Challenges in Discovering Drugs That Target the Protein–Protein Interactions of Disordered Proteins
Abstract
1. Protein–Protein Interactions (PPIs) as Drug Targets
2. Characteristics of Molecules Targeting PPIs
3. Disordered Proteins in PPIs as Attractive Drug Targets
4. Strategies for Treating Neurodegenerative Disorders
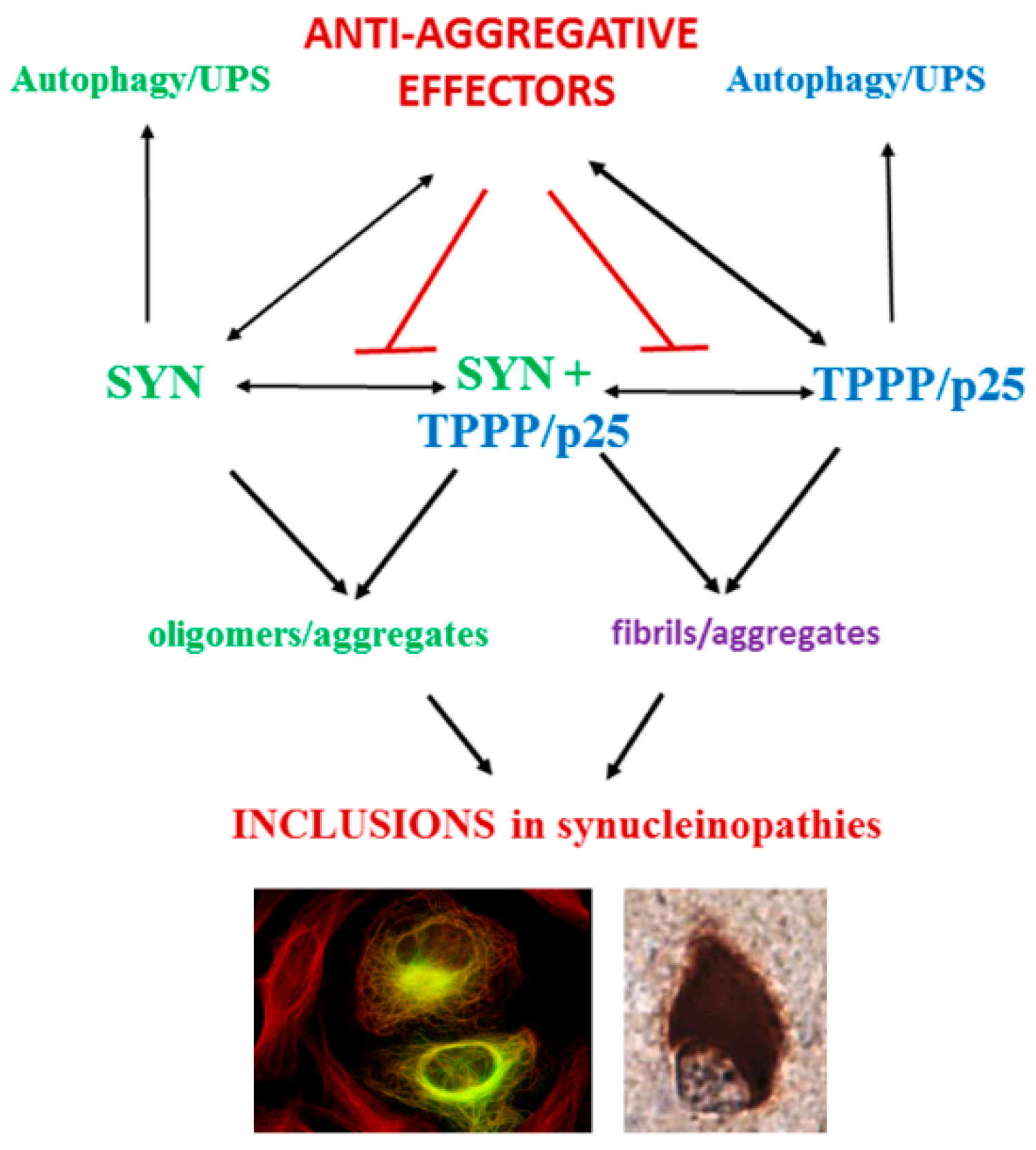
4.1. Direct PPI Targeting
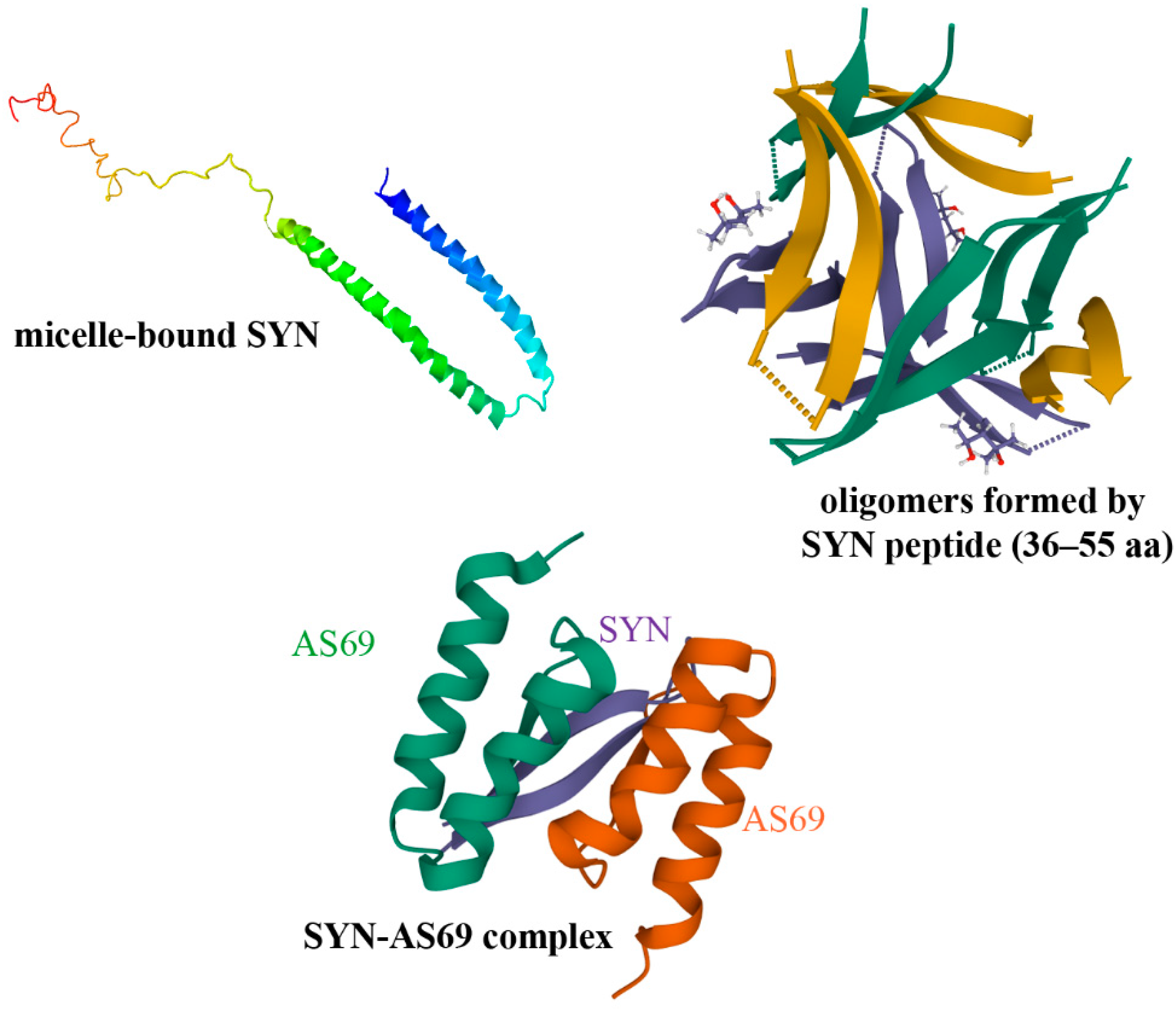
4.1.1. Small Molecules, Peptides
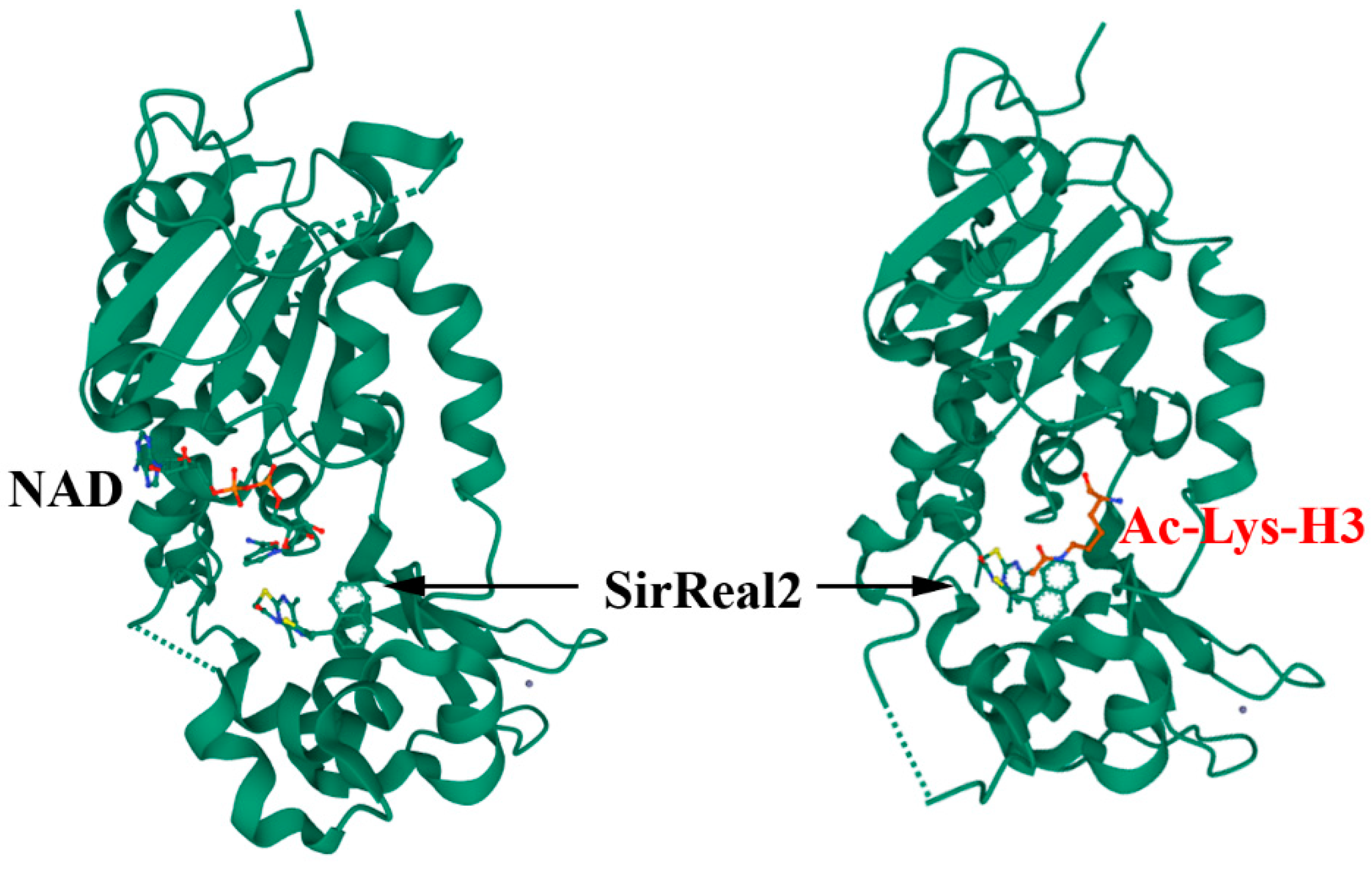
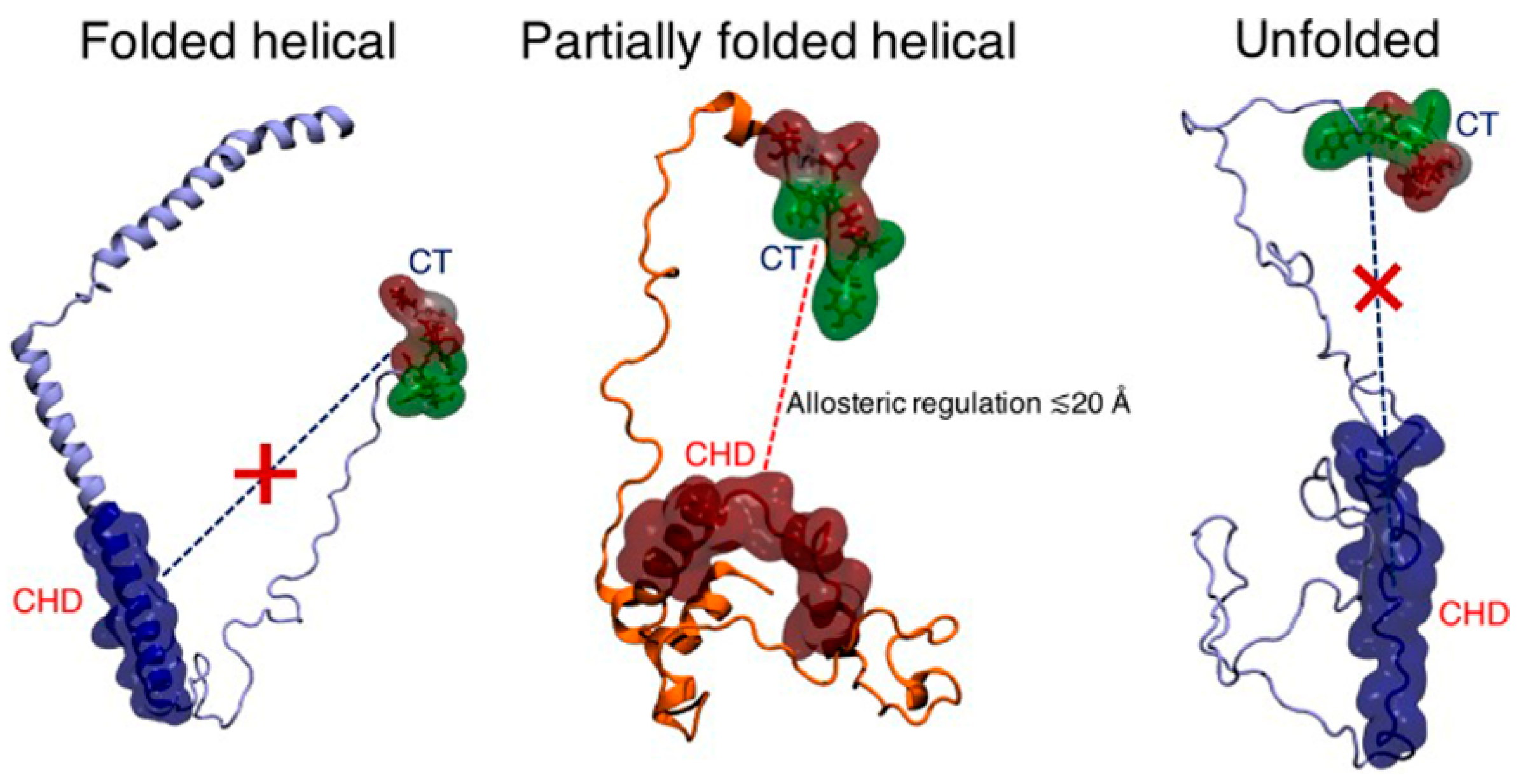
4.1.2. Allosteric Regulation
4.1.3. Oligonucleotide Aptamers
4.1.4. PROteolysis TArgeting Chimera (PROTAC) Technology
4.2. Indirect PPI Targeting
4.2.1. Regulation at Gene Level: Transcription Factors
4.2.2. Post-Translational Modifications (PTMs)
4.2.3. Other Interactions
4.3. Multi-Targeting of PPIs
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent advances in the development of protein-protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.G.; Scott, D.E.; Abell, C. Drugging challenging targets using fragment-based approaches. Curr. Opin. Chem. Biol. 2010, 14, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Pons-Espinal, M.; Blasco-Agell, L.; Consiglio, A. Dissecting the non-neuronal cell contribution to Parkinson’s disease pathogenesis using induced pluripotent stem cells. Cell. Mol. Life Sci. CMLS 2021, 78, 2081–2094. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Higueruelo, A.P.; Marsh, M.; Sigurdardottir, A.; Pitt, W.R.; Blundell, T.L. Biophysical and computational fragment-based approaches to targeting protein-protein interactions: Applications in structure-guided drug discovery. Q. Rev. Biophys. 2012, 45, 383–426. [Google Scholar] [CrossRef] [PubMed]
- Martino, E.; Chiarugi, S.; Margheriti, F.; Garau, G. Mapping, Structure and Modulation of PPI. Front. Chem. 2021, 9, 718405. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Gestwicki, J.E. Features of protein-protein interactions that translate into potent inhibitors: Topology, surface area and affinity. Expert Rev. Mol. Med. 2012, 14, e16. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.A.; Khuri, F.R.; Fu, H. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol. Sci. 2013, 34, 393–400. [Google Scholar] [CrossRef]
- Shin, W.H.; Kumazawa, K.; Imai, K.; Hirokawa, T.; Kihara, D. Current Challenges and Opportunities in Designing Protein-Protein Interaction Targeted Drugs. Adv. Appl. Bioinform. Chem. AABC 2020, 13, 11–25. [Google Scholar] [CrossRef]
- Torchet, R.; Druart, K.; Ruano, L.C.; Moine-Franel, A.; Borges, H.; Doppelt-Azeroual, O.; Brancotte, B.; Mareuil, F.; Nilges, M.; Menager, H.; et al. The iPPI-DB initiative: A Community-centered database of Protein-Protein Interaction modulators. Bioinformatics 2021, 37, 89–96. [Google Scholar] [CrossRef]
