ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress
Abstract
1. Introduction
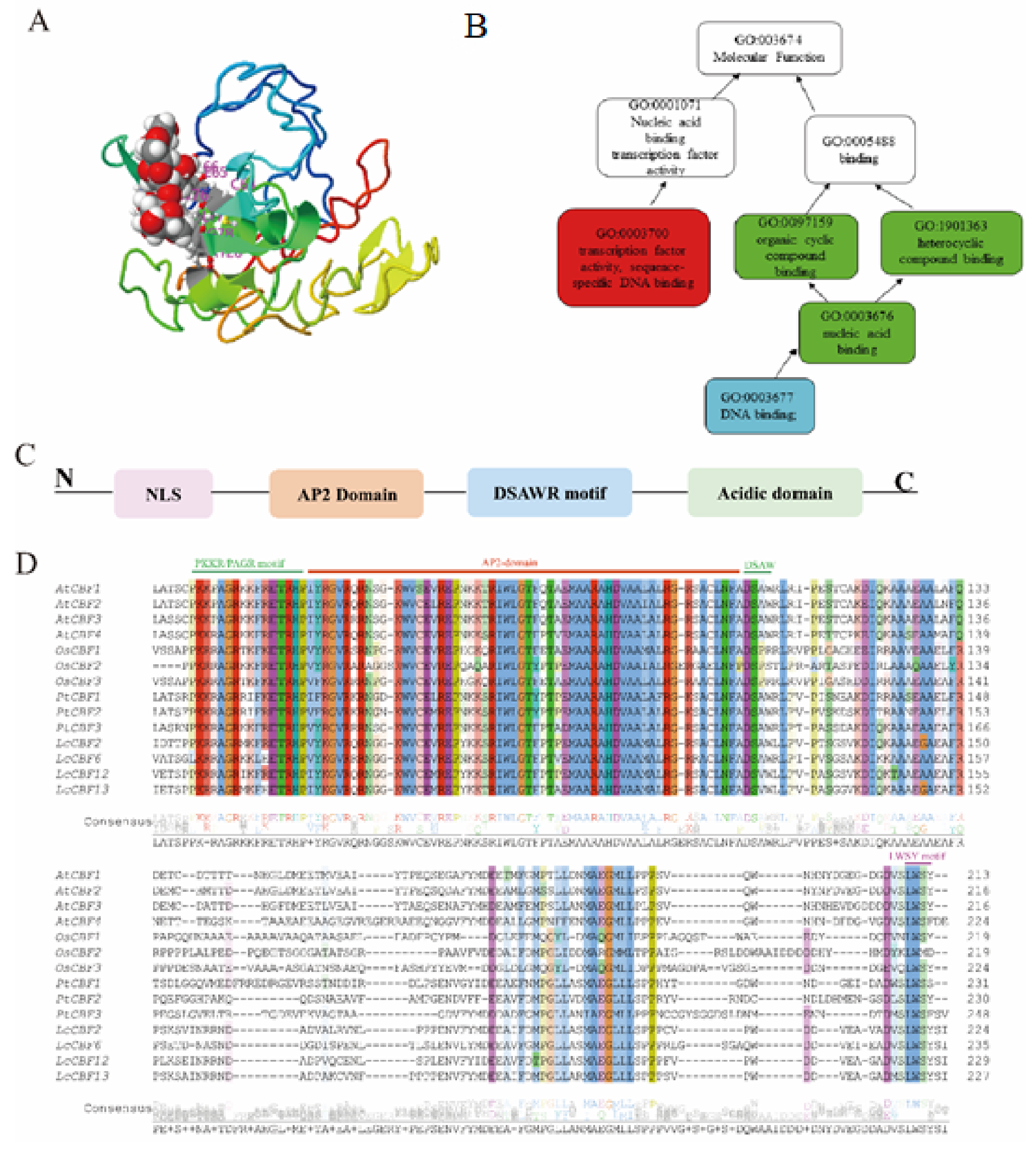
2. Conserved Motifs and Their Functionality in CBF, ICE, and COR Genes
2.1. C-Repeat Binding Factor/Dehydration-Responsive Elements Binding 1 (CBF/DREB1)
2.2. Inducer of CBF Expression (ICE)
2.3. Cold Regulated (COR) Genes
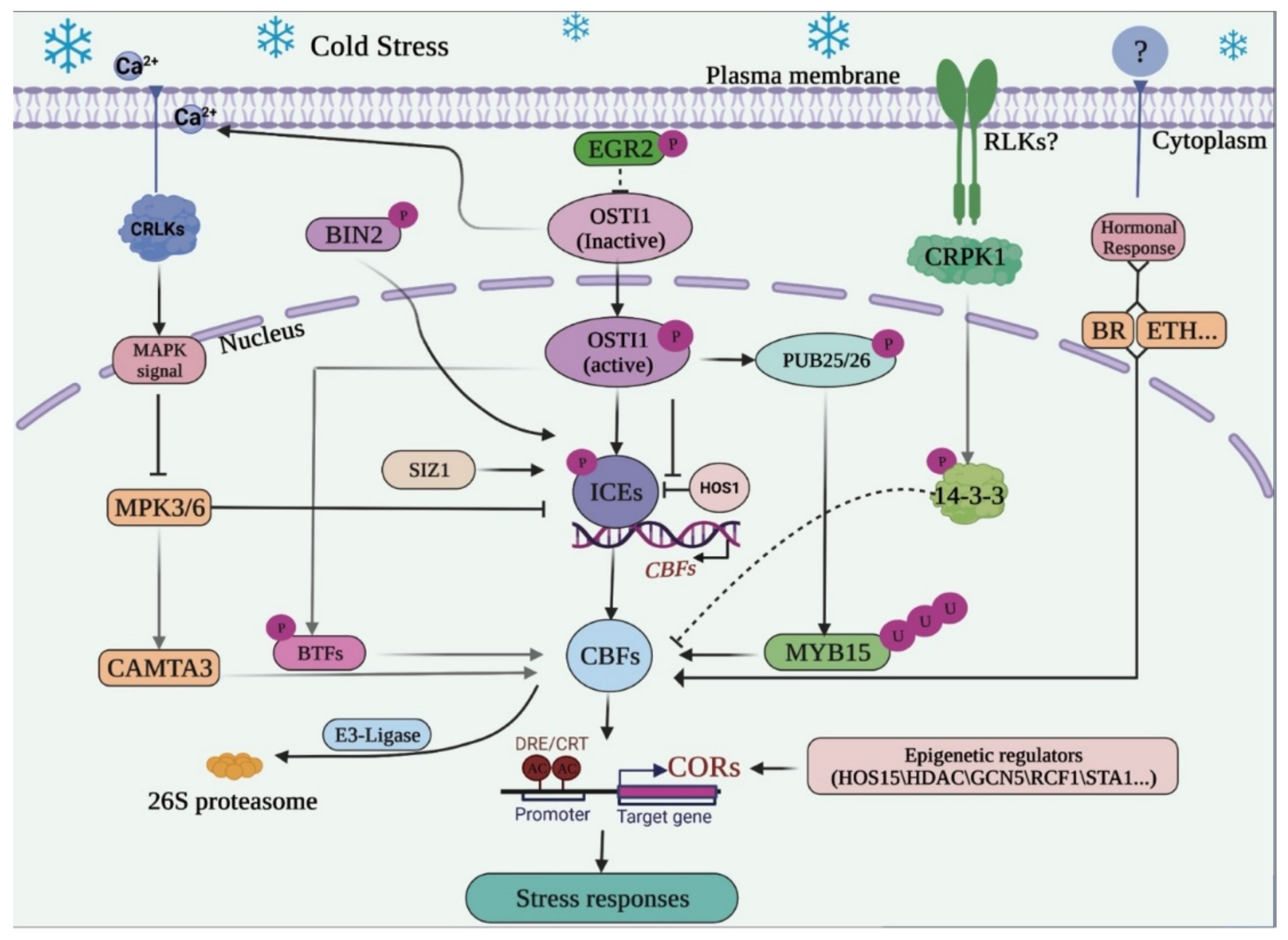
3. Mitogen-Activated Protein Kinase (MAPK) Cascade and Hormonal Responses Regulating the ICE-CBF-COR
4. Post-Transcriptional Regulations and Post-Translational Modification
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ritonga, F.N.; Chen, S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Oshunsanya, S.O.; Nwosu, N.J.; Li, Y. Abiotic Stress in Agricultural Crops Under Climatic Conditions. In Sustainable Agriculture, Forest and Environmental Management; Jhariya, M.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; Volume 9, pp. 71–100. [Google Scholar]
- Yang, C.; Yang, H.; Xu, Q. Comparative metabolomics analysis of the response to cold stress of resistant and susceptible Tibetan hulless barley (Hordeum distichon). Phytochemistry 2020, 174, 112. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sommer, M.L.; Hochholdinger, F. Cold response and tolerance in cereal roots. J. Exp. Bot. 2021, 72, 7474–7481. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, X.; Guo, Y. Identification of CBF Transcription Factors in Tea Plants and a Survey of Potential CBF Target Genes under Low Temperature. Int. J. Mol. Sci. 2019, 20, 5137. [Google Scholar] [CrossRef]
- Meng, X.; Liang, Z.; Dai, Y. Predicting transcriptional responses to cold stress across plant species. Proc. Natl. Acad. Sci. USA 2021, 118, e2026330118. [Google Scholar] [CrossRef]
- Mehrotra, S.; Verma, S. Transcriptional regulation and signalling of cold stress response in plants: An overview of current understanding. Environ. Exp. Bot. 2020, 180, 104243. [Google Scholar] [CrossRef]
- Shu, Y.; Li, W.; Zhao, J.; Zhang, S.; Xu, H.; Liu, Y.; Guo, C. Transcriptome sequencing analysis of alfalfa reveals CBF genes potentially playing important roles in response to freezing stress. Genet. Mol. Biol. 2017, 40, 824–833. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant Cold Acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Guo, J.; Ren, Y.; Tang, Z. Characterization and expression profiling of the ICE-CBF-COR genes in wheat. Peer J. 2019, 7, e8190. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Zhao, Z. COR27 and COR28 are Novel Regulators of the COP1–HY5 Regulatory Hub and Photomorphogenesis in Arabidopsis. Plant Cell 2020, 32, 1393154. [Google Scholar] [CrossRef]
- Wang, D.Z.; Jin, Y.N.; Ding, X.H. Gene Regulation and Signal Transduction in the ICE-CBF-COR Signaling Pathway during Cold Stress in Plants. Biochemistry 2017, 82, 1103–1117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dang, P.; He, C. Cold acclimation by the CBF–COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Bremer, A.; Kent, B. Intrinsically Disordered Stress Protein COR15A Resides at the Membrane Surface during Dehydration. Biophys. J. 2017, 113, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Badawi, M.; Reddy, Y.M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 2008, 49, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Wang, L. Response and adaptation mechanisms of tea plant to low-temperature stress. In Stress Physiology of Tea in the Face of Climate Change; Springer: Berlin/Heidelberg, Germany, 2018; pp. 39–61. [Google Scholar]
