang, A.H.J.; Chang, H.Y. In search of tail-anchored protein machinery in plants: Reevaluating the role of arsenite transporters. Sci. Rep. 2017, 7, 46022. [Google Scholar] [CrossRef]
- Anderson, S.A.; Satyanarayan, M.B.; Wessendorf, R.L.; Lu, Y.; Fernandez, D.E. A homolog of GuidedEntry of Tail-anchored proteins3 functions in membrane-specific protein targeting in chloroplasts of Arabidopsis. Plant Cell 2021, 33, 2812–2833. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Abell, B.M. Chloroplast envelope protein targeting fidelity is independent of cytosolic components in dual organelle assays. Front. Plant Sci. 2012, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.L.; Chen, L.J.; Smith, M.D.; Su, Y.S.; Schnell, D.J.; Li, H.M. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 2004, 16, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Jores, T.; Rapaport, D. Early stages in the biogenesis of eukaryotic β-barrel proteins. FEBS Lett. 2017, 591, 2671–2681. [Google Scholar] [CrossRef]
- Klinger, A.; Gosch, V.; Bodensohn, U.; Ladig, R.; Schleiff, E. The signal distinguishing between targeting of outer membrane β-barrel protein to plastids and mitochondria in plants. Biochim. Biophys. Acta-Mol. Cell Res. 2019, 1866, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Walther, D.M.; Rapaport, D. Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta-Mol. Cell Res. 2009, 1793, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Jores, T.; Klinger, A.; Groß, L.E.; Kawano, S.; Flinner, N.; Duchardt-Ferner, E.; Wöhnert, J.; Kalbacher, H.; Endo, T.; Schleiff, E.; et al. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat. Commun. 2016, 7, 12036. [Google Scholar] [CrossRef] [PubMed]
- Höhr, A.I.C.; Lindau, C.; Wirth, C.; Qiu, J.; Stroud, D.A.; Kutik, S.; Guiard, B.; Hunte, C.; Becker, T.; Pfanner, N.; et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science 2018, 359, eaah6834. [Google Scholar] [CrossRef] [PubMed]
- Horne, J.E.; Brockwell, D.J.; Radford, S.E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. 2020, 295, 10340–10367. [Google Scholar] [CrossRef]
- Gessmann, D.; Chung, Y.H.; Danoff, E.J.; Plummer, A.M.; Sandlin, C.W.; Zaccai, N.R.; Fleming, K.G. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc. Natl. Acad. Sci. USA 2014, 111, 5878–5883. [Google Scholar] [CrossRef]
- Day, P.M.; Inoue, K.; Theg, S.M. Chloroplast outer membrane b-barrel proteins use components of the general import apparatus. Plant Cell 2019, 31, 1845–1855. [Google Scholar] [CrossRef]
- Gross, L.E.; Klinger, A.; Spies, N.; Ernst, T.; Flinner, N.; Simm, S.; Ladig, R.; Bodensohn, U.; Schleiff, E. Insertion of plastidic b-barrel proteins into the outer envelopes of plastids involves an intermembrane space intermediate formed with Toc75-V/OEP80. Plant Cell 2021, 33, 1657–1681. [Google Scholar] [CrossRef]
- Huang, W.; Ling, Q.; Bédard, J.; Lilley, K.; Jarvis, P. In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol. 2011, 157, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Hsu, S.C.; Bédard, J.; Inoue, K.; Jarvis, P. The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 2008, 148, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Tranel, P.J.; Keegstra, K. A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 1996, 8, 2093–2104. [Google Scholar] [CrossRef]
- Kouranov, A.; Chen, X.; Fuks, B.; Schnell, D.J. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 1998, 143, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Day, P.M.; Potter, D.; Inoue, K. Evolution and targeting of omp85 homologs in the chloroplast outer envelope membrane. Front. Plant Sci. 2014, 5, 535. [Google Scholar] [CrossRef]