- Li, Q. Application of Fragment-Based Drug Discovery to Versatile Targets. Front. Mol. Biosci. 2020, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Philippe, G.; Huang, Y.H.; Cheneval, O.; Lawrence, N.; Zhang, Z.; Fairlie, D.P.; Craik, D.J.; de Araujo, A.D.; Henriques, S.T. Development of cell-penetrating peptide-based drug leads to inhibit MDMX:p53 and MDM2:p53 interactions. Biopolymers 2016, 106, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, P.; Zhang, L.; Huang, J.; Zhu, K.; Luo, C. Current Strategies and Applications for Precision Drug Design. Front. Pharmacol. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, M.; Colombo, G. Targeting Difficult Protein-Protein Interactions with Plain and General Computational Approaches. Molecules 2018, 23, 2256. [Google Scholar] [CrossRef] [PubMed]
- Shangary, S.; Wang, S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: A novel approach for cancer therapy. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L.; The Consortium for Top Down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Bhattacharjee, A.; Ajmal Ali, M.; Al-Hemaid, F.; Lee, J. Binding of small molecules at interface of protein-protein complex—A newer approach to rational drug design. Saudi J. Biol. Sci. 2017, 24, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Shen, Q.; Zhang, J. Allosteric Methods and Their Applications: Facilitating the Discovery of Allosteric Drugs and the Investigation of Allosteric Mechanisms. Acc. Chem. Res. 2019, 52, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.P. The concept of allosteric modulation: An overview. Drug Discov. Today Technol. 2013, 10, e223–e228. [Google Scholar] [CrossRef] [PubMed]
- Ni, D.; Liu, N.; Sheng, C. Allosteric Modulators of Protein-Protein Interactions (PPIs). Adv. Exp. Med. Biol. 2019, 1163, 313–334. [Google Scholar] [CrossRef] [PubMed]
- Mannes, M.; Martin, C.; Menet, C.; Ballet, S. Wandering beyond small molecules: Peptides as allosteric protein modulators. Trends Pharmacol. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Santofimia-Castano, P.; Rizzuti, B.; Xia, Y.; Abian, O.; Peng, L.; Velazquez-Campoy, A.; Neira, J.L.; Iovanna, J. Targeting intrinsically disordered proteins involved in cancer. Cell. Mol. Life Sci. CMLS 2020, 77, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Laszlo, L.; Kovacs, J.; Jensen, P.H.; Lindersson, E.; Botond, G.; Molnar, T.; Perczel, A.; Hudecz, F.; Mezo, G.; et al. Natively unfolded tubulin polymerization promoting protein TPPP/p25 is a common marker of alpha-synucleinopathies. Neurobiol. Dis. 2004, 17, 155–162. [Google Scholar] [CrossRef]
- Trejo-Lopez, J.A.; Yachnis, A.T.; Prokop, S. Neuropathology of Alzheimer’s Disease. Neurotherapeutics 2021, 1–13. [Google Scholar] [CrossRef]
- Mochizuki, H.; Choong, C.J.; Masliah, E. A refined concept: Alpha-synuclein dysregulation disease. Neurochem. Int. 2018, 119, 84–96. [Google Scholar] [CrossRef]
- Ono, K. The Oligomer Hypothesis in alpha-Synucleinopathy. Neurochem. Res. 2017, 42, 3362–3371. [Google Scholar] [CrossRef]
- Winner, B.; Jappelli, R.; Maji, S.K.; Desplats, P.A.; Boyer, L.; Aigner, S.; Hetzer, C.; Loher, T.; Vilar, M.; Campioni, S.; et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA 2011, 108, 4194–4199. [Google Scholar] [CrossRef] [PubMed]