- Chandler, J.W. Class VIIIb APETALA2 Ethylene Response Factors in Plant Development. Trends Plant Sci. 2018, 23, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Yu, S.L.; Park, J. Accession-Dependent CBF Gene Deletion by CRISPR/Cas System in Arabidopsis. Front. Plant Sci. 2017, 8, 1910. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhai, S.; Wang, W. Ideentification of genes from the ICE-CBF-COR pathway under cold stress in Aegilops-Triticum composite group and the evolution analysis with those from Triticeae. Physiol. Mol. Biol. Plants 2018, 24, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.K.; Kumar, V.S. Overxpression of Arabidopsis ICE1 enhances yield and multiple abiotic stress tolerance in indica rice. Plant Signal Behav. 2020, 15, 1814547. [Google Scholar] [CrossRef]
- Vyse, K.; Faivre, L. Transcriptional and post-transcriptional regulation and transcriptional memory of chromatin regulators in response to low temperature. Front. Plant Sci. 2020, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Hirsz, D.; Dixon, L.E. The Roles of Temperature-Related Post-Transcriptional Regulation in Cereal Floral Development. Plants 2021, 10, 2230. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Yi, H.; Chen, X. Post-translational modifications of proteins have versatile roles in regulating plant immune responses. Int. J. Mol. Sci. 2019, 20, 2807. [Google Scholar] [CrossRef] [PubMed]
- Damaris, R.N.; Yang, P. Protein Phosphorylation Response to Abiotic Stress in Plants. Methods Mol. Biol. 2021, 2358, 635–674. [Google Scholar]
- Praat, M.; De Smet, I.; van Zanten, M. Protein kinase and phosphatase control of plant temperature responses. J. Exp. Bot. 2021, 72, 7459–7473. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, L. The mitogen-activated protein kinase kinase MKK2 positively regulates constitutive cold resistance in the potato. Environ. Exp. Bot. 2022, 19, 104702. [Google Scholar] [CrossRef]
- Sharma, S.; Prasd, A. Role of ubiquitination enzymes in abiotic environmental interactions with plants. Int. J. Biol. Macromol. 2021, 181, 494–507. [Google Scholar] [CrossRef]
- Yao, W.; Wang, L.; Wang, J. VpPUB24, a novel gene from Chinese grapevine, Vitis pseudoreticulata, targets VpICE1 to enhance cold tolerance. J. Exp. Bot. 2017, 68, 2933–2949. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, Y.; Li, Z. PUB25 and PUB26 Promote Plant Freezing Tolerance by Degrading the Cold Signaling Negative Regulator MYB15. Dev. Cell 2019, 51, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.Q.; Chen, J.Y.; Kuang, J.F. The Banana Fruit SINA Ubiquitin Ligase MaSINA1 Regulates the Stability of MaICE1 to be Negatively Involved in Cold Stress Response. Front. Plant Sci. 2017, 8, 995. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Unterholzner, S.J.; Rathnayake, A.I. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 2016, 113, e5982–e5991. [Google Scholar] [CrossRef]
- Lantzouni, O.; Alkofer, A.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef] [PubMed]
- Wi, S.D.; Lee, E.S. Redox-mediated structural and functional switching of C-repeat binding factors enhances plant cold tolerance. New Phytol. 2022, 233, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Liou, Y.C.; Daley, M.E. Folding and structural characterization of highly disulfide-bonded beetle antifreeze protein produced in bacteria. Protein Expr. Purif. 2000, 19, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, R.A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar]
- Okamuro, J.K.; Caster, B.; Villarroel, R. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. Embo J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef]
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci. 2011, 180, 3–11. [Google Scholar] [CrossRef]
- Warren, G.J. Cold stress: Manipulating freezing tolerance in plants. Curr. Biol. 1998, 8, 514–516. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, J.; Sun, T. The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant. Biol. 2017, 59, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ban, Q.; Hao, J. Genome-Wide Characterization of the C-repeat Binding Factor (CBF) Gene Family Involved in the Response to Abiotic Stresses in Tea Plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 921. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Liu, S.; Wu, W. Genome-wide identification and cold stress-induced expression analysis of the CBF gene family in Liriodendron chinense. J. For. Res. 2021, 32, 2531–2543. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005, 1, e26. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Kidokoro, S.; Hayashi, K.; Haraguchi, H. Post-translational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, 3349234. [Google Scholar] [CrossRef]