- Schleiff, E.; Becker, T. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 2011, 12, 48–59. [Google Scholar] [CrossRef]
- Inoue, K.; Baldwin, A.J.; Shipman, R.L.; Matsui, K.; Theg, S.M.; Ohme-Takagi, M. Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 2005, 171, 425–430. [Google Scholar] [CrossRef]
- Endow, J.K.; Rocha, A.G.; Baldwin, A.J.; Roston, R.L.; Yamaguchi, T.; Kamikubo, H.; Inoue, K. Polyglycine acts as a rejection signal for protein transport at the chloroplast envelope. PLoS ONE 2016, 11, e0167802. [Google Scholar] [CrossRef]
- Inoue, K.; Potter, D. The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J. 2004, 39, 354–365. [Google Scholar] [CrossRef]
- Gross, L.E.; Spies, N.; Simm, S.; Schleiff, E. Toc75-V/OEP80 is processed during translocation into chloroplasts, and the membrane-embedded form exposes its POTRA domain to the intermembrane space. FEBS Open Bio 2020, 10, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Eckart, K.; Eichacker, L.; Sohrt, K.; Schleiff, E.; Heins, L.; Soll, J. A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep. 2002, 3, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Hwang, I. Direct targeting of proteins from the cytosol to organelles: The ER versus endosymbiotic organelles. Traffic 2013, 14, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Schleiff, E.; Klösgen, R.B. Without a little help from “my” friends: Direct insertion of proteins into chloroplast membranes? Biochim. Biophys. Acta-Mol. Cell Res. 2001, 1541, 22–33. [Google Scholar] [CrossRef]
- Qbadou, S.; Tien, R.; Soll, J.; Schleiff, E. Membrane insertion of the chloroplast outer envelope protein, Toc34: Constrains for insertion and topology. J. Cell Sci. 2003, 116, 837–846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wallas, T.R.; Smith, M.D.; Sanchez-Nieto, S.; Schnell, D.J. The roles of Toc34 and Toc75 in targeting the Toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 2003, 278, 44289–44297. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, N.R.; Theg, S.M. Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci. 2005, 10, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.J.; Forouhar, F.; Li, H.M.; Tu, S.L.; Yeh, Y.H.; Kao, S.; Shr, H.L.; Chou, C.C.; Chen, C.; Hsiao, C.D. Crystal structure of pea toc34, a novel gtpase of the chloroplast protein translocon. Nat. Struct. Biol. 2002, 9, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Wang, F.; Schnell, D.J. Toc receptor dimerization participates in the initiation of membrane translocation during protein import into chloroplasts. J. Biol. Chem. 2009, 284, 31130–31141. [Google Scholar] [CrossRef]
- Ivanova, Y.; Smith, M.D.; Chen, K.; Schnell, D.J. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 2004, 15, 3379–3392. [Google Scholar] [CrossRef]
- Smith, M.D.; Hiltbrunner, A.; Kessler, F.; Schnell, D.J. The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 2002, 159, 833–843. [Google Scholar] [CrossRef]
- Bauer, J.; Hiltbrunner, A.; Weibel, P.; Vidi, P.A.; Alvarez-Huerta, M.; Smith, M.D.; Schnell, D.J.; Kessler, F. Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol. 2002, 159, 845–854. [Google Scholar] [CrossRef]