- Hijaz, B.A.; Volpicelli-Daley, L.A. Initiation and propagation of alpha-synuclein aggregation in the nervous system. Mol. Neurodegener. 2020, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Greffard, S.; Verny, M.; Bonnet, A.M.; Seilhean, D.; Hauw, J.J.; Duyckaerts, C. A stable proportion of Lewy body bearing neurons in the substantia nigra suggests a model in which the Lewy body causes neuronal death. Neurobiol. Aging 2010, 31, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Orosz, F.; Kovacs, G.G.; Lehotzky, A.; Olah, J.; Vincze, O.; Ovadi, J. TPPP/p25: From unfolded protein to misfolding disease: Prediction and experiments. Biol. Cell 2004, 96, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Lehotzky, A.; Tirián, L.; Tőkési, N.; Lénárt, P.; Szabó, B.; Kovács, J.; Ovádi, J. Dynamic targeting of microtubules by TPPP/p25 affects cell survival. J. Cell Sci. 2004, 117, 6249–6259. [Google Scholar] [CrossRef] [PubMed]
- Olah, J.; Lehotzky, A.; Szunyogh, S.; Szenasi, T.; Orosz, F.; Ovadi, J. Microtubule-Associated Proteins with Regulatory Functions by Day and Pathological Potency at Night. Cells 2020, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. A protein-chameleon: Conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn. 2003, 21, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Protein moonlighting: What is it, and why is it important? Philos. Trans. R. Soc. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef]
- Olah, J.; Ovadi, J. Pharmacological targeting of alpha-synuclein and TPPP/p25 in Parkinson’s disease: Challenges and opportunities in a Nutshell. FEBS Lett. 2019, 593, 1641–1653. [Google Scholar] [CrossRef] [PubMed]
- Oertel, W.; Schulz, J.B. Current and experimental treatments of Parkinson disease: A guide for neuroscientists. J. Neurochem. 2016, 139, 325–337. [Google Scholar] [CrossRef]
- Upcott, M.; Chaprov, K.D.; Buchman, V.L. Toward a Disease-Modifying Therapy of Alpha-Synucleinopathies: New Molecules and New Approaches Came into the Limelight. Molecules 2021, 26, 7351. [Google Scholar] [CrossRef] [PubMed]
- Vassilakopoulou, V.; Karachaliou, C.E.; Evangelou, A.; Zikos, C.; Livaniou, E. Peptide-Based Vaccines for Neurodegenerative Diseases: Recent Endeavors and Future Perspectives. Vaccines 2021, 9, 1278. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.; Maass, S.; Schuberth, M.; Respondek, G.; Paul, F.; Mansmann, U.; Oertel, W.H.; Lorenzl, S.; Krismer, F.; Seppi, K.; et al. The PROMESA-protocol: Progression rate of multiple system atrophy under EGCG supplementation as anti-aggregation-approach. J. Neural Transm. 2016, 123, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Ryazanov, S.; Leonov, A.; Levin, J.; Shi, S.; Schmidt, F.; Prix, C.; Pan-Montojo, F.; Bertsch, U.; Mitteregger-Kretzschmar, G.; et al. Anle138b: A novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 2013, 125, 795–813. [Google Scholar] [CrossRef]
- Moussaud, S.; Malany, S.; Mehta, A.; Vasile, S.; Smith, L.H.; McLean, P.J. Targeting alpha-synuclein oligomers by protein-fragment complementation for drug discovery in synucleinopathies. Expert Opin. Ther. Targets 2015, 19, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and dynamics of micelle-bound human alpha-synuclein. J. Biol. Chem. 2005, 280, 9595–9603. [Google Scholar] [CrossRef]