- Li, W.; Chen, Y.; Ye, M. Evolutionary history of the C-repeat binding factor/dehydration-responsive element-binding 1 (CBF/DREB1) protein family in 43 plant species and characterization of CBF/DREB1 proteins in Solanum tuberosum. BMC Evol. Biol. 2020, 20, 142. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S. The Cold-Inducible CBF1 Factor–Dependent Signaling Pathway Modulates the Accumulation of the Growth-Repressing DELLA Proteins via Its Effect on Gibberellin Metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef]
- Jiang, F.; Wang, F.; Wu, Z. Components of the Arabidopsis CBF cold-response pathway are conserved in non-heading Chinese cabbage. Plant Mol. Biol. Rep. 2011, 29, 525–532. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Jing, H. Three Novel C-Repeat Binding Factor Genes of Dimocarpus longan Regulate Cold Stress Response in Arabidopsis. Front. Plant Sci. 2020, 11, 1026. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Li, J.; Yang, Q. Phylogenetic, molecular, and functional characterization of PpyCBF proteins in Asian pears (Pyrus pyrifolia). Int. J. Mol. Sci. 2019, 20, 2074. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Kim, B.H.; Ji, C.Y. Overexpressing IbCBF3 increases low temperature and drought stress tolerance in transgenic sweetpotato. Plant Physiol. Biochem. 2017, 118, 45–54. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Ma, Q.; Yan, W. Divergent Regulation of CBF Regulon on Cold Tolerance and Plant Phenotype in Cassava Overexpressing Arabidopsis CBF3 Gene. Front. Plant Sci. 2016, 7, 1866. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Abdullah, S.N. Oil palm EgCBF3 conferred stress tolerance in transgenic tomato plants through modulation of the ethylene signaling pathway. J. Plant Physiol. 2016, 202, 107–120. [Google Scholar] [CrossRef]
- Zhuang, L.; Yuan, X.; Chen, Y. PpCBF3 from Cold-Tolerant Kentucky Bluegrass Involved in Freezing Tolerance Associated with Up-Regulation of Cold-Related Genes in Transgenic Arabidopsis thaliana. PLoS ONE 2015, 10, e0132928. [Google Scholar] [CrossRef][Green Version]
- Kidokoro, S.; Watanabe, K.; Ohori, T. Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 2015, 81, 505–518. [Google Scholar] [CrossRef]
- Byun, M.Y.; Lee, J.; Cui, L.H. Constutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 2015, 236, 61–74. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Bassett, C. Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 2011, 233, 971–983. [Google Scholar] [CrossRef]
- Pino, M.T.; Skinner, J.S. Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant Cell Env. 2008, 31, 393–406. [Google Scholar] [CrossRef]
- Gutha, L.R.; Reddy, A.R. Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol. Biol. 2008, 68, 533–555. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Kwon, C.W.; Choi, D.W. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J. 2007, 5, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.S.; von Zitzewitz, J.; Szucs, P. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 2005, 59, 533–551. [Google Scholar] [CrossRef]
- Gao, M.J.; Allard, G.; Byass, L. Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol. Biol. 2002, 49, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Salvo, M.; Rey, F.; Arruabarrena, A.; Gambetta, G.; Rodrigo, M.J.; Zacarías, L.; Lado, J. Transcriptional Analysis of C-Repeat Binding Factors in Fruit of Citrus Species with Differential Sensitivity to Chilling Injury during Postharvest Storage. Int. J. Mol. Sci. 2021, 22, 804. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, P.; Yan, Y. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Tang, K.; Zhao, L.; Ren, Y. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 2020, 62, 258–263. [Google Scholar] [CrossRef]