- Muckel, E.; Soll, J. A protein import receptor of chloroplasts is inserted into the outer envelope membrane by a novel pathway. J. Biol. Chem. 1996, 271, 23846–23852. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, S.J.; Lee, Y.J.; Jin, J.B.; Hwang, I. The M domain of atToc159 plays an essential role in the import of proteins into chloroplasts and chloroplast biogenesis. J. Biol. Chem. 2003, 278, 36794–36805. [Google Scholar] [CrossRef]
- Lung, S.C.; Smith, M.D.; Weston, J.K.; Gwynne, W.; Secord, N.; Chuong, S.D.X. The C-terminus of Bienertia sinuspersici Toc159 contains essential elements for its targeting and anchorage to the chloroplast outer membrane. Front. Plant Sci. 2014, 5, 722. [Google Scholar] [CrossRef]
- Lung, S.C.; Chuong, S.D.X. A transit peptide-like sorting signal at the C terminus directs the bienertia sinuspersici preprotein receptor toc159 to the chloroplast outer membrane. Plant Cell 2012, 24, 1560–1578. [Google Scholar] [CrossRef]
- Bauer, J.; Chen, K.; Hiltbunner, A.; Wehrli, E.; Eugster, M.; Schnell, D.; Kessler, F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 2000, 403, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, X.; Schnell, D.J. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 2000, 122, 813–822. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Bennewitz, B.; Klösgen, R.B. Dual or not dual?—comparative analysis of fluorescence microscopy-based approaches to study organelle targeting specificity of nuclear-encoded plant proteins. Front. Plant Sci. 2018, 9, 1350. [Google Scholar] [CrossRef]
- Garg, S.G.; Gould, S.B. The role of charge in protein targeting evolution. Trends Cell Biol. 2016, 26, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Spånning, E.; Glaser, E.; Wieslander, Å. Import determinants of organelle-specific and dual targeting peptides of mitochondria and chloroplasts in Arabidopsis thaliana. Mol. Plant 2014, 7, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Carrie, C.; Small, I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim. Biophys. Acta-Mol. Cell Res. 2013, 1833, 253–259. [Google Scholar] [CrossRef]
- Chen, Y.L.; Chen, L.J.; Chu, C.C.; Huang, P.K.; Wen, J.R.; Li, H. min TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 2018, 564, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Rossig, C.; Reinbothe, C.; Gray, J.; Valdes, O.; Von Wettstein, D.; Reinbothe, S. Three proteins mediate import of transit sequence-less precursors into the inner envelope of chloroplasts in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2013, 110, 19962–19967. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- Ling, Q.; Jarvis, P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 2015, 25, 2527–2534. [Google Scholar] [CrossRef]
- Liu, H.; Stone, S.L. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 2010, 22, 2630–2641. [Google Scholar] [CrossRef]
- Kachroo, A.; Venugopal, S.C.; Lapchyk, L.; Falcone, D.; Hildebrand, D.; Kachroo, P. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 5152–5157. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A.D. Characterization of a Novel Carotenoid Cleavage Dioxygenase from Plants. J. Biol. Chem. 2001, 276, 25208–25211. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Zhang, X.; Schläppi, M.; Xu, Z.Q. Cold-inducible expression of AZI1 and its function in improvement of freezing tolerance of Arabidopsis thaliana and Saccharomyces cerevisiae. J. Plant Physiol. 2011, 168, 1576–1587. [Google Scholar] [CrossRef]