- Salveson, P.J.; Spencer, R.K.; Nowick, J.S. X-ray Crystallographic Structure of Oligomers Formed by a Toxic beta-Hairpin Derived from alpha-Synuclein: Trimers and Higher-Order Oligomers. J. Am. Chem. Soc. 2016, 138, 4458–4467. [Google Scholar] [CrossRef]
- Mirecka, E.A.; Shaykhalishahi, H.; Gauhar, A.; Akgul, S.; Lecher, J.; Willbold, D.; Stoldt, M.; Hoyer, W. Sequestration of a beta-hairpin for control of alpha-synuclein aggregation. Angew. Chem. 2014, 53, 4227–4230. [Google Scholar] [CrossRef]
- Yeboah, F.; Kim, T.E.; Bill, A.; Dettmer, U. Dynamic behaviors of alpha-synuclein and tau in the cellular context: New mechanistic insights and therapeutic opportunities in neurodegeneration. Neurobiol. Dis. 2019, 132, 104543. [Google Scholar] [CrossRef]
- Daniels, M.J.; Nourse, J.B., Jr.; Kim, H.; Sainati, V.; Schiavina, M.; Murrali, M.G.; Pan, B.; Ferrie, J.J.; Haney, C.M.; Moons, R.; et al. Cyclized NDGA modifies dynamic alpha-synuclein monomers preventing aggregation and toxicity. Sci. Rep. 2019, 9, 2937. [Google Scholar] [CrossRef]
- Braun, A.R.; Liao, E.E.; Horvath, M.; Kalra, P.; Acosta, K.; Young, M.C.; Kochen, N.N.; Lo, C.H.; Brown, R.; Evans, M.D.; et al. Potent inhibitors of toxic alpha-synuclein identified via cellular time-resolved FRET biosensors. NPJ Park. Dis. 2021, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Lima, V.A.; Esquinelato, R.; Carmo-Goncalves, P.; Nascimento, L.A.D.; Lee, H.; Eliezer, D.; Romao, L.; Follmer, C. The dopamine receptor agonist apomorphine stabilizes neurotoxic alpha-synuclein oligomers. FEBS Lett. 2021. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B.; Woerman, A.L.; Mordes, D.A.; Watts, J.C.; Rampersaud, R.; Berry, D.B.; Patel, S.; Oehler, A.; Lowe, J.K.; Kravitz, S.N.; et al. Evidence for alpha-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. USA 2015, 112, E5308–E5317. [Google Scholar] [CrossRef] [PubMed]
- Rahayel, S.; Misic, B.; Zheng, Y.Q.; Liu, Z.Q.; Abdelgawad, A.; Abbasi, N.; Caputo, A.; Zhang, B.; Lo, A.; Kehm, V.; et al. Differentially targeted seeding reveals unique pathological alpha-synuclein propagation patterns. Brain 2021. [Google Scholar] [CrossRef] [PubMed]
- Schwarzman, A.L.; Senkevich, K.A.; Emelyanov, A.K.; Pchelina, S.N. Prion Properties of Alpha-Synuclein. Mol. Biol. 2019, 53, 380–387. [Google Scholar] [CrossRef]
- Menendez-Gonzalez, M.; Padilla-Zambrano, H.S.; Tomas-Zapico, C.; Garcia, B.F. Clearing Extracellular Alpha-Synuclein from Cerebrospinal Fluid: A New Therapeutic Strategy in Parkinson’s Disease. Brain Sci. 2018, 8, 52. [Google Scholar] [CrossRef]
- Valdinocci, D.; Radford, R.A.W.; Goulding, M.; Hayashi, J.; Chung, R.S.; Pountney, D.L. Extracellular Interactions of Alpha-Synuclein in Multiple System Atrophy. Int. J. Mol. Sci. 2018, 19, 4129. [Google Scholar] [CrossRef]
- Tokesi, N.; Olah, J.; Hlavanda, E.; Szunyogh, S.; Szabo, A.; Babos, F.; Magyar, A.; Lehotzky, A.; Vass, E.; Ovadi, J. Identification of motives mediating alternative functions of the neomorphic moonlighting TPPP/p25. BBA Mol. Basis Dis. 2014, 1842, 547–557. [Google Scholar] [CrossRef]