- Kurbidaeva, A.; Ezhova, T.; Novokreshchenova, M. Arabidopsis thaliana ICE2 gene: Phylogeny, structural evolution and functional diversification from ICE1. Plant Sci. 2014, 229, 10–22. [Google Scholar] [CrossRef]
- Kashyap, P.; Deswal, R. Two ICE isoforms showing differential transcriptional regulation by cold and hormones participate in Brassica juncea cold stress signaling. Gene 2019, 695, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 CASCADE and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.L.; Lin, L.C.F.; Hsu, H.C. Saussurea involucrata (Snow Lotus) ICE1 and ICE2 Orthologues Involved in Regulating Cold Stress Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10850. [Google Scholar] [CrossRef] [PubMed]
- Wani, U.M.; Majeed, S.T.; Raja, V.; Wani, Z.A.; Jan, N.; Andrabi, K.I.; John, R. Ectopic expression of a novel cold-resistance protein 1 from Brassica oleracea promotes tolerance to chilling stress in transgenic tomato. Sci. Rep. 2021, 11, 16574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, L.; Song, A. Chrysanthemum (Chrysanthemum morifolium) CmICE2 conferred freezing tolerance in Arabidopsis. Plant Physiol. Biochem. 2020, 146, 31–41. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Kang, H.G.; Park, M.Y. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019, 289, 110254. [Google Scholar] [CrossRef]
- Chen, W.J.; Wang, X.; Yan, S. The ICE-like transcription factor HbICE2 is involved in jasmonate-regulated cold tolerance in the rubber tree (Hevea brasiliensis). Plant Cell Rep. 2019, 38, 699–714. [Google Scholar] [CrossRef]
- Man, L.; Xiang, D.; Wang, L. Stress-responsive gene RsICE1 from Raphanus sativus increases cold tolerance in rice. Protoplasma 2017, 254, 945–956. [Google Scholar] [CrossRef]
- Deng, C.; Ye, H.; Fan, M. The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis. Plant Signal Behav. 2017, 12, e1316442. [Google Scholar] [CrossRef]
- Lu, X.; Yang, L.; Yu, M. A novel Zea mays ssp. mexicana L. MYC-type ICE-like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 113, 78–88. [Google Scholar] [CrossRef]
- Feng, H.L.; Ma, N.N.; Meng, X. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 2013, 73, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Leuendorf, J.E.; Frank, M.; Schmülling, T. Acclimation, priming and memory in the response of Arabidopsis thaliana seedlings to cold stress. Sci. Rep. 2020, 10, 689. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Kilma, K. COR/LEA Proteins as Indicators of Frost Tolerance in Triticeae: A Comparison of Controlled versus Field Conditions. Plants 2021, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, R.; Li, M. Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J. Biol. Chem. 2008, 283, 461–468. [Google Scholar] [CrossRef]
- Welti, R.; Li, W. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Okawa, K.; Nakayama, K.; Kakizaki, T. Identification and characterization of Cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Environ. 2008, 31, 1470–1483. [Google Scholar] [CrossRef]
- Zhou, A.; Liu, E.; Li, H. PsCor413pm2, a Plasma Membrane-Localized, Cold-Regulated Protein from Phlox subulata, Confers Low Temperature Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2579. [Google Scholar] [CrossRef]
- Hu, X.; Liu, J.; Liu, E.; Qioa, K. Arabidopsis cold-regulated plasma membrane protein Cor413pm1 is a regulator of ABA response. Biochem. Biophys. Res. Commun. 2021, 561, 88–92. [Google Scholar] [CrossRef]