- Higashi, Y.; Okazaki, Y.; Takano, K.; Myouga, F.; Shinozaki, K.; Knoch, E.; Fukushima, A.; Saito, K. HEAT INDUCIBLE LIPASE1 remodels chloroplastic monogalactosyldiacylglycerol by liberating α-linolenic acid in arabidopsis leaves under heat stress. Plant Cell 2018, 30, 1887–1905. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Nakanishi, H.; Suzuki, K.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Miyagishima, S. ya The YlmG protein has a conserved function related to the distribution of nucleoids in chloroplasts and cyanobacteria. BMC Plant Biol. 2010, 10, 57. [Google Scholar] [CrossRef]
- Gao, H.; Kadirjan-Kalbach, D.; Froehlicht, J.E.; Osteryoung, K.W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 2003, 100, 4328–4333. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, J. Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 2009, 57, 146–159. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Heijne, G. Von ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Nugent, T.; Jones, D.T. Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinform. 2012, 13, 169. [Google Scholar] [CrossRef]
- Bagos, P.G.; Liakopoulos, T.D.; Spyropoulos, I.C.; Hamodrakas, S.J. PRED-TMBB: A web server for predicting the topology of β-barrel outer membrane proteins. Nucleic Acids Res. 2004, 32, 400–404. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007, 35, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Sperschneider, J.; Catanzariti, A.M.; Deboer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed]
- Small, I.; Peeters, N.; Legeai, F.; Lurin, C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 2004, 4, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 4049. [Google Scholar] [CrossRef]


| AGI 1 | Name 2 | N TP 3 | C TP-Like 4 |
|---|---|---|---|
| Signal Anchored (SA) Proteins | |||
| At1g67690 | M3 Protease | ✓ | |
| At2g19860 | HXK2 | ||
| At2g34585 * | - | ||
| At2g38670 | PECT1 | ||
| At3g17970 | TOC64-III | ✓ | |
| At3g21865 | PEX22 | ✓ | |
| At3g52420 | OEP7 | ||
| At3g63170 | FAP1 | ✓ | |
| At4g12470 * | pEARLI1-Like Lipid Transfer Protein | ✓ | |
| At4g27680 | NTPase | ||
| At4g29130 | HXK1 | ||
| At5g17770 | CBR | ||
| At5g20520 | WAV2 | ✓ | |
| At5g25900 | KO1/GA3 | ||
| At5g51020 | CRL | ||
| At5g64816 | - | ✓ | |
| Tail Anchored (TA) Proteins | |||
| At1g02280 | TOC33 | ||
| At1g09920 | - | ||
| At1g13900 | PAP2 | ||
| At1g26340 * | Cytochrome b5 | ||
| At1g27300 | - | ||
| At1g27390 | Tom20-2 | ||
| At2g16070 | PDV2 | ||
| At2g32240 | DUF869 | ||
| At3g03870 * | F20H23.8 Protein | ✓ | |
| At3g27820 | MDAR4 | ||
| At3g57090 * | Mitochondrial Fission 1 Protein | ||
| At3g63150 | MIRO2 | ||
| At4g32250 | Tyrosine Kinase | ||
| At4g35000 | APX3 | ||
| At5g05000 | TOC34 | ✓ | |
| At5g11560 | - | ✓ | |
| At5g21990 | OEP61-TPR | ||
| At5g27330 | - | ||
| At5g27540 | MIRO1 | ||
| At5g56730 | Peptidase M16 | ||
| Other Single Passα-Helical Proteins | |||
| At1g16000 | OEP9 | ||
| At1g80890 | OEP9.2 | ||
| At2g25660 * | TIC236 | ✓ | ✓ |
| At3g52230 | OMP24 Homolog | ||
| At3g63160 | OEP6 | ||
| Multi-Passα-Helical Proteins | |||
| At1g12230 | Transaldolase | ✓ | |
| At1g34430 | PDC E2 | ✓ | |
| At1g44170 | ALDH3H1 | ✓ | |
| At1g54150 * | E3 Ubiquitin-Protein Ligase SPL2 | ✓ | |
| At1g59560 * | E3 Ubiquitin-Protein Ligase SPL1 | ||
| At1g63900 | SP1 | ✓ | |
| At1g64850 | - | ✓ | |
| At1g68680 | - | ||
| At1g77590 | LACS9 | ✓ | |
| At2g01320 | WBC7 | ||
| At2g11810 | MGD3 | ✓ | |
| At2g28900 | OEP16-1 | ||
| At2g34590 | PDC E1 Beta | ✓ | |
| At2g40690 * | G3P Dehydrogenase | ✓ | |
| At2g44640 | - | ||
| At2g47770 | TSPO | ||
| At3g07430 * | YlmG Homolog | ✓ | |
| At3g49560 * | TIM Protein | ||
| At3g51870 | PAPST1 Homolog | ||
| At3g62880 | OEP16-4 | ||