- Szenasi, T.; Olah, J.; Szabo, A.; Szunyogh, S.; Lang, A.; Perczel, A.; Lehotzky, A.; Uversky, V.N.; Ovadi, J. Challenging drug target for Parkinson’s disease: Pathological complex of the chameleon TPPP/p25 and alpha-synuclein proteins. BBA Mol. Basis Dis. 2017, 1863, 310–323. [Google Scholar] [CrossRef]
- Koshland, D.E. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. USA 1958, 44, 98–104. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Hong, T.; Chen, X.; Cui, L. SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res. Rev. 2019, 55, 100961. [Google Scholar] [CrossRef] [PubMed]
- Rumpf, T.; Schiedel, M.; Karaman, B.; Roessler, C.; North, B.J.; Lehotzky, A.; Olah, J.; Ladwein, K.I.; Schmidtkunz, K.; Gajer, M.; et al. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun. 2015, 6, 6263. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Xu, L.; Thompson, D. Long-range Regulation of Partially Folded Amyloidogenic Peptides. Sci. Rep. 2020, 10, 7597. [Google Scholar] [CrossRef] [PubMed]
- Szunyogh, S.; Olah, J.; Szenasi, T.; Szabo, A.; Ovadi, J. Targeting the interface of the pathological complex of alpha-synuclein and TPPP/p25. BBA Mol. Basis Dis. 2015, 1852, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Preeti, K.; Fernandes, V.; Khatri, D.K.; Singh, S.B. Role of MicroRNAs, Aptamers in Neuroinflammation and Neurodegenerative Disorders. Cell. Mol. Neurobiol. 2021, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qu, J.; Xue, F.; Zheng, Y.; Yang, B.; Chang, Y.; Yang, H.; Zhang, J. Novel DNA Aptamers for Parkinson’s Disease Treatment Inhibit alpha-Synuclein Aggregation and Facilitate its Degradation. Mol. Ther. Nucleic Acids 2018, 11, 228–242. [Google Scholar] [CrossRef]
- Ren, X.; Zhao, Y.; Xue, F.; Zheng, Y.; Huang, H.; Wang, W.; Chang, Y.; Yang, H.; Zhang, J. Exosomal DNA Aptamer Targeting alpha-Synuclein Aggregates Reduced Neuropathological Deficits in a Mouse Parkinson’s Disease Model. Mol. Ther. Nucleic Acids 2019, 17, 726–740. [Google Scholar] [CrossRef]
- Hughes, G.R.; Dudey, A.P.; Hemmings, A.M.; Chantry, A. Frontiers in PROTACs. Drug Discov. Today 2021, 26, 2377–2383. [Google Scholar] [CrossRef]
- Hyun, S.; Shin, D. Chemical-Mediated Targeted Protein Degradation in Neurodegenerative Diseases. Life 2021, 11, 607. [Google Scholar] [CrossRef]
- Fan, X.; Jin, W.Y.; Lu, J.; Wang, J.; Wang, Y.T. Rapid and reversible knockdown of endogenous proteins by peptide-directed lysosomal degradation. Nat. Neurosci. 2014, 17, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Kargbo, R.B. PROTAC Compounds Targeting alpha-Synuclein Protein for Treating Neurogenerative Disorders: Alzheimer’s and Parkinson’s Diseases. ACS Med. Chem. Lett. 2020, 11, 1086–1087. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Drakulic, D.; Lazic, A.; Ninkovic, D.S.; Schwirtlich, M.; Mojsin, M. SOX Transcription Factors as Important Regulators of Neuronal and Glial Differentiation During Nervous System Development and Adult Neurogenesis. Front. Mol. Neurosci. 2021, 14, 654031. [Google Scholar] [CrossRef] [PubMed]