- Strimbeck, R.G. Hiding in plain sight: The F segment and other conserved features of seed plant SK(n) dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Pan, Y.; Li, J. Heterologous expression of Arabidopsis C-repeat binding factor 3 (AtCBF3) and cold-regulated 15A (AtCOR15A) enhanced chilling tolerance in transgenic eggplant (Solanum melongena L.). Plant Cell Rep. 2014, 33, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Hernandez, M.; Romero, I.; Escribano, I.; Merodio, C.; Sanchez-Ballesta, M.T. Deciphering the Role of CBF/DREB Transcription Factors and Dehydrins in Maintaining the Quality of Table Grapes cv. Autumn Royal Treated with High CO(2) Levels and Stored at 0°C. Front Plant Sci. 2017, 8, 1591. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhou, H.; Lin, F. Cold-Regulated Gene27 Integrates Signals from Light and the Circadian Clock to Promote Hypocotyl Growth in Arabidopsis. Plant Cell 2020, 32, 3155–3169. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, Z. Cold-regulated gene LeCOR413PM2 confers cold stress tolerance in tomato plants. Gene 2021, 764, 145097. [Google Scholar] [CrossRef]
- Shi, H.; He, X.; Zhao, Y. Constitutive expression of a group 3 LEA protein from Medicago falcata (MfLEA3) increases cold and drought tolerance in transgenic tobacco. Plant Cell Rep. 2020, 39, 851–860. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Dong, G. A novel cold-regulated protein isolated from Saussurea involucrata confers cold and drought tolerance in transgenic tobacco (Nicotiana tabacum). Plant Sci. 2019, 289, 110246. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Wang, X. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef]
- Peng, Y.; Reyes, J.L.; Wei, H. RcDhn5, a cold acclimation-responsive dehydrin from Rhododendron catawbiense rescues enzyme activity from dehydration effects in vitro and enhances freezing tolerance in RcDhn5-overexpressing Arabidopsis plants. Physiol. Plant 2008, 134, 583–597. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef]
- Liu, Q.; Ding, Y.; Shi, Y. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. Embo J. 2021, 40, e104559. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S. Ca(2+)/Calmodulin Complex Triggers CAMTA Transcriptional Machinery Under Stress in Plants: Signaling Cascade and Molecular Regulation. Front. Plant Sci. 2020, 11, 598327. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, L.; Wang, X.; Leng, X. Identification and bioinformatic analysis of signal responsive/calmodulin-binding transcription activators gene models in Vitis vinifera. Mol. Biol. Rep. 2014, 41, 2937–2949. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Li, Z. Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 2021, 12, 4713. [Google Scholar] [CrossRef]
- Wang, G.; Zeng, H.; Hu, X. Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil 2015, 386, 205–221. [Google Scholar] [CrossRef]
- Rahman, H.; Xu, Y.P.; Zhnag, X.R. Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 581. [Google Scholar] [CrossRef]
- Wei, M.; Xu, X.; Li, C. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci. Rep. 2017, 7, 17910. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, X.; Ge, T. Genome-wide identification of citrus CAMTA genes and their expression analysis under stress and hormone treatments. J. Hort. Sci. Biotech. 2019, 94, 331–340. [Google Scholar] [CrossRef]
- Pant, P.; Iqbal, Z.; Pandey, B. Genome-wide comparative and evolutionary analysis of Calmodulin-binding Transcription Activator (CAMTA) family in Gossypium species. Sci. Rep. 2018, 8, 5573. [Google Scholar] [CrossRef]
- Wang, Z.; Wan, Y.; Meng, X. Genome-Wide Identification and Analysis of MKK and MAPK Gene Families in Brassica Species and Response to Stress in Brassica napus. Int. J. Mol. Sci. 2021, 22, 544. [Google Scholar] [CrossRef]
- Wang, H.; Gong, M. Genome-wide Identification of Jatropha curcas MAPK, MAPKK, and MAPKKK Gene Families and Their Expression Profile Under Cold Stress. Sci. Rep. 2018, 8, 16163. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Liu, Z.; Wang, Y. Comparative transcriptomic analysis provides insights into the coordinated mechanisms of leaves and roots response to cold stress in Common Vetch. Ind. Crops. Prod. 2020, 158, 112949. [Google Scholar] [CrossRef]