| At4g15440 | HPL Homolog | ||
| At4g15810 | NTPase | ||
| At4g16160 | OEP16-2 | ✓ | |
| At4g16450 | Complex I Subunit | ✓ | |
| At4g26670 * | TIM Protein | ||
| At4g27990 | YGGT-B Protein | ✓ | |
| At4g31780 | MGD1 | ✓ | |
| At4g38920 | Vacuolar ATPase Subunit | ||
| At5g06290 | Prx B | ✓ | |
| At5g13530 * | E3 Ubiquitin-Protein Ligase KEG | ||
| At5g16010 * | Dehydrogenase | ||
| At5g21920 | YGGT-A Protein | ||
| At5g24650 * | TIM Protein | ||
| At5g35210 | PTM | ||
| At5g55510 * | TIM Protein | ||
| β-Barrel Proteins | |||
| At1g20816 | OEP21-1 | ||
| At1g45170 | OEP24-1 | ||
| At1g76405 | OEP21-2 | ||
| At2g06010 | - | ||
| At2g43950 | OEP37 | ✓ | |
| At3g01280 | VDAC1 | ||
| At3g44160 | P39/OEP80 TR1 | ||
| At3g46740 | TOC75-III | ✓ | |
| At3g48620 | P36/OEP80 TR2 | ||
| At4g09080 | TOC75-IV | ||
| At5g15090 | VDAC3 | ||
| At5g19620 | OEP80/TOC75-V | ✓ | |
| At5g42960 | OEP24-2 | ||
| Other Proteins | |||
| At1g02560 | ClpP5 | ✓ | |
| At1g07930 | E-Tu | ||
| At1g09340 | CRB | ||
| At1g70480 | DUF220 | ✓ | |
| At2g16640 | TOC132 | ✓ | |
| At2g17390 | AKR2B | ✓ | |
| At2g17695 | OEP23 | ||
| At2g20890 | THF1/PSB29 | ✓ | |
| At2g24440 | - | ✓ | |
| At2g27490 | ATCOAE | ||
| At2g32290 * | Beta-Amylase 6 | ✓ | |
| At2g32650 | PTAC18-Like | ✓ | |
| At3g01500 | Beta CA1 | ✓ | |
| At3g06510 | SFR2/GGGT | ✓ | |
| At3g06960 | TGD4 | ||
| At3g11670 | DGD1 | ✓ | |
| At3g12580 | Hsc70-4 | ✓ | |
| At3g16620 | TOC120 | ✓ | |
| At3g16950 | PDC E3 | ✓ | |
| At3g19720 * | ARC5 | ||
| At3g25690 | CHUP1 | ||
| At3g25860 | PDC E2 | ✓ | |
| At3g26070 | PAP/FBN3a | ✓ | |
| At3g26740 | CCL | ✓ | |
| At3g46030 | Histone H2B | ✓ | |
| At3g46780 | pTAC16 | ✓ | |
| At3g49350 | - | ✓ | |
| At3g53560 | TPR Protein | ✓ | |
| At3g63520 * | Carotenoid Cleaving Protein | ||
| At4g00550 | DGD2 | ✓ | |
| At4g02482 | Putative GTPase | ✓ | |
| At4g02510 | TOC159 | ✓ | |
| At4g05050 | UBQ11 | ||
| At4g13550 * | Putative Triglyceride Lipase | ✓ | |
| At4g14430 | Enoyl-CoA Isomerase | ✓ | |
| At4g17170 | RAB2 | ✓ | |
| At4g36650 | pBrP | ||
| At5g02500 | Hsc70-1 | ✓ | |
| At5g02580 * | Argininosuccinate Lyase | ||
| At5g16870 | PTH2 Family Protein | ||
| At5g20300 | TOC90 | ✓ | |
| At5g20410 | MGD2 | ✓ | |
| At5g23190 | CYP86B1 | ✓ | |
| At5g35360 | CAC2/BC | ✓ | |
| At5g42070 | - | ✓ | |
| At5g43070 | WPP1 | ✓ | |
| At5g53280 | PDV1 | ||
| At5g58140 | PHOT2 | ✓ | |
| At5g59840 | RAB8A-Like Protein | ✓ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fish, M.; Nash, D.; German, A.; Overton, A.; Jelokhani-Niaraki, M.; Chuong, S.D.X.; Smith, M.D. New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways. Int. J. Mol. Sci. 2022, 23, 1571. https://doi.org/10.3390/ijms23031571
Fish M, Nash D, German A, Overton A, Jelokhani-Niaraki M, Chuong SDX, Smith MD. New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways. International Journal of Molecular Sciences. 2022; 23(3):1571. https://doi.org/10.3390/ijms23031571
Chicago/Turabian StyleFish, Michael, Delaney Nash, Alexandru German, Alyssa Overton, Masoud Jelokhani-Niaraki, Simon D. X. Chuong, and Matthew D. Smith. 2022. "New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways" International Journal of Molecular Sciences 23, no. 3: 1571. https://doi.org/10.3390/ijms23031571
APA StyleFish, M., Nash, D., German, A., Overton, A., Jelokhani-Niaraki, M., Chuong, S. D. X., & Smith, M. D. (2022). New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways. International Journal of Molecular Sciences, 23(3), 1571. https://doi.org/10.3390/ijms23031571