- Winner, B.; Regensburger, M.; Schreglmann, S.; Boyer, L.; Prots, I.; Rockenstein, E.; Mante, M.; Zhao, C.; Winkler, J.; Masliah, E.; et al. Role of alpha-synuclein in adult neurogenesis and neuronal maturation in the dentate gyrus. J. Neurosci. 2012, 32, 16906–16916. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Tiwari, P.C.; Nath, R.; Pant, K.K. Role of neuroinflammation and latent transcription factors in pathogenesis of Parkinson’s disease. Neurol. Res. 2016, 38, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Mizuta, I.; Takafuji, K.; Ando, Y.; Satake, W.; Kanagawa, M.; Kobayashi, K.; Nagamori, S.; Shinohara, T.; Ito, C.; Yamamoto, M.; et al. YY1 binds to alpha-synuclein 3’-flanking region SNP and stimulates antisense noncoding RNA expression. J. Hum. Genet. 2013, 58, 711–719. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, C.; Chen, L.; Zhang, J.J.; Ge, W.L.; Yuan, H.; Meng, L.D.; Huang, X.M.; Shen, P.; Miao, Y.; et al. YY1 targets tubulin polymerisation-promoting protein to inhibit migration, invasion and angiogenesis in pancreatic cancer via p38/MAPK and PI3K/AKT pathways. Br. J. Cancer 2019, 121, 912–921. [Google Scholar] [CrossRef]
- Saraiva, C.; Paiva, J.; Santos, T.; Ferreira, L.; Bernardino, L. MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control. Release 2016, 235, 291–305. [Google Scholar] [CrossRef]
- Khodr, C.E.; Becerra, A.; Han, Y.; Bohn, M.C. Targeting alpha-synuclein with a microRNA-embedded silencing vector in the rat substantia nigra: Positive and negative effects. Brain Res. 2014, 1550, 47–60. [Google Scholar] [CrossRef]
- Yang, J.; Luo, S.; Zhang, J.; Yu, T.; Fu, Z.; Zheng, Y.; Xu, X.; Liu, C.; Fan, M.; Zhang, Z. Exosome-mediated delivery of antisense oligonucleotides targeting alpha-synuclein ameliorates the pathology in a mouse model of Parkinson’s disease. Neurobiol. Dis. 2021, 148, 105218. [Google Scholar] [CrossRef]
- Song, J.X.; Liu, J.; Jiang, Y.; Wang, Z.Y.; Li, M. Transcription factor EB: An emerging drug target for neurodegenerative disorders. Drug Discov. Today 2021, 26, 164–172. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Li, J.D. The Roles of Post-translational Modifications on alpha-Synuclein in the Pathogenesis of Parkinson’s Diseases. Front. Neurosci. 2019, 13, 381. [Google Scholar] [CrossRef] [PubMed]
- Braithwaite, S.P.; Stock, J.B.; Mouradian, M.M. α-Synuclein phosphorylation as a therapeutic target in Parkinson’s disease. Rev. Neurosci. 2012, 23, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Klopstock, T.; Jackowski, S.; Kuscer, E.; Tricta, F.; Videnovic, A.; Jinnah, H.A. Rational Design of Novel Therapies for Pantothenate Kinase-Associated Neurodegeneration. Mov. Disord. 2021, 36, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Renko, J.M.; Mahato, A.K.; Visnapuu, T.; Valkonen, K.; Karelson, M.; Voutilainen, M.H.; Saarma, M.; Tuominen, R.K.; Sidorova, Y.A. Neuroprotective Potential of a Small Molecule RET Agonist in Cultured Dopamine Neurons and Hemiparkinsonian Rats. J. Park. Dis. 2021, 11, 1023–1046. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; D’Amore, C.; Sarno, S.; Salvi, M.; Ruzzene, M. Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021, 6, 183. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, E.; Paudel, Y.N.; Julian, T.; Shaikh, M.F.; Piperi, C. Pivotal Role of Fyn Kinase in Parkinson’s Disease and Levodopa-Induced Dyskinesia: A Novel Therapeutic Target? Mol. Neurobiol. 2021, 58, 1372–1391. [Google Scholar] [CrossRef] [PubMed]