- Wei, X.; Liu, S.; Sun, C. Convergence and Divergence: Signal Perception and Transduction Mechanisms of Cold Stress in Arabidopsis and Rice. Plants 2021, 10, 1864. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Wu, J.; Wang, Y. Plant Mitogen-Activated Protein Kinase Cascades in Environmental Stresses. Int. J. Mol. Sci. 2021, 22, 1543. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chaudhuri, S.; Yang, L. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 2010, 285, 7119–7126. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Euglem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Jiang, X.; Hoehenwarter, W.; Scheel, D. Phosphorylation of the CAMTA3 Transcription Factor Triggers Its Destabilization and Nuclear Export. Plant Physiol. 2020, 184, 1056–1071. [Google Scholar] [CrossRef] [PubMed]
- Townley, H.E.; Knight, M.R. Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 2002, 128, 1169–1172. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, P. MAP Kinase Cascades Regulate the Cold Response by Modulating ICE1 Protein Stability. Dev. Cell 2017, 43, 618–629. [Google Scholar] [CrossRef]
- Li, H.; Ding, Y. MPK3-and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jia, X.; Shi, Y. OST 1-mediated BTF 3L phosphorylation positively regulates CBF s during plant cold responses. EMBO J. 2018, 37, e98228. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X. W Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Zheng, Y.; Lv, Y. Multi-omics analysis reveals specific modifications associated with reduced chilling injury in bell pepper fruit by methyl jamonate. Postharvest Biol. Technol. 2022, 185, 111799. [Google Scholar] [CrossRef]
- ZHAO, M.L.; Wang, J.N. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Enviro. 2013, 36, 30–51. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [PubMed]
- Robison, J.D.; Yamasaki, Y.; Randall, S.K. The ethylene signaling pathway negatively impacts CBF/DREB-regulated cold response in soybean (Glycine max). Front. Plant Sci. 2019, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, T.; Gan, S. Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 2016, 6, 24066. [Google Scholar] [CrossRef]
- Shi, Y.; Tian, S.; Hou, L. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, W.; Xia, X. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physi. Plant 2014, 152, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, H.; Mao, Z. Ethylene increases the cold tolerance of apple via the MdERF1B-MdCIbHLH1 regulatory module. Plant J. 2021, 106, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Lv, K.; Zhao, K.; Chen, S. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 2020, 292, 110375. [Google Scholar] [CrossRef]
- Sadura, I.; Janeczko, A. Brassinosteroids and the Tolerance of Cereals to Low and High Temperature Stress: Photosynthesis and the Physicochemical Properties of Cell Membranes. Int. J. Mol. Sci. 2021, 23, 342. [Google Scholar] [CrossRef]
- Ye, K.; Li, H.; Ding, Y. Brassinosteroid-Insensitive2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 2019, 31, 2682–2696. [Google Scholar] [CrossRef]
- Fang, P.; Wang, Y.; Wang, M. Crosstalk between Brassinosteroid and Redox Signaling Contributes to the Activation of CBF Expression during Cold Responses in Tomato. Antioxidants 2021, 10, 509. [Google Scholar] [CrossRef]
- Moon, J.; Park, Y.J.; Son, S.H. Brassinosteroids signaling via BZR1 down-regulates expression of ACC oxidase 4 to control growth of Arabidopsis thaliana seedlings. Plant Signal Behav. 2020, 15, 1734333. [Google Scholar] [CrossRef]
- Chi, C.; Li, X.; Fang, P. Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J. Exp. Bot. 2020, 71, 1092–1106. [Google Scholar] [CrossRef]
- Ohnishi, T.; Watanabe, B. CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080. [Google Scholar] [CrossRef]
- Calixto, C.P.G.; Guo, W. Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.J. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 2007, 50, 439–451. [Google Scholar] [CrossRef]
- Owttrim, G.W. RNA helicases and abiotic stress. Nucleic Acids Res. 2006, 34, 3220–3230. [Google Scholar] [CrossRef] [PubMed]
- Egawa, C.; Kobayashi, E. Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 2006, 81, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Guo, L. A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J. Exp. Bot. 2006, 57, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mi, X.; Zhao, S. Comprehensive profiling of alternative splicing landscape during cold acclimation in tea plant. BMC Genom. 2020, 21, 65. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, A.B.; Zhong, X. Functions and mechanisms of plant histone deacetylases. Sci. China Life Sci. 2020, 63, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Bhadouriya, S.L.; Mehrotra, S.; Basantani, M.K. Role of Chromatin Architecture in Plant Stress Responses: An Update. Front. Plant Sci. 2020, 11, 603380. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, P.G.H.; Perrella, G. The diverse and unanticipated roles of histone deacetylase 9 in coordinating plant development and environmental acclimation. J. Exp. Bot. 2020, 71, 6211–6225. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Meng, X.; Liu, Y.; Wang, X.; Wang, T.J.; Zhang, A.; Li, N.; Qi, X.; Liu, B.; Xu, Z.Y. The chromatin remodeler ZmCHB101 impacts alternative splicing contexts in response to osmotic stress. Plant Cell Rep. 2019, 38, 131–145. [Google Scholar] [CrossRef]
- Park, J.; Lim, C.J.; Shen, M. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 2018, 115, e5400–e5409. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Jin, J.B.; Hasegawa, P.M. Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 2007, 10, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, H.; Zhang, X. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Wang, Q.; Yuan, H. Chilling-induced DNA Demethylation is associated with the cold tolerance of Hevea brasiliensis. BMC Plant Biol. 2018, 18, 70. [Google Scholar] [CrossRef]
- Liu, Z.; Jia, Y.; Ding, Y. Plasma membrane CRPK1-mediated phosphorylation of 14–3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 2017, 66, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Mazzucotelli, E.; Belloni, S.; Marone, D. The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006, 7, 509–522. [Google Scholar] [CrossRef]
- Dong, C.H.; Agarwal, M.; Zhang, Y. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Lim, C.J.; Park, J.; Shen, M. The Histone-Modifying Complex PWR/HOS15/HD2C Epigenetically Regulates Cold Tolerance. Plant Physiol. 2020, 184, 1097–1111. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, W.; Hu, J. Characterization of the key region and putative phosphorylation sites of EcaICE1 in its molecular interaction with the EcaHOS1 protein in Eucalyptus camaldulensis. Plant Biol. 2021, 23, 400–406. [Google Scholar] [CrossRef]
- Castro, P.H.; Tavares, R.M.; Bejarano, E.R. SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol. Life Sci. 2012, 69, 3269–3283. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Jin, B.J.; Lee, J. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.; Nassuth, A.; Arora, R. Cold hardiness in trees: A mini-review. Front. Plant Sci. 2018, 9, 1394. [Google Scholar] [CrossRef] [PubMed]


| Gene | Species | Transgenic Technique | Transgenic Plant | Effect | References |
|---|---|---|---|---|---|
| DlCBF1-3 | D. longan | Agrobacterium-mediated transfer | A. thaliana | cold stress tolerance | [53] |
| PpyCBF1-3 | P. pyrifolia | Agrobacterium-mediated transfer | A. thaliana | cold tolerance | [54] |
| IbCBF3 | Sweet potato | Agrobacterium-mediated transfer | S. tuberosum | cold tolerance | [55] |