- Quist, A.; Doudevski, I.; Lin, H.; Azimova, R.; Ng, D.; Frangione, B.; Kagan, B.; Ghiso, J.; Lal, R. Amyloid ion channels: A common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA 2005, 102, 10427–10432. [Google Scholar] [CrossRef] [PubMed]
- El-Battari, A.; Rodriguez, L.; Chahinian, H.; Delezay, O.; Fantini, J.; Yahi, N.; Di Scala, C. Gene Therapy Strategy for Alzheimer’s and Parkinson’s Diseases Aimed at Preventing the Formation of Neurotoxic Oligomers in SH-SY5Y Cells. Int. J. Mol. Sci. 2021, 22, 11550. [Google Scholar] [CrossRef] [PubMed]
- Yahi, N.; Di Scala, C.; Chahinian, H.; Fantini, J. Innovative treatment targeting gangliosides aimed at blocking the formation of neurotoxic alpha-synuclein oligomers in Parkinson’s disease. Glycoconj. J. 2021, 1–11. [Google Scholar] [CrossRef]
- Reusch, R.N. The role of short-chain conjugated poly-(R)-3-hydroxybutyrate (cPHB) in protein folding. Int. J. Mol. Sci. 2013, 14, 10727–10748. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Lengweiler, U.D.; Seebach, D.; Reusch, R.N. Proof for a nonproteinaceous calcium-selective channel in Escherichia coli by total synthesis from (R)-3-hydroxybutanoic acid and inorganic polyphosphate. Proc. Natl. Acad. Sci. USA 1997, 94, 9075–9079. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Reusch, R.N.; Igarashi, K.; Root-Bernstein, R. Molecular complementarity between simple, universal molecules and ions limited phenotype space in the precursors of cells. Biol. Direct 2014, 10, 28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brundin, P.; Dave, K.D.; Kordower, J.H. Therapeutic approaches to target alpha-synuclein pathology. Exp. Neurol. 2017, 298, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Lehotzky, A.; Olah, J.; Fekete, J.T.; Szenasi, T.; Szabo, E.; Gyorffy, B.; Varady, G.; Ovadi, J. Co-Transmission of Alpha-Synuclein and TPPP/p25 Inhibits Their Proteolytic Degradation in Human Cell Models. Front. Mol. Biosci. 2021, 8, 666026. [Google Scholar] [CrossRef] [PubMed]
- Loscher, W.; Klein, P. New approaches for developing multi-targeted drug combinations for disease modification of complex brain disorders. Does epilepsy prevention become a realistic goal? Pharmacol. Ther. 2021, 229, 107934. [Google Scholar] [CrossRef] [PubMed]
- Van Bulck, M.; Sierra-Magro, A.; Alarcon-Gil, J.; Perez-Castillo, A.; Morales-Garcia, J.A. Novel Approaches for the Treatment of Alzheimer’s and Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 719. [Google Scholar] [CrossRef]
- Rascol, O.; Fabbri, M.; Poewe, W. Amantadine in the treatment of Parkinson’s disease and other movement disorders. Lancet Neurol. 2021, 20, 1048–1056. [Google Scholar] [CrossRef]
- Demuro, S.; Di Martino, R.M.C.; Ortega, J.A.; Cavalli, A. GSK-3beta, FYN, and DYRK1A: Master Regulators in Neurodegenerative Pathways. Int. J. Mol. Sci. 2021, 22, 9098. [Google Scholar] [CrossRef]
- Soleimani Zakeri, N.S.; Pashazadeh, S.; MotieGhader, H. Drug Repurposing for Alzheimer’s Disease Based on Protein-Protein Interaction Network. BioMed Res. Int. 2021, 2021, 1280237. [Google Scholar] [CrossRef]
- Norris, V.; Amar, P.; Legent, G.; Ripoll, C.; Thellier, M.; Ovadi, J. Sensor potency of the moonlighting enzyme-decorated cytoskeleton: The cytoskeleton as a metabolic sensor. BMC Biochem. 2013, 14, 3. [Google Scholar] [CrossRef]
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in macromolecular complexes: The roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.L.; Kavallaris, M.; McCarroll, J.A. Microtubules and their role in cellular stress in cancer. Front. Oncol. 2014, 4, 153. [Google Scholar] [CrossRef] [PubMed]
- Eira, J.; Silva, C.S.; Sousa, M.M.; Liz, M.A. The cytoskeleton as a novel therapeutic target for old neurodegenerative disorders. Prog. Neurobiol. 2016, 141, 61–82. [Google Scholar] [CrossRef] [PubMed]
- Gestwicki, J.E.; Marinec, P.S. Chemical control over protein-protein interactions: Beyond inhibitors. Comb. Chem. High Throughput Screen. 2007, 10, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Field, J.J.; Singh, A.J.; Kanakkanthara, A.; Halafihi, T.; Northcote, P.T.; Miller, J.H. Microtubule-stabilizing activity of zampanolide, a potent macrolide isolated from the Tongan marine sponge Cacospongia mycofijiensis. J. Med. Chem. 2009, 52, 7328–7332. [Google Scholar] [CrossRef]
- Field, J.J.; Northcote, P.T.; Paterson, I.; Altmann, K.H.; Diaz, J.F.; Miller, J.H. Zampanolide, a Microtubule-Stabilizing Agent, Is Active in Resistant Cancer Cells and Inhibits Cell Migration. Int. J. Mol. Sci. 2017, 18, 971. [Google Scholar] [CrossRef]
- Prota, A.E.; Bargsten, K.; Zurwerra, D.; Field, J.J.; Diaz, J.F.; Altmann, K.H.; Steinmetz, M.O. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science 2013, 339, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Jubb, H.; Blundell, T.L.; Ascher, D.B. Flexibility and small pockets at protein-protein interfaces: New insights into druggability. Prog. Biophys. Mol. Biol. 2015, 119, 2–9. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oláh, J.; Szénási, T.; Lehotzky, A.; Norris, V.; Ovádi, J. Challenges in Discovering Drugs That Target the Protein–Protein Interactions of Disordered Proteins. Int. J. Mol. Sci. 2022, 23, 1550. https://doi.org/10.3390/ijms23031550
Oláh J, Szénási T, Lehotzky A, Norris V, Ovádi J. Challenges in Discovering Drugs That Target the Protein–Protein Interactions of Disordered Proteins. International Journal of Molecular Sciences. 2022; 23(3):1550. https://doi.org/10.3390/ijms23031550
Chicago/Turabian StyleOláh, Judit, Tibor Szénási, Attila Lehotzky, Victor Norris, and Judit Ovádi. 2022. "Challenges in Discovering Drugs That Target the Protein–Protein Interactions of Disordered Proteins" International Journal of Molecular Sciences 23, no. 3: 1550. https://doi.org/10.3390/ijms23031550
APA StyleOláh, J., Szénási, T., Lehotzky, A., Norris, V., & Ovádi, J. (2022). Challenges in Discovering Drugs That Target the Protein–Protein Interactions of Disordered Proteins. International Journal of Molecular Sciences, 23(3), 1550. https://doi.org/10.3390/ijms23031550