| AtCBF3 | A. thaliana | Agrobacterium-mediated transfer | S. melongena L. | cold stress tolerance | [56] |
| EgCBF3 | E. guineensi | Agrobacterium-mediated transfer | L. esculenta | freezing tolerance | [57] |
| PpCBF3 | P. pratensis L. | Agrobacterium-mediated transfer | A. thaliana | freezing tolerance | [58] |
| GmDREB1B | G. max | Agrobacterium-mediated transfer | G. max | cold tolerance | [59] |
| DaCBF7 | D. antarctica | Agrobacterium-mediated transfer | O. sativa | cold tolerance | [60] |
| PpCBF1V | P. pratensis L. | Agrobacterium-mediated transfer | M. domestica | cold tolerance | [61] |
| AtCBF1 | A. thaliana | Agrobacterium-mediated transfer | S. lycopersicum | freezing tolerance cold tolerance | [62] |
| OsDREB1B | O. sativa | Agrobacterium-mediated transfer | N. tabacum | cold tolerance | [63] |
| HvCBF4 | H. vulgare | Agrobacterium-mediated transfer | O. sativa | Regulates cold stress | [64] |
| TaDREB2 | T. aestivum | Agrobacterium-mediated transfer | Hordeum vulgare | freezing tolerance | [65] |
| BnCBF5/17 | B. napus | Agrobacterium-mediated transfer | Brassica napus | freezing tolerance | [66] |
| Gene | Species | Transgenic Technique | Transgenic Plant | Effect | References |
|---|---|---|---|---|---|
| SiICE1/2 | S. involucrata | Agrobacterium-mediated transfer | Arabidopsis | cold tolerance | [75] |
| AtICE1 | A. thaliana | Agrobacterium-mediated transfer | Indica rice | cold regulation | [21] |
| CmICE2 | C. morifolium | Agrobacterium-mediated transfer | Arabidopsis | cold tolerance | [77] |
| BjICE46/53 | B. juncea | Agrobacterium-mediated transfer | Arabidopsis | cold tolerance | [73] |
| HbICE1/2 | H. brasiliens | Agrobacterium-mediated transfer | Arabidopsis | cold tolerance | [79] |
| ZjICE2 | Z. japonica | Agrobacterium-mediated transfer | Arabidopsis | cold tolerance | [78] |
| RsICE1 | R. sativus | Agrobacterium-mediated transfer | Rice | cold tolerance | [80] |
| OsICE1/2 | O. sativa | Agrobacterium-mediated transfer | Arabidopsis | cold tolerance | [81] |
| ZmmICE1 | Z. mays | Agrobacterium-mediated transfer | Arabidopsis | freezing tolerance | [82] |
| SlICE1a | S. lycopersicum | Agrobacterium-mediated transfer | Tobacco | cold tolerance | [83] |
| TaICE41/87 | T. aestivum | Agrobacterium-mediated transfer | Arabidopsis | freezing tolerance | [20] |
| Gene | Species | Transgenic Technique | Transgenic Plant | Effect | References |
|---|---|---|---|---|---|
| LeCOR413PM2 | L. esculanta | A. tumefaciens | Tomato | cold tolerance | [97] |
| AtCOR27/28 | A. thaliana | A. tumefaciens | Arabidopsis | freezing tolerance | [11] |
| MfLEA3 | M. falcata | A. tumefaciens | Tobacco | cold tolerance | [98] |
| SikCOR413PM1 | S. involucrate | A. tumefaciens | Tobacco | cold tolerance | [99] |
| SiDHN | S. involucrata | A. tumefaciens | Tomato | cold tolerance | [100] |
| PsCOR413PM2 | P. subulate | A. tumefaciens | Arabidopsis | cold tolerance | [91] |
| RcDhn5 | R. catawbiense | A. tumefaciens | Arabidopsis | freezing tolerance | [101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. Int. J. Mol. Sci. 2022, 23, 1549. https://doi.org/10.3390/ijms23031549
Hwarari D, Guan Y, Ahmad B, Movahedi A, Min T, Hao Z, Lu Y, Chen J, Yang L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. International Journal of Molecular Sciences. 2022; 23(3):1549. https://doi.org/10.3390/ijms23031549
Chicago/Turabian StyleHwarari, Delight, Yuanlin Guan, Baseer Ahmad, Ali Movahedi, Tian Min, Zhaodong Hao, Ye Lu, Jinhui Chen, and Liming Yang. 2022. "ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress" International Journal of Molecular Sciences 23, no. 3: 1549. https://doi.org/10.3390/ijms23031549
APA StyleHwarari, D., Guan, Y., Ahmad, B., Movahedi, A., Min, T., Hao, Z., Lu, Y., Chen, J., & Yang, L. (2022). ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. International Journal of Molecular Sciences, 23(3), 1549. https://doi.org/10.3390/ijms23031549







